Abstract
t(12;21)(p13;q22)[ETV6-RUNX1] is the most common chromosomal translocation in childhood acute lymphoblastic leukemia, and it can often be backtracked to Guthrie cards supporting prenatal initiation and high levels of circulating t(12;21)-positive cells at birth. To explore the prevalence of ETV6-RUNX1–positive cells in healthy neonates, mononuclear cells from 1417 umbilical cord blood samples were isolated within 24 hours from birth and subsequently screened for ETV6-RUNX1 transcripts using a highly sensitive real-time reverse transcription polymerase chain reaction assay. In first-run polymerase chain reaction, 14 samples were positive at levels below 10−5, of which specific hybridization reflecting the relevant genetic region was positive in 9 cases. Repeated analyses using stored mRNA and flowcytometric sorting of a CD19+, CD8+, and CD19−/CD8− subpopulations from cryopreserved mononuclear cells from the same cord blood samples (mean sorted: 18 × 106 cells) revealed no positive findings, which demonstrates that the level and/or frequency of ETV6-RUNX1–positive cells is markedly lower than suggested in previous studies.
Introduction
The development of childhood B-cell lineage acute lymphoblastic leukemia (ALL) involves (at least) 2 genetic events (hits),1 the first of which frequently arises prenatally.2-11 Unless the postnatal genetic hit(s) is inevitable, the prevalence of newborns harboring preleukemic first hit–cells should exceed the cumulative incidence of the corresponding leukemia. Accordingly, gene transcripts from the most common childhood ALL chromosomal translocation, t(12;21)(p13;q22)[ETV6-RUNX1], were demonstrated in approximately 1% of healthy newborns in one British study,12 corresponding to 100-fold the cumulative incidence of ETV6-RUNX1–positive ALL in childhood.13 Noteworthy, the positive cells were reported to occur at levels of 10−3-10−4, and a threshold of 10−5 was used for classification of t(12;2)-positive samples with no report of subthreshold frequencies.12 Since these results are important for mapping the natural history of t(12;21)-positive ALL and furthermore limits the options for future screening, we assessed the prevalence of ETV6-RUNX–positive cells in 1417 newborns.
Methods
Mononucleated cells (MNCs) were isolated from umbilical cord blood (UCB) samples from healthy, full-term newborns through Ficoll density centrifugation14 within 24 hours after birth to minimize cellular and/or mRNA degradation.15 This study was approved by the Danish Data Protection Agency and the Danish Scientific Ethics Committee.
mRNA from ≥ 2.5 × 106 MNCs was extracted using a KingFisher mL robot (ThermoFisher) and the MagAttrackDirect mRNA-M48 Kit (QIAGEN): 750 μL of lysis solution, 75 μL of magnetic beads, and 550 μL of washing solution I+II. mRNA was eluted in 50 μL of RNase-free water, divided into 2 tubes, and stored at −80°C until use. Surplus MNCs were cryopreserved in liquid nitrogen. For subsequent flow cytometrically sorted subpopulations, mRNA was collected in 25 μL.
cDNA was generated from 18 μL of mRNA as previously described16 for the initial screening or using 5μM random hexamers for the sorted subpopulations. Each of the 1417 samples was screened in triplicates for ETV6-RUNX1 by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The ABL gene was amplified simultaneously to test the cDNA quality. In a 30-μL PCR mixture, 10 μL of cDNA was mixed with 1× PCR Universal Mastermix (Applied Biosystems), 10μM of each primer, and 6μM TaqMan probe (DNA Technology). Primers are described previously.17 A 50-cycle PCR was performed in a 7500 Fast Real-Time PCR System cycler (Applied Biosystems). In case of positive results, the reaction was repeated using stored mRNA from the initial mRNA isolation. All positive products were verified using previously described dot-blot method.16
A functional fusion gene should occur in the same cell lineages as the corresponding leukemia.18 Accordingly, to confirm the initial qRT-PCR findings, cryopreserved MNCs from the same UCB sample were thawed and flow cytometrically sorted into a CD19+-subpopulation, a CD8+ subpopulation (as negative control), and a remaining CD19−/CD8− population. The cells were incubated with anti–human CD19(PC5), anti-human CD8(RFE), anti–human CD4(fluorescein isothiocyanate [FITC]) according to the manufacturer's instructions (DAKO), and subpopulations were positively sorted using a MoFlo XDP Cell Sorter (Beckman-Coulter). Gates were set according to an isotype control. The subpopulation purity was verified by reanalysis.
Subpopulations were analyzed as described above. A parallel qRT-PCR assay amplifying the CD19 transcript using previously described primer/TaqMan probes19 confirmed the predicted ΔCtCD19, which should reflect the potential ΔCtETV6-RUNX1, if the ETV6-RUNX1–positive cells were restricted to the CD19+ subpopulation. As the first sorted samples proved negative, total RNA from the flow cytometrically sorted cell subpopulations was further purified in the remaining 6 of 14 cases using the RNeasy Plus Mini Kit (QIAGEN).
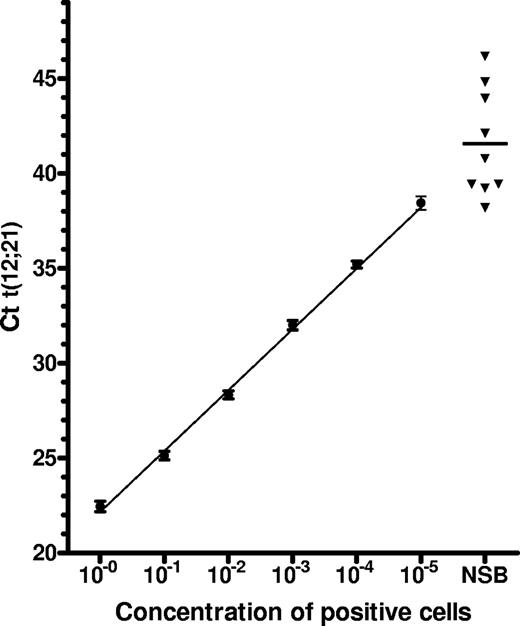
Standard cell concentration curves from serial dilutions in cord blood mononuclear cells of ETV6-RUNX1–positive diagnostic bone marrow cells from 3 children with ALL had detection rates of 100% at frequencies of 10−4 and 80% for frequencies at 10−5 (Figure 1).
Results and discussion
UCB samples from 1417 neonates were collected. Mean gestational age was 40 weeks plus 0.6 days (range, 37 weeks + 0 days to 42 weeks + 5 days). Mean time from birth to sample processing (ie, MNC harvest, mRNA isolation, and storage of frozen MNCs) was 12.34 hours (range, 0.5-23.5 hours). The mean number of cells obtained was 26.8 × 106 (interquartile range [IQR], 18.2 × 106 to 32.8 × 106). The mean number of MNCs stored in liquid nitrogen was 18.0 × 106 (IQR, 12.2 × 106-22.0 × 106). The mean cycle threshold for ABL (CtABL) was 24.01 (95% interval: 21.49-26.53), which is within the expected range of this control gene in < 24-hour-old samples.16,17
In the first screening, 14 individuals were ETV6-RUNX1 transcript–positive at a mean Ct(ETV6-RUNX1) of 43.3 (range, 38.2-49.0) (Table 1). Two cases were positive in 2 of 3 triplicates. Using standardized dilution curves, the levels of ETV6-RUNX1–positive cells were estimated to be < 10−5 (Figure 1). Dot-blot analyses confirmed the presence of the ETV6-RUNX1 transcript in only 9 of the 14 cases. The positive signals could not be confirmed by rescreening of stored mRNA in any of the 14 cases.
Results of screening of healthy neonates
| Patient no. . | Maternal age (y + mo) . | GA (wk + d) . | Time to processing (h) . | Initial screening . | Second screening, all negative for ETV6-RUNX1, no. of cells (/106) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ABL . | ETV6-RUNX1 . | No. positive/total no. . | Dot-blot . | CD19 . | CD8 . | Remaining . | ||||
| 48 900 | 38 + 8 | 39 + 1 | 1.25 | 23.81 | 39.24 | 1/6 | + | 1.032 | 0.518 | n/a |
| 49 248 | 27 + 10 | 41 + 1 | 4.75 | 23.56 | 39.43 | 1/6 | + | 0.166 | 0.414 | n/a |
| 49 915 | 29 + 9 | 41 + 3 | 19 | 22.12 | 43.26 | 1/6 | + | 0.648 | 0.862 | n/a |
| 49 975 | 24 + 5 | 38 + 0 | 4 | 24.10 | 38.20 | 1/6 | + | 0.930 | 0.594 | n/a |
| 50 220* | 36 + 0 | 40 + 2 | 9.5 | 23.29 | 43.96 | 1/6 | + | 1.979 | 1.090 | 12.703 |
| 50 262* | 30 + 1 | 39 + 0 | 8.75 | 23.52 | 42.11 | 1/6 | − | 1.018 | 0.605 | 5.620 |
| 50 902 | 26 + 5 | 40 + 3 | 21.75 | 25.18 | 44.82 / 45.07 | 2/6 | − | 0.821 | 0.277 | n/a |
| 51 255 | 37 + 6 | 40 + 0 | 2.5 | 24.69 | 49.02 | 1/6 | − | 0.892 | 0.342 | 6.395 |
| 51 455* | 33 + 6 | 41 + 3 | 5.25 | 25.31 | 48.83 | 1/6 | − | 1.335 | 1.114 | 9.939 |
| 51 522 | 29 + 11 | 41 + 2 | 4.5 | 24.78 | 46.18 | 1/6 | + | 0.891 | 0.342 | 6.395 |
| 51 530* | 28 + 4 | 37 + 0 | 23.5 | 24.08 | 44.67 | 1/6 | + | 0.635 | 0.515 | 2.452 |
| 51 885* | 33 + 8 | 41 + 1 | 9 | 24.00 | 39.45 | 1/6 | + | 1.387 | 0.590 | 6.493 |
| 52 084* | 40 + 6 | 37 + 0 | 6.5 | 24.42 | 40.79 | 1/6 | + | 0.785 | 0.474 | 4.001 |
| 52 254* | 28 + 5 | 41 + 5 | 19.75 | 23.01 | 42.57 / 45.65 | 2/6 | − | 1.467 | 0.840 | 6.795 |
| Mean | 31 + 0.9 | 39 + 6.5 | 10 | 23.99 | 43.33 | n/a | n/a | 0.999 | 0.613 | 6.755 |
| Patient no. . | Maternal age (y + mo) . | GA (wk + d) . | Time to processing (h) . | Initial screening . | Second screening, all negative for ETV6-RUNX1, no. of cells (/106) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ABL . | ETV6-RUNX1 . | No. positive/total no. . | Dot-blot . | CD19 . | CD8 . | Remaining . | ||||
| 48 900 | 38 + 8 | 39 + 1 | 1.25 | 23.81 | 39.24 | 1/6 | + | 1.032 | 0.518 | n/a |
| 49 248 | 27 + 10 | 41 + 1 | 4.75 | 23.56 | 39.43 | 1/6 | + | 0.166 | 0.414 | n/a |
| 49 915 | 29 + 9 | 41 + 3 | 19 | 22.12 | 43.26 | 1/6 | + | 0.648 | 0.862 | n/a |
| 49 975 | 24 + 5 | 38 + 0 | 4 | 24.10 | 38.20 | 1/6 | + | 0.930 | 0.594 | n/a |
| 50 220* | 36 + 0 | 40 + 2 | 9.5 | 23.29 | 43.96 | 1/6 | + | 1.979 | 1.090 | 12.703 |
| 50 262* | 30 + 1 | 39 + 0 | 8.75 | 23.52 | 42.11 | 1/6 | − | 1.018 | 0.605 | 5.620 |
| 50 902 | 26 + 5 | 40 + 3 | 21.75 | 25.18 | 44.82 / 45.07 | 2/6 | − | 0.821 | 0.277 | n/a |
| 51 255 | 37 + 6 | 40 + 0 | 2.5 | 24.69 | 49.02 | 1/6 | − | 0.892 | 0.342 | 6.395 |
| 51 455* | 33 + 6 | 41 + 3 | 5.25 | 25.31 | 48.83 | 1/6 | − | 1.335 | 1.114 | 9.939 |
| 51 522 | 29 + 11 | 41 + 2 | 4.5 | 24.78 | 46.18 | 1/6 | + | 0.891 | 0.342 | 6.395 |
| 51 530* | 28 + 4 | 37 + 0 | 23.5 | 24.08 | 44.67 | 1/6 | + | 0.635 | 0.515 | 2.452 |
| 51 885* | 33 + 8 | 41 + 1 | 9 | 24.00 | 39.45 | 1/6 | + | 1.387 | 0.590 | 6.493 |
| 52 084* | 40 + 6 | 37 + 0 | 6.5 | 24.42 | 40.79 | 1/6 | + | 0.785 | 0.474 | 4.001 |
| 52 254* | 28 + 5 | 41 + 5 | 19.75 | 23.01 | 42.57 / 45.65 | 2/6 | − | 1.467 | 0.840 | 6.795 |
| Mean | 31 + 0.9 | 39 + 6.5 | 10 | 23.99 | 43.33 | n/a | n/a | 0.999 | 0.613 | 6.755 |
+ indicates positive; −, negative; and n/a, not available.
Indicates that sample had total RNA analyzed.
Results of first screening versus sensitivity assay. ■ Mean Ct value of 10-fold dilutions of 2 × 106ETV6-RUNX1–positive cells diluted in lymphocytes from UCB samples. Error bars represent 1 SD. The linear regression line is shown. ▾ Ct value of samples found positive in the first screening round (only dot-blot–positive samples shown).
Results of first screening versus sensitivity assay. ■ Mean Ct value of 10-fold dilutions of 2 × 106ETV6-RUNX1–positive cells diluted in lymphocytes from UCB samples. Error bars represent 1 SD. The linear regression line is shown. ▾ Ct value of samples found positive in the first screening round (only dot-blot–positive samples shown).
Cryopreserved cells from all 14 cases were sorted for CD19+ and CD8+ cells. After the first 5 samples were found negative, the CD19−/CD8− population was also analyzed in the remaining 9 cases. Renewed 50-cycle qRT-PCR analyses for the ETV6-RUNX1 transcript were negative in all the subpopulations. The mean CtABL from the CD19+ cells was 25.0.
The commonly held belief of approximately 1% of newborns harboring functional t(12;21) transcripts detectable at levