Abstract
Src homology 2 domain-containing phosphatase 2 (Shp2), encoded by Ptpn11, is a member of the nonreceptor protein-tyrosine phosphatase family, and functions in cell survival, proliferation, migration, and differentiation in many tissues. Here we report that loss of Ptpn11 in murine hematopoietic cells leads to bone marrow aplasia and lethality. Mutant mice show rapid loss of hematopoietic stem cells (HSCs) and immature progenitors of all hematopoietic lineages in a gene dosage-dependent and cell-autonomous manner. Ptpn11-deficient HSCs and progenitors undergo apoptosis concomitant with increased Noxa expression. Mutant HSCs/progenitors also show defective Erk and Akt activation in response to stem cell factor and diminished thrombopoietin-evoked Erk activation. Activated Kras alleviates the Ptpn11 requirement for colony formation by progenitors and cytokine/growth factor responsiveness of HSCs, indicating that Ras is functionally downstream of Shp2 in these cells. Thus, Shp2 plays a critical role in controlling the survival and maintenance of HSCs and immature progenitors in vivo.
Introduction
Hematopoiesis is maintained by a small number of multipotent long-term hematopoietic stem cells (LT-HSCs) with extensive self-renewal capability. These cells give rise to lineage-committed progenitors, which produce various types of mature blood cells. Cytokine and growth factor signaling is critical for the sustained production of HSCs and progenitors and directs cellular survival, migration, and differentiation.1,2
Src homology 2 (SH2) domain-containing phosphatase 2 (Shp2) is a ubiquitously expressed nonreceptor protein-tyrosine phosphatase (PTP), encoded by the Ptpn11 gene, which regulates signaling networks and cell fates in many organisms. Shp2 is composed of 2 SH2 domains (N-SH2 and C-SH2), a PTP domain, a C-terminal tail with tyrosyl phosphorylation sites that modulate some RTK signaling pathways, and a proline-rich motif of unknown function. In multiple tissues, Shp2 positively regulates Ras/Erk signaling downstream of receptor tyrosine kinases (RTKs) and cytokine receptors.3 Shp2 also appears to have cell-type- and/or agonist-dependent effects on the activation of Akt, Jnk, NF-κB, Rho, and Nfat.3 Germline PTPN11 mutations underlie approximately 50% of Noonan syndrome, which is characterized by short stature, skeletal abnormalities, cardiac defects, learning disabilities, and a predisposition to hematologic abnormalities, particularly juvenile myelomonocytic leukemia. Somatic gain-of-function mutations in PTPN11 also are the most common cause of sporadic juvenile myelomonocytic leukemia.4
Although Shp2 is required for normal Ras/Erk activation in many contexts,3 the underlying mechanism shows great diversity and depends on the precise cell type and stimulus involved. Epistasis studies in Caenorhabditis elegans and Drosophila show that activated Ras/let-60/ras1 suppresses the effects of Shp2/ptp-2/csw loss,5,6 arguing that Shp2 acts upstream of Ras in these signaling pathways. However, in C elegans vulva development and Drosophila R7 photoreceptors and muscle precursors, Shp2/ptp-2/csw appears to act downstream of Ras/let-60/ras1 in some contexts.5,7-9 Furthermore, in the EGL-15/Egfr signaling pathway, ptp-2 may act parallel to let-60.10 In mice, expression of activated Kras (KrasG12D) does not completely restore defective lens proliferation and lacrimal gland development caused by the loss of Ptpn11.11 Collectively, these results indicate that Shp2 can act upstream, downstream, or parallel to Ras in different systems, and imply that the consequences and the mediators of Shp2 action must be specifically delineated in particular biologic contexts.
Homozygous inactivation of murine Shp2 results in early embryonic lethality12-14 because of a critical role of Shp2 in trophoblast stem cell survival.14 Loss of Shp2 also reduces proliferation and causes aberrant differentiation and death of neural stem and early progenitor cells15,16 ; by contrast, Ptpn11 deficiency enhances embryonic stem cell self-renewal.17,18 Studies using in vitro embryonic stem cell differentiation,18-20 Rag2-deficient blastocyst complementation,21 embryonic stem cell aggregation,22 and bone marrow (BM) transplantation23 assays have suggested a role for Shp2 in promoting mesoderm differentiation and hematopoiesis. However, most of these studies used cells and mice expressing an allele of Ptpn11 that encodes a truncated Shp2 variant with increased PTP activity, which could have neomorphic effects in some cell contexts. Furthermore, the role of Shp2 in adult hematopoiesis remains to be established.
Here we show that Shp2 is required for the maintenance of HSCs in a gene dosage-dependent and cell-autonomous manner. We also tested the epistatic relationship of Shp2 and Ras in murine hematopoiesis, and find that constitutive Ras activity suppresses the requirement for Shp2 in myeloid progenitors and HSCs. Together, these data support an essential role of Shp2 upstream of Ras in hematopoietic stem and progenitor cell survival.
Methods
Mice and reconstitution experiments
Ptpn11flox/flox mice were generated previously24 and were bred to Mx1-Cre (The Jackson Laboratory) and KrasLSL-G12D (NCI Mouse Repository) mice, as indicated. To induce Ptpn11 deletion, newborn Mx1-Cre;Ptpn11flox/flox mice were injected intraperitoneally with 3 doses of polyinosinic-polycytidylic acid (pIpC, 150 μg/dose). Control mice were injected in parallel. Mice were monitored and killed at the indicated times or when moribund. For reconstitution experiments, 106 BM cells from 6- to 8-week-old Mx1-Cre;Ptpn11+/+, Mx1-Cre;Ptpn11flox/+, and Mx1-Cre;Ptpn11flox/flox CD45.2+ C57BL/6 mice were injected intravenously, along with 105 CD45.1+ wild-type BM cells, into 6- to 8-week-old lethally irradiated (950 cGy) B6.SJL-Ptprca Pep3b/BoyJ CD45.1+ recipients. After 5 weeks of engraftment, chimeric mice were treated with 5 doses of pIpC (200 μg/dose). Reconstitution by donor cells was detected by staining peripheral white blood cells with anti-CD45.1 (A20) and anti-CD45.2 (104) antibodies. Mice were maintained in accord with guidelines approved by the animal welfare committees of Harvard Medical School and University Health Network.
Histology and pathology examination
Blood counts were determined using a Hemavet 950FS (Drew Scientific). Tissues and organs were collected in 10% formalin and processed by the Specialized Histopathology Services at Brigham and Women's Hospital in Boston, MA.
Retroviral and lentiviral constructs
To generate pMSCV IRES GFP-Cre (pMIGC), a 1.8-kb fragment containing an EGFP-Cre fusion gene was polymerase chain reaction (PCR)-amplified from plasmid pEGFPCREfrtpgktn5neofrt (a gift of Nigel Killeen, University of California, San Francisco), which contains an NLS-Cre sequence in frame with an N-terminal GFP Cre fusion gene in pEGFP-C1 (Clontech). To create pMIGC, the EGFP-Cre amplicon was subcloned as an EcoRI-BamHI fragment into pSP72 (Promega) and then excised as a NcoI-SalI fragment and used to replace the NcoI-SalI GFP segment of pMIG. For Gateway cloning, pMIG-GW and pMIGC-GW were made by inserting the RfA Gateway conversion cassette (Invitrogen) into the unique HpaI cloning site upstream of the IRES sequence in pMIG and pMIGC, respectively. To generate lentivirus expressing the dominant negative p53 mutant GSE56, the truncated NGFR reporter gene was excised from the bidirectional lentiviral construct MA1 and replaced with the GSE56 cDNA.25 Details of these constructs are available from the authors on request.
Retroviral and lentiviral infection
Retroviral supernatants were derived by transfection of 293T cells at 60% to 80% confluence in 100-mm plates using 30 μL Lipofectamine 2000 (Invitrogen) and 10 μg each of retroviral vector DNA and Ecopac packaging construct (gift of R. Van Etten) combined in a total of 1.5 mL Optimem medium (University of California, San Francisco Cell Culture Facility). Supernatant medium was harvested 48 to 72 hours later and used fresh. Fetal liver cells harvested from E14.5 embryos underwent a 24-hour stimulation period in Stemspan medium (StemCell Technologies) supplemented with 15% fetal calf serum, interleukin-11 100 ng/mL (R&D Systems), FLT3 ligand 50 ng/mL (PeproTech), and stem cell factor (SCF, 100 ng/mL, PeproTech), followed by spin infection at 1000g for 1.5 to 2 hours in fresh viral supernatant with polybrene 5 μg/mL. Cells recovered at 37°C for 1 to 3 hours, and the viral supernatant was replaced with stimulation medium. Twenty-four hours later, the spin infection was repeated with fresh virus. The following day, 10 000 green fluorescent protein (GFP)+ cells were sorted directly into 1 mL of methylcellulose medium. For lentiviral infection experiments, lin− BM cells harvested from control and Ptpn11Δ/Δ mice 10 days after pIpC induction were incubated with control lentivirus or lentivirus expressing the dominant negative p53 mutant GSE56 at a multiplicity of infection of 50 to 100 for 16 hours. Infected cells were washed and cultured for an additional 48 hours in vitro before staining with the indicated antibodies, and then were sorted as described.
Flow cytometry
Single-cell suspensions of BM, spleen, and peripheral blood were resuspended in phosphate-buffered saline with 2% fetal bovine serum and stained with directly conjugated antibodies specific for c-Kit (2B8), Sca1 (D7), CD127 (A7R34), CD34 (RAM34), CD16/32, (93) and antibodies against lineage (Lin) markers, including CD3 (145–2C11), CD4 (RM4–5), CD8α (53–6.7), CD19 (6D5), CD45/B220 (RA3–6B2), Gr1(Ly-6G), and Ter119. Antibodies were purchased from BD Biosciences PharMingen, eBioscience, or BioLegend. Flow cytometry was performed with an LSRII (BD Biosciences), and data were analyzed with FlowJo software Version 7.5.5 (TreeStar). For cell cycle analyses, cells were stained with Hoechst 33342 (H; Invitrogen) and pyronin Y (PY; Sigma-Aldrich) at 37°C in the presence of verapamil (Sigma-Aldrich), and the mean percentages of cells in G0 (H−PY−), G1(H−PY+), and S/G2M (H+PY+) were quantified by flow cytometry as indicated in the text. To analyze signaling in LK and Lin−Sca1+c-Kit+ (LSK) cells, Lin− cells were fluorescence-activated cell sorter (FACS)-purified and starved for 1 hour before stimulation with SCF (50 ng/mL) or thrombopoietin (TPO, 50 ng/mL) for 5 or 10 minutes. Cells were fixed with 1.5% paraformaldehyde, permeabilized with acetone, and then stained with anti–c-kit, anti-Sca1, and either anti-pErk or anti-pAkt antibodies, as described previously.26,27 Phospho-specific antibodies were purchased from Cell Signaling Technology; cytokines were from PeproTech.
Colony assays
Fetal liver cells were harvested, transduced with retrovirus, and subjected to colony (granulocyte-macrophage colony-forming units [CFU-GM]) assays in M3231 methylcellulose (StemCell Technologies) supplemented with granulocyte-macrophage colony-stimulating factor 10 ng/mL (PeproTech). Individual colonies were harvested with a 10-μL pipette under direct microscopic visualization and suspended in 10 μL of water. For genotyping, 4 μL of this suspension was amplified in a multiplex PCR reaction using primers MPC-F (5′-ATGGACGTGGGTGCCTGCAT-3′), MPRC-R (5′-GTCCGCGGTCGCGACGTA-3′), and MPFL-R (5′-CCCAACTAGCAGCCTGGCAAA-3′) and 35 cycles of 94°C for 30 seconds, 66°C for 90 seconds, and 72°C for 60 seconds. These conditions produce distinct bands for wild-type (81 bp), flox (157 bp), and Δ (184 bp) alleles of Ptpn11. Detailed PCR conditions are available from the authors on request.
Quantitative reverse-transcribed PCR analysis
Total RNA was isolated from FACS-purified BM populations using the PicoPure Kit (Arcturus Bioscience) and subjected to reverse transcription with SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR assays were performed on an ABI 7500 Fast Real-Time PCR System using the TaqMan Universal PCR master mixture (Applied Biosystems). Predesigned primers and probes used in quantitative PCR assay are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Single-cell proliferation assays
SLAM HSCs (Lin−c-kit+Sca1+CD150+CD48−) were FACS-purified and deposited in 60-well Terasaki plates (1 cell/well) in Iscove modified Dulbecco medium in the presence of SCF (50 ng/mL), TPO (50 ng/ml), Flt3L (50 ng/mL), interleukin-11 (20 ng/mL), transferrin (5 μg/mL), insulin (5 μg/mL), bovine serum albumin (0.1%), and fetal bovine serum (4%). Cell number in each well was evaluated microscopically on a daily basis for the indicated times.
Statistical analysis
Data are presented as mean ± SEM and analyzed by log-rank test, Student t test, or analysis of variance with Bonferroni post-hoc test, as indicated.
Results
Disruption of Shp2 causes BM failure
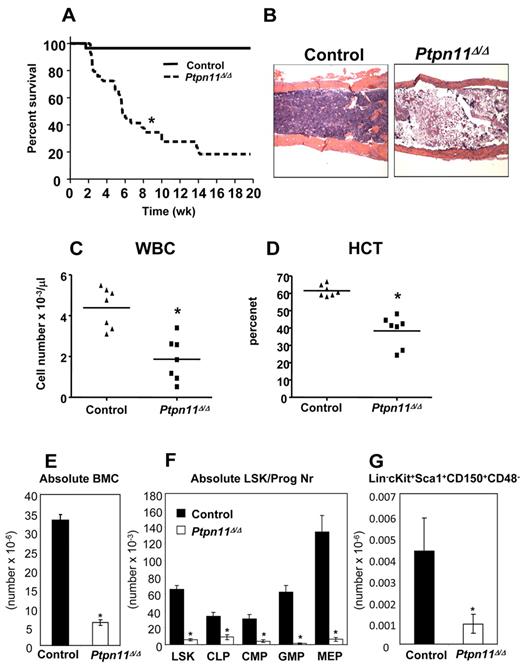
To begin to assess the role of Shp2 in adult hematopoiesis, we determined its pattern of expression in HSCs and immature hematopoietic progenitors. Quantitative reverse-transcribed PCR analysis (supplemental Figure 1A) revealed similar levels of Ptpn11 expression in adult LT-HSCs, short-term HSCs, multipotent progenitors, common myeloid progenitors, granulocyte-macrophage progenitors, megakaryocytic-erythroid progenitors, and common lymphoid progenitors. To assess Shp2 function, we bred a conditional mutant (floxed) Ptpn11 allele24 onto the Mx1-Cre mouse strain, which expresses Cre recombinase (Cre) in response to interferon exposure.28 Ptpn11flox/flox (hereafter, control) and Mx1-Cre; Ptpn11flox/flox mice were injected with pIpC to induce endogenous interferon production and hence, Cre expression. Most of the induced Mx1-Cre; Ptpn11flox/flox (hereafter, Ptpn11Δ/Δ) mice became moribund or died by 6 to 8 weeks (Figure 1A). At this time, mutant mice were leukopenic, severely anemic, and had profoundly reduced numbers of BM cells (Figure 1B-E). The remaining surviving Ptpn11Δ/Δ mice probably have incomplete deletion of the floxed Ptpn11 allele.
Deletion of Ptpn11 leads to fatal BM failure and loss of HSCs. (A) Kaplan-Meier survival analysis of a cohort of Mx1-Cre;Ptpn11flox/flox (n = 30) and littermate control (n = 28) mice after pIpC treatment. Ptpn11Δ/Δ mice have a median life span of 5 weeks; control mice remain healthy for > 10 months. *P < .05, log-rank test. (B) Hematoxylin and eosin-stained sections from humeri of control and Ptpn11Δ/Δ animals 4 to 5 weeks after receiving their last dose of pIpC. The samples were analyzed using an Olympus BX41 microscope with the objective lens 4×/0.75 Olympus UPlanFL (Olympus). The pictures were taken using Olympus QColor5 and analyzed with acquisition software QCapture Pro v6.0 (QImaging) and Adobe Photoshop 6.0 (Adobe). (C-D) White blood cell count (WBC; C) and hematocrit (HCT; D) of control and Ptpn11Δ/Δ mice. *P < .05, Student t test. (E) BM cellularity (per 2 hind limbs) in control and Ptpn11Δ/Δ animals shown as mean ± SEM. *P < .05, Student t test. (F) Absolute number (mean ± SEM). *P < .05, Student t test. LSK, common lymphoid progenitors (Lin− Sca1+ c-kitloIL7Rα+), common myeloid progenitors (Lin−Sca1−c-kit+CD34+FcγRlo), granulocyte-macrophage progenitors (Lin−Sca1− c-kit+CD34+FcγRhi), megakaryocytic-erythroid progenitors (Lin−Sca1−c-kit+CD34−FcγR−/lo) of control (n = 7) and Ptpn11Δ/Δ mice (n = 7). (G) Absolute number (mean ± SEM). *P < .05, Student t test. SLAM-HSCs from control (n = 3) and Ptpn11Δ/Δ mice (n = 3).
Deletion of Ptpn11 leads to fatal BM failure and loss of HSCs. (A) Kaplan-Meier survival analysis of a cohort of Mx1-Cre;Ptpn11flox/flox (n = 30) and littermate control (n = 28) mice after pIpC treatment. Ptpn11Δ/Δ mice have a median life span of 5 weeks; control mice remain healthy for > 10 months. *P < .05, log-rank test. (B) Hematoxylin and eosin-stained sections from humeri of control and Ptpn11Δ/Δ animals 4 to 5 weeks after receiving their last dose of pIpC. The samples were analyzed using an Olympus BX41 microscope with the objective lens 4×/0.75 Olympus UPlanFL (Olympus). The pictures were taken using Olympus QColor5 and analyzed with acquisition software QCapture Pro v6.0 (QImaging) and Adobe Photoshop 6.0 (Adobe). (C-D) White blood cell count (WBC; C) and hematocrit (HCT; D) of control and Ptpn11Δ/Δ mice. *P < .05, Student t test. (E) BM cellularity (per 2 hind limbs) in control and Ptpn11Δ/Δ animals shown as mean ± SEM. *P < .05, Student t test. (F) Absolute number (mean ± SEM). *P < .05, Student t test. LSK, common lymphoid progenitors (Lin− Sca1+ c-kitloIL7Rα+), common myeloid progenitors (Lin−Sca1−c-kit+CD34+FcγRlo), granulocyte-macrophage progenitors (Lin−Sca1− c-kit+CD34+FcγRhi), megakaryocytic-erythroid progenitors (Lin−Sca1−c-kit+CD34−FcγR−/lo) of control (n = 7) and Ptpn11Δ/Δ mice (n = 7). (G) Absolute number (mean ± SEM). *P < .05, Student t test. SLAM-HSCs from control (n = 3) and Ptpn11Δ/Δ mice (n = 3).
The dramatic phenotype of Ptpn11Δ/Δ mice led us to examine the HSC-enriched LSK compartment and early progenitors in the mutant BM. Indeed, the frequencies and absolute numbers of LSK, common lymphoid progenitors, common myeloid progenitors, granulocyte-macrophage progenitors, and megakaryocytic-erythroid progenitors were all markedly reduced in Ptpn11Δ/Δ BM (Figure 1F, supplemental Figure 1B; and data not shown). Analysis of the HSC compartment (LSKCD150+CD48−, also known as SLAM-HSCs) within the LSK population also revealed a marked reduction in cell number, indicating a defect at the earliest stage of adult hematopoietic development (Figure 1G). To assess progenitor cell function, we performed colony assays. Consistent with the flow cytometric data, Ptpn11Δ/Δ BM cells produced significantly lower numbers of colony-forming units-erythroid, burst-forming units-erythroid, CFU-GM, and colony-forming units granulocyte-erythrocyte-macrophage-megakaryocyte colonies compared with control BM cells (supplemental Figure 1C). Together, these data suggest that the rapid demise of Ptpn11Δ/Δ mice is probably the result of severe BM aplasia and the loss of almost all of hematopoietic stem cells and progenitors of the lymphoid, erythroid, and myeloid lineages.
Requirement for Shp2 in HSCs is cell-autonomous
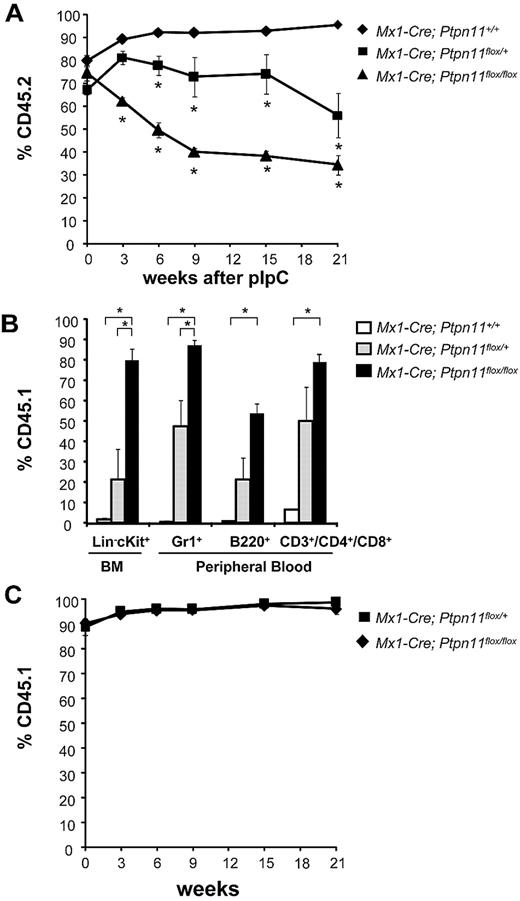
To ask whether the loss of hematopoietic cells in Ptpn11 mutant animals is cell-autonomous, we transferred uninduced Mx1-Cre;Ptpn11+/+, Mx1-Cre;Ptpn11flox/+, and Mx1-Cre;Ptpn11flox/flox BM cells expressing the CD45.2 cell surface marker, together with wild-type CD45.1-expressing carrier BM cells, into lethally irradiated congenic CD45.1 animals. Recipients were maintained for 5 weeks to allow sufficient time for steady-state hematopoiesis to be reestablished. Mice were then injected with pIpC, and the ratio of CD45.1+ versus CD45.2+ cells was measured in the peripheral blood. All mice showed comparable levels of CD45.2 engraftment before pIpC treatment. Yet, whereas recipients of Mx1-Cre;Ptpn11+/+ BM maintained more than 90% CD45.2+ peripheral blood cells for up to 21 weeks after pIpC treatment, the percentage of CD45.2+ cells in peripheral blood was reduced to 50% and 40% in mice that had received Mx1-Cre;Ptpn11flox/+ and Mx1-Cre;Ptpn11flox/flox cells, respectively (Figure 2A). At 21 weeks, donor CD45.2+ cells in recipients of Mx1-Cre;Ptpn11flox/+ BM cells accounted for 50% of blood neutrophils (Gr1+), 50% of T lymphocytes (CD3+), and 75% of B lymphocytes (B220+) (Figure 2B). By contrast, there was marked attrition of Mx1-Cre;Ptpn11flox/flox cells in vivo with 90% of Gr1+ neutrophils, 80% of CD3+ T cells, and 50% of B220+ B cells expressing the recipient CD45.1 marker. Whereas CD45.1+ Lin−c-kit+ competitor cells composed only approximately 20% of BM cells in Mx1-Cre;Ptpn11flox/+ BM recipients, they represented more than 80% of cells in recipients of Mx1-Cre;Ptpn11flox/flox BM. These results indicate that most donor-derived Mx1-Cre;Ptpn11flox/flox HSCs were lost by 21 weeks after pIpC induction, as the remaining 40% CD45.2 cells in the peripheral blood were mostly B220+ cells. Interestingly, CD19-Cre;Ptpn11flox/flox mice show normal B-cell development and maintenance, suggesting that loss of Ptpn11 can be tolerated in B-cell lineage (W.Y., L. Pao, and B.G.N., manuscript in preparation).
Cell-autonomous requirement for Ptpn11 in HSC maintenance. (A) Lethally irradiated CD45.1+ recipients were transplanted with 106 BM cells from Mx1-Cre;Ptpn11+/+ (n = 6), Mx1-Cre;Ptpn11flox/+ (n = 4), or Mx1-Cre;Ptpn11flox/flox (n = 4) mice, along with 105 wild-type CD45.1+ cells. After 5 weeks of engraftment, chimeric mice were treated with 5 doses of pIpC, and the percentage of peripheral blood cells expressing CD45.1 and CD45.2 was quantified by flow cytometry at the indicated times. *P < .05, analysis of variance. (B) Levels of CD45.1+ BM Lin−c-kit+ progenitors (LK) and peripheral blood B220+, CD3+/CD4+/CD8+, and Gr1+ cells were quantified 21 weeks after pIpC induction. *P < .05, analysis of variance. (C) Lethally irradiated Mx1-Cre;Ptpn11flox/+ (n = 4) and Mx1-Cre;Ptpn11flox/flox (n = 4) control recipients were transplanted with 106 BM cells from wild-type CD45.1+ donors. Mice were treated and analyzed as described in panel A.
Cell-autonomous requirement for Ptpn11 in HSC maintenance. (A) Lethally irradiated CD45.1+ recipients were transplanted with 106 BM cells from Mx1-Cre;Ptpn11+/+ (n = 6), Mx1-Cre;Ptpn11flox/+ (n = 4), or Mx1-Cre;Ptpn11flox/flox (n = 4) mice, along with 105 wild-type CD45.1+ cells. After 5 weeks of engraftment, chimeric mice were treated with 5 doses of pIpC, and the percentage of peripheral blood cells expressing CD45.1 and CD45.2 was quantified by flow cytometry at the indicated times. *P < .05, analysis of variance. (B) Levels of CD45.1+ BM Lin−c-kit+ progenitors (LK) and peripheral blood B220+, CD3+/CD4+/CD8+, and Gr1+ cells were quantified 21 weeks after pIpC induction. *P < .05, analysis of variance. (C) Lethally irradiated Mx1-Cre;Ptpn11flox/+ (n = 4) and Mx1-Cre;Ptpn11flox/flox (n = 4) control recipients were transplanted with 106 BM cells from wild-type CD45.1+ donors. Mice were treated and analyzed as described in panel A.
Induced Mx1-Cre mice also show gene deletion in various nonhematopoietic tissues after pIpC treatment. To examine whether loss of Ptpn11 in these interferon-responsive tissues contributes to the loss of HSCs, we injected wild-type CD45.1+ BM cells into lethally irradiated Mx1-Cre;Ptpn11flox/+ and Mx1-Cre;Ptpn11flox/flox animals. Five weeks after engraftment, the mice were treated with pIpC and peripheral blood cell counts were monitored (Figure 2C). Both Mx1-Cre;Ptpn11flox/+ and Mx1-Cre;Ptpn11flox/flox hosts remained healthy and maintained normal blood counts for more than 21 weeks after pIpC treatment (Figure 2C; and data not shown). Therefore, deletion of Ptpn11 in nonhematopoietic tissues did not compromise steady-state hematopoiesis of normal BM cells. Together, these data suggest that Ptpn11 is essential for steady-state hematopoiesis and HSC maintenance in a gene dose-dependent and cell-autonomous manner.
Aberrant cell cycle profile and cell death of Ptpn11-deficient HSCs and progenitors
Loss of HSCs and progenitor cells could result from several mechanisms, which include failure to maintain quiescence (loss of self-renewal), decreased proliferation, and/or cell death. We determined the effects of Ptpn11 deficiency on proliferation by assessing cell-cycle parameters in immature progenitors (lin−c-kit+Sca1− [LK]) and HSC-enriched LSKCD150+ cells. At early times after pIpC treament, control and Ptpn11Δ/Δ cells had similar cell cycle profiles. However, compared with controls, Ptpn11Δ/Δ LK and LSKCD150+ cells consistently exhibited a decreased G0 population at day 23 after pIpC exposure (Figure 3A-B; supplemental Figure 3). In addition, compared with controls, mutant LSKCD150+ cells showed a significantly higher proportion of cells in G1. The proportion of mutant LK cells in G1 also trended higher, although this did not reach statistical significance. Ptpn11Δ/Δ LK and LSKCD150+ cells also showed significantly increased apoptosis, as revealed by annexin V staining, at 20 and 23 days after pIpC treatment (Figure 3C-D). We also sorted SLAM-HSCs from control and mutant mice 10 days after pIpC induction and placed them at one cell per well in a medium that allows preservation of multipotency in vitro29 and contains saturating doses of SCF and TPO, 2 cytokines critical for HSC maintenance in vivo.2 After4 days, more than 80% of control cells were viable and showed substantial proliferation (Figure 3E). By contrast, more than 60% of Ptpn11Δ/Δ cells had died, whereas the rest had undergone only one or 2 cell divisions (Figure 3F). Together, these data suggest the loss of Ptpn11 leads to rapid death of HSCs and immature progenitors.
Ptpn11 deletion leads to decreased quiescence and increased cell death. (A-B) Cell cycle analysis of Ptpn11Δ/Δ HSCs and progenitors. LK (A) and LSKCD150+ (B) cells were subjected to 2-parameter analysis of DNA content (Hoechst 33342 [H]) vs RNA content (Pyronin [PY]). The mean percentages (± SEM) of cells in G0 (H−PY−), G1(H−PY+), and S/G2M (H+PY+) are indicated. *P < .05, Student t test. (C) Lin-c-kit+Sca1− and (D) LSKCD150+ cells harvested from control (n = 3) and Ptpn11Δ/Δ (n = 3) mice were stained with annexin V to assess cell death at the indicated times after pIpC induction. The average percentage (± SEM) of annexin V+ cells is shown. *P < .05, Student t test. (E-F) Single SLAM-HSCs were sorted into 60-well plates (1 cell/well) and cultured in media containing SCF, TPO, Flt3L, and interleukin-11. Proliferation of each clone was evaluated microscopically over 4 days. Representative data from one of 4 experiments with similar results are shown. The average percentages (± SEM) of surviving clones from the 4 experiments are shown at the top of each panel. *P < .05, Student t test.
Ptpn11 deletion leads to decreased quiescence and increased cell death. (A-B) Cell cycle analysis of Ptpn11Δ/Δ HSCs and progenitors. LK (A) and LSKCD150+ (B) cells were subjected to 2-parameter analysis of DNA content (Hoechst 33342 [H]) vs RNA content (Pyronin [PY]). The mean percentages (± SEM) of cells in G0 (H−PY−), G1(H−PY+), and S/G2M (H+PY+) are indicated. *P < .05, Student t test. (C) Lin-c-kit+Sca1− and (D) LSKCD150+ cells harvested from control (n = 3) and Ptpn11Δ/Δ (n = 3) mice were stained with annexin V to assess cell death at the indicated times after pIpC induction. The average percentage (± SEM) of annexin V+ cells is shown. *P < .05, Student t test. (E-F) Single SLAM-HSCs were sorted into 60-well plates (1 cell/well) and cultured in media containing SCF, TPO, Flt3L, and interleukin-11. Proliferation of each clone was evaluated microscopically over 4 days. Representative data from one of 4 experiments with similar results are shown. The average percentages (± SEM) of surviving clones from the 4 experiments are shown at the top of each panel. *P < .05, Student t test.
Next, we surveyed the expression of genes previously implicated in the control of cell survival in control and Ptpn11Δ/Δ LK and HSC-enriched, LSKCD150+ cells. Control and mutant cells expressed similar levels of Puma, BclXL, and Mcl1 (Figure 4A; supplemental Figure 4). Although Bcl2 expression was comparable in control and Ptpn11Δ/Δ LSKCD150+ cells, it was down-regulated slightly in Ptpn11Δ/Δ LK cells; this decrease might contribute to the compromised survival of these cells. More strikingly, however, Noxa expression was increased in Ptpn11-deficient LK and LSKCD150+ cells, coincident with the onset of cell death. LSK cells harvested from control and Ptpn11 mutant mice 10 days after pIpC treatment showed no signs of apoptosis (Figure 3D) or elevated Noxa expression (data not shown); however, on in vitro culture for 48 hours, mutant cells showed significant cell death (Figure 3F; and data not shown) and markedly elevated Noxa levels (Figure 4B).
p53-independent up-regulation of Noxa in Ptpn11Δ/Δ HSCs and progenitors. (A) Quantitative PCR of Noxa and Puma in LSKCD150+ and LK cells from control and Ptpn11Δ/Δ mice 23 days after pIpC induction. Results present mean ± SEM relative to Gapdh expression. *P < .05, Student t test. Representative data from one of 4 experiments with similar results are shown. (B) Lin− BM cells, harvested from control and Ptpn11Δ/Δ mice 10 days after pIpC induction, were infected with control lentivirus or lentivirus expressing a dominant negative p53 mutant (GSE56). Cells were cultured for 48 hours in vitro before staining with anti–c-kit and anti-Sca1 antibodies. GFP+ LSK cells were sorted and Noxa expression (mean ± SEM) was determined by quantitative PCR. *P < .05, Student t test. NS indicates not significant. Representative data from one of 3 experiments with similar results are shown. (C) Lin− BM cells were infected with control lentivirus or lentivirus expressing GSE56. Cells were cultured for 48 hours in vitro and were left untreated or irradiated (IR, 300 cGy), harvested, and subjected to quantitative PCR as described in panel B. The levels of Puma and Noxa normalized to Gapdh expression are shown (mean ± SEM). *P < .05, Student t test.
p53-independent up-regulation of Noxa in Ptpn11Δ/Δ HSCs and progenitors. (A) Quantitative PCR of Noxa and Puma in LSKCD150+ and LK cells from control and Ptpn11Δ/Δ mice 23 days after pIpC induction. Results present mean ± SEM relative to Gapdh expression. *P < .05, Student t test. Representative data from one of 4 experiments with similar results are shown. (B) Lin− BM cells, harvested from control and Ptpn11Δ/Δ mice 10 days after pIpC induction, were infected with control lentivirus or lentivirus expressing a dominant negative p53 mutant (GSE56). Cells were cultured for 48 hours in vitro before staining with anti–c-kit and anti-Sca1 antibodies. GFP+ LSK cells were sorted and Noxa expression (mean ± SEM) was determined by quantitative PCR. *P < .05, Student t test. NS indicates not significant. Representative data from one of 3 experiments with similar results are shown. (C) Lin− BM cells were infected with control lentivirus or lentivirus expressing GSE56. Cells were cultured for 48 hours in vitro and were left untreated or irradiated (IR, 300 cGy), harvested, and subjected to quantitative PCR as described in panel B. The levels of Puma and Noxa normalized to Gapdh expression are shown (mean ± SEM). *P < .05, Student t test.
Because Noxa can be regulated by p53,30 and Shp2 inhibits p53-dependent apoptosis in neural crest-derived cells,31 we asked whether cell death in Ptpn11Δ/Δ hematopoietic progenitor cells was p53-dependent. Immunoblots of lin−c-kit+ BM cells from control and Ptpn11Δ/Δ mice showed that p53 was not present at a detectable level (data not shown). Furthermore, overexpressing GSE56, a dominant negative mutant of p53,25 in mutant LSK cells failed to rescue their proliferative response to SCF and TPO (supplemental Figure 5) and did not prevent elevated expression of Noxa (Figure 4B). By contrast, GSE56 expression in wild-type lin− BM cells completely blocked the Puma and Noxa up-regulation in response to irradiation (Figure 4C), indicating that GSE56 was expressed at a level sufficient to block p53 activity in these cells. Together, these data strongly suggest that apoptosis induced by Ptpn11 deficiency in HSCs/progenitors is p53-independent.
Perturbed SCF and TPO signaling in Ptpn11Δ/Δ stem/progenitor cells
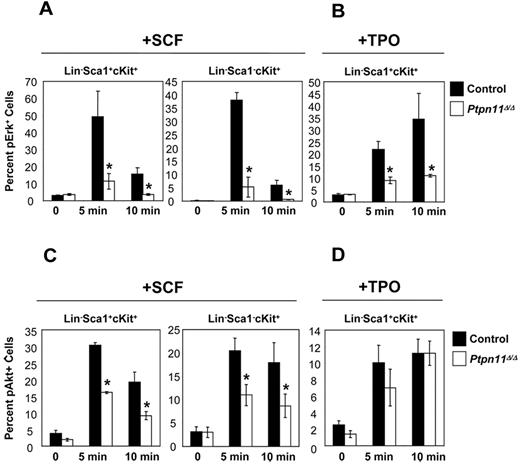
The failure of Ptpn11Δ/Δ HSCs to expand in response to cytokine and growth factor stimulation led us to interrogate signaling pathways downstream of key receptors, such as Kit and Mpl, in these cells. Multiparameter flow cytometric analysis showed that SCF-evoked Erk and Akt activation was reduced substantially in Ptpn11Δ/Δ LSK and LK cells (Figure 5A,C). Although TPO-evoked Akt activation was unaffected by Ptpn11 deficiency (Figure 5D), TPO-evoked Erk activation was markedly reduced in Ptpn11Δ/Δ LSK cells (Figure 5B). STAT3 and STAT5 activation were similar in control and and Ptpn11 mutant cells, and there was no detectable activation of Erk or Akt in response to TPO in either control or mutant LK cells (data not shown). Notably, control and Ptpn11-deficient LK and LSK cells express similar levels of Mpl RNA (supplemental Figure 6). Together, these data demonstrate that mutant LSK and LK cells show compromised growth factor and cytokine receptor signaling, which probably accounts for their impaired proliferation and survival in vitro and in vivo.
Signaling defects in Ptpn11Δ/Δ LSK and LK cells. Lin− BM cells from control (n = 4) and Ptpn11Δ/Δ (n = 4) mice were purified by FACS and starved for 1 hour in serum-free media, before they were either left untreated or stimulated with SCF (50 ng/mL) or TPO (50 ng/mL) for the indicated times. Cells were fixed, permeabilized, and stained with anti–c-kit, anti-Sca1, and anti-pErk (A-B) or pAkt (C-D). Levels of phospho-specific antigens in the LSK and LK population were determined by flow cytometry. The percentages of pErk+ or pAkt+ cells from 4 experiments are shown as mean ± SEM. *P < .05, Student t test.
Signaling defects in Ptpn11Δ/Δ LSK and LK cells. Lin− BM cells from control (n = 4) and Ptpn11Δ/Δ (n = 4) mice were purified by FACS and starved for 1 hour in serum-free media, before they were either left untreated or stimulated with SCF (50 ng/mL) or TPO (50 ng/mL) for the indicated times. Cells were fixed, permeabilized, and stained with anti–c-kit, anti-Sca1, and anti-pErk (A-B) or pAkt (C-D). Levels of phospho-specific antigens in the LSK and LK population were determined by flow cytometry. The percentages of pErk+ or pAkt+ cells from 4 experiments are shown as mean ± SEM. *P < .05, Student t test.
Kras activation is epistatic to Ptpn11 disruption in HSCs and myeloid progenitors
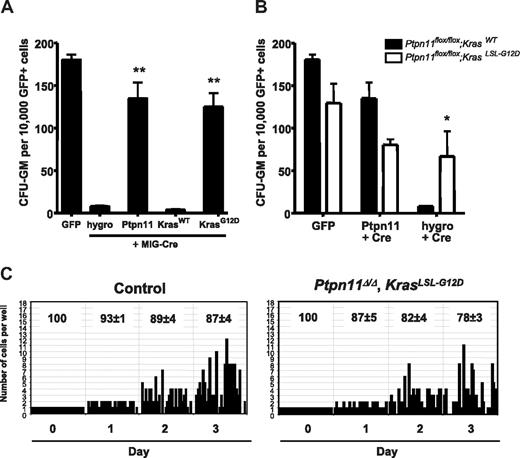
We tested whether Ras activation is the required signal downstream of Shp2 in HSCs/progenitors by asking whether a gain-of-function Ras allele could suppress the effects of Ptpn11 deletion. Embryonic day 14.5 fetal liver cells were transduced with retroviral vectors expressing GFP or a GFP-Cre fusion protein. GFP+ cells were isolated by FACS and plated in methylcellulose containing granulocyte-macrophage colony-stimulating factor (Figure 6A). We observed nearly complete loss of CFU-GM in Ptpn11flox/flox cells expressing GFP-Cre and the hygromycin resistance gene. Colony growth was rescued by coexpression of Ptpn11 cDNA, indicating that the lack of colony formation was the result of the loss of Ptpn11. These data indicate that, in addition to its role in adult hematopoiesis, Shp2 also is required for the survival and/or growth of fetal liver myeloid progenitors. To test the ability of activated Ras to rescue this phenotype, we constructed a vector coexpressing GFP-Cre and a constitutively active isoform of Kras. Ptpn11flox/flox cells expressing GFP-Cre and the oncogenic KrasG12D allele were able to form CFU-GM. Importantly, these cells showed complete deletion of Ptpn11 (supplemental Figure 7A). These data demonstrate that overexpression of activated Kras can alleviate the requirement for Ptpn11 in fetal liver myeloid progenitors.
Expression of activated Kras compensates for loss of Ptpn11 in HSCs and progenitors. (A) Primary myeloid progenitors from embryonic day 14.5 Ptpn11flox/flox fetal livers were transduced with the indicated retroviruses, and FACS-purified GFP+ cells were cultured in methylcellulose medium containing granulocyte-macrophage colony-stimulating factor. GFP virus served as a negative control. MIG-Cre vectors were designed to coexpress the hygromycin resistance gene, WT Ptpn11, WT Kras, or activated KrasG12D. Results are shown as mean ± SEM of 3 independent experiments. *P < .05, Student t test. **P < .01, Student t test. (B) Effect of Ptpn11 loss in Ptpn11flox/flox fetal liver cells with or without an activated KrasLSL-G12D allele. Cells of the indicated genotype were transduced with GFP virus control or with MIG-Cre virus coexpressing exogenous Ptpn11 or the hygromycin resistance gene. Results are shown as mean ± SEM of 3 independent experiments. *P < .05, Student t test. (C) SLAM-HSCs from control (n = 2) and Mx1-Cre; KrasLSL-G12D; Ptpn11flox/flox (n = 4) mice were purified by FACS and deposited into 60-well plates (1 cell/well), and proliferation of each clone was evaluated microscopically over 3 days. The average percentages (± SEM) of surviving clones are shown at the top of each panel. Representative data from these experiments with similar results are shown.
Expression of activated Kras compensates for loss of Ptpn11 in HSCs and progenitors. (A) Primary myeloid progenitors from embryonic day 14.5 Ptpn11flox/flox fetal livers were transduced with the indicated retroviruses, and FACS-purified GFP+ cells were cultured in methylcellulose medium containing granulocyte-macrophage colony-stimulating factor. GFP virus served as a negative control. MIG-Cre vectors were designed to coexpress the hygromycin resistance gene, WT Ptpn11, WT Kras, or activated KrasG12D. Results are shown as mean ± SEM of 3 independent experiments. *P < .05, Student t test. **P < .01, Student t test. (B) Effect of Ptpn11 loss in Ptpn11flox/flox fetal liver cells with or without an activated KrasLSL-G12D allele. Cells of the indicated genotype were transduced with GFP virus control or with MIG-Cre virus coexpressing exogenous Ptpn11 or the hygromycin resistance gene. Results are shown as mean ± SEM of 3 independent experiments. *P < .05, Student t test. (C) SLAM-HSCs from control (n = 2) and Mx1-Cre; KrasLSL-G12D; Ptpn11flox/flox (n = 4) mice were purified by FACS and deposited into 60-well plates (1 cell/well), and proliferation of each clone was evaluated microscopically over 3 days. The average percentages (± SEM) of surviving clones are shown at the top of each panel. Representative data from these experiments with similar results are shown.
Next, we asked whether Ptpn11-deficient CFU-GM could be rescued by activated Kras expressed at normal levels, using fetal liver cells from Ptpn11flox/flox mice that also contained the KrasLSL-G12D conditional knock-in allele. In these cells, Cre expression simultaneously causes deletion of Ptpn11 and expression of activated KrasG12D under the control of its endogenous promoter. Ptpn11flox/flox; KrasLSL-G12D fetal liver cells transduced with the hygro GFP-Cre vector formed robust colonies, whereas cells lacking the KrasLSL-G12D allele could not (Figure 6B). PCR analysis of individual colonies verified the KrasG12D Ptpn11Δ/Δ genotype (supplemental Figure 7B). These results confirm that constitutive K-Ras activation overcomes the requirement for Shp2 in myeloid progenitors.
Finally, we asked whether KrasG12D expression could rescue the growth factor/cytokine proliferation of adult Ptpn11-deficient HSCs. SLAM-HSCs were harvested from control and Mx1-Cre; Ptpn11flox/flox KrasLSL-G12D mice and subjected to single-cell proliferation assays. As shown in Figure 6C, the number of mutant clones that proliferated was similar to that of control cells. Analysis by quantitative PCR indicated complete absence of Ptpn11 mRNA in LK mutant cells (supplemental Figure 7C). Together, these data demonstrate that hyperactive Ras is epistatic to Shp2 loss in primary hematopoietic stem and progenitor cells, consistent with a model in which Ras activation is a major function provided by Shp2 in these cells.
Discussion
Genetic data that directly address the role of the Ras/Erk pathway in HSCs function are limited. In this report, we identify a cell-autonomous requirement for Shp2 in the survival and maintenance of HSCs and immature hematopoietic progenitors in vivo. A previous study showed that mice with heterozygous expression of a Ptpn11 truncation allele have normal BM cellularity and peripheral blood counts but impaired HSC function.23 Our data reveal a much more profound requirement for Shp2, as complete absence of both Ptpn11 alleles leads to rapid and severe attrition of the hematopoietic system. Because differentiated B cells and macrophages are largely unaffected in CD19 Cre;Ptpn11flox/flox (W.Y., L. Pao, and B.G.N., manuscript in preparation) and LysM Cre;Ptpn11flox/flox mice, respectively (W.Y. and B.G.N, manuscript in revision), our data further suggest that the loss of these lineages in pIpC-treated MxCre; Ptpn11flox/flox mice is largely the result of depletion of immature progenitors and/or HSCs.
Loss of Ptpn11-deficient LSKCD150+ and LK populations is accompanied by apoptosis (Figure 3C-D) and a reduction of G0 cells, concomitant with accumulation of G1 cells (Figure 3A-B). The latter might reflect impaired G1/S progression in the absence of Ptpn11. However, as the aberrant cell cycle profile only becomes evident 23 days after pIpC treatment, it probably reflects an attempt of mutant HSCs/progenitors to respond to the rapid depletion of mature BM cells in Ptpn11Δ/Δ mice. Ptpn11-deficient HSCs also could be prone to enter the cell cycle because of defective Tpo/Mpl signaling (Figure 5), which is crucial for maintaining quiescence of postnatal HSCs.32,33 Together, our data show that the loss of Ptpn11-mutant HSCs and progenitor cells is accompanied by the loss of quiescence and increased cell death.
Death of Ptpn11 mutant hematopoietic progenitor cells is detectable only 20 days after pIpC treatment, whereas the absolute number of mutant progenitors begins to decline by day 14 (data not shown). Yet, complete absence of Ptpn11 mRNA was observed by day 10 (supplemental Figure 2; and data not shown), and LSK cells harvested at this time show defective Erk activation after SCF stimulation, consistent with the complete absence of Shp2 protein as well. A possible explanation for the lag between loss of Ptpn11 expression and the onset of apoptosis is that dead cells are phagocytosed rapidly by myeloid cells, so that increased apoptosis cannot be detected at early times after Ptpn11 deletion. As Ptpn11-deficient myeloid progenitors and mature myeloid cells are progressively depleted, apoptotic cells accumulate. Consistent with this notion, Ptpn11Δ/Δ SLAM-HSCs harvested at day 10 after pIpC treatment undergo rapid cell death in vitro (Figure 3E-F) when they are cultured in a cytokine cocktail that preserves WT HSC multipotency in vitro.29
Noxa expression is markedly up-regulated in Ptpn11-deficient HSC-enriched LSKCD150+ cells and immature hematopoietic progenitors, correlating with the onset of apoptosis in these cells. Noxa is a transcriptional target of p53,30,34 and Shp2 can suppress p53-dependent apoptosis.31 Surprisingly, however, Noxa up-regulation and death of Ptpn11Δ/Δ immature hematopoietic progenitors are p53-independent. Shp2 also is known to suppress p73-dependent apoptosis,35 but p73 was not expressed at detectable levels in control or Ptpn11 mutant HSCs and progenitors (data not shown). Interestingly, Noxa is up-regulated in a p53-independent manner in T cells after CD3 stimulation and helps limit T-cell survival.36,37 Increased Noxa levels in Ptpn11Δ/Δ cells might promote cell death by antagonizing Mcl1, which is critical for HSC survival.30,37,38 Genetic experiments to address the requirement of Noxa and the role of Mcl1 in triggering apoptosis in Ptpn11-deficient HSCs and progenitors are ongoing.
In conclusion, we find that endogenous expression of a constitutively active Kras allele rescues Ptpn11Δ/Δ HSCs and myeloid progenitor cells in vivo, placing Ras downstream of or parallel to Shp2 in a pathway controlling hematopoietic stem and progenitor cell survival. Thus, hematopoietic progenitors behave similarly to C elegans oocytes or Drosophila melanogaster embryos, in which activated Ras alleles suppress Shp2 deficiency (“Introduction”). By contrast, activated K-Ras does not fully rescue Ptpn11 deficiency in lens and lacrimal gland development,11 a finding attributed to the loss of Shp2-mediated antagonism of Sprouty2 (a negative regulator of the Ras/Erk pathway). Our data suggest that Sprouty-independent regulation of Ras signaling is more dominant in murine HSCs and progenitors. Together, our observations are consistent with the genetics of juvenile myelomonocytic leukemia and Noonan syndrome,4 which place mutant PTPN11 in the RAS signaling pathway. Our data show that introducing KrasG12D can suppress the requirement for Ptpn11 in HSCs and immature progenitors. It is not clear whether this reflects a specific requirement for K-Ras and whether N-Ras or H-Ras would have similar effects. Future experiments will also establish which pathways downstream of KrasG12D are required to suppress the Ptpn11Δ/Δ phenotype. Such studies might inform therapeutic approaches and minimize hematologic side effects in treating patients with somatic or germline mutant PTPN11.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Pier-André Pentillä for expert assistance with flow cytometry and David Tuveson and Tyler Jacks for providing LSL-KrasG12D mice.
This work was supported by the National Institutes of Health (grant RO1 CA114945, B.G.N.; grant R01 CA104282, K.M.S.; and in part, grant P20 RR025179, W.Y.), the Ontario Ministry of Health and Long Term Care, Concern Foundation, and the American Society of Hematology (Scholar award; B.S.B.).
The views expressed do not necessarily reflect those of the Ontario Ministry of Health and Long Term Care.
National Institutes of Health
Authorship
Contribution: G.C. and B.S.B. designed and performed research, analyzed data, and wrote the paper; L.S.C., W.Y., M.M., A.D.S., S.G., W.X.H., A.X.L., X.W., M.B., T.S., J.G., and J.L.K. performed research and analyzed data; N.N.I. and J.E.D. analyzed data; K.M.S. designed research and analyzed data; and B.G.N. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of W.Y. is Department of Orthopaedics, and COBRE Center for Stem Cell Biology, Brown University Alpert Medical School, and Rhode Island Hospital, Providence, RI.
Correspondence: Gordon Chan, Campbell Family Cancer Research Institute, Ontario Cancer Institute, 101 College St, TMDT Rm 8-301, Toronto, ON M5G 1L7, Canada; e-mail: gordon.chan@uhnresearch.ca; and Benjamin G. Neel, Campbell Family Cancer Research Institute, Ontario Cancer Institute, 101 College St, TMDT Rm 8-301, Toronto, ON M5G 1L7, Canada; e-mail: bneel@uhnresearch.ca.
References
Author notes
L.S.C. and W.Y. contributed equally to this study.

![Figure 3. Ptpn11 deletion leads to decreased quiescence and increased cell death. (A-B) Cell cycle analysis of Ptpn11Δ/Δ HSCs and progenitors. LK (A) and LSKCD150+ (B) cells were subjected to 2-parameter analysis of DNA content (Hoechst 33342 [H]) vs RNA content (Pyronin [PY]). The mean percentages (± SEM) of cells in G0 (H−PY−), G1(H−PY+), and S/G2M (H+PY+) are indicated. *P < .05, Student t test. (C) Lin-c-kit+Sca1− and (D) LSKCD150+ cells harvested from control (n = 3) and Ptpn11Δ/Δ (n = 3) mice were stained with annexin V to assess cell death at the indicated times after pIpC induction. The average percentage (± SEM) of annexin V+ cells is shown. *P < .05, Student t test. (E-F) Single SLAM-HSCs were sorted into 60-well plates (1 cell/well) and cultured in media containing SCF, TPO, Flt3L, and interleukin-11. Proliferation of each clone was evaluated microscopically over 4 days. Representative data from one of 4 experiments with similar results are shown. The average percentages (± SEM) of surviving clones from the 4 experiments are shown at the top of each panel. *P < .05, Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/16/10.1182_blood-2010-11-319517/4/m_zh89991169760003.jpeg?Expires=1766026209&Signature=PQJHAEZ6FuJ7ecml2WVgry-CPR6CvFxFhkOUJAjR68HuLg6dFTE78a4Jwy41a3zjbtFx4LwIQ0yBHQ9BgvMUJc45DdN6CgniMCLZDrx6zuNsEvNIWvxV0lxbZue4Qh8aFQQlokvly-gXsq2ISSXCe6aDz~gXmR~tg48UxSYgnSj00YjdxqG~GD94Q7z5Nf6NlbFlhIF8vUDm5J1~DOkxy1DgbaSkgygrgRgyrW8z0bcKT3u-vy61DBRDpHPQh0b6d-aLA~uUpY7lwhRdvzotAeIZv1ls78PneUCP~m2D6aos3AV1oaE9R83-BelCfDv4eyNX0Rg3PediXMqgP-7FuA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal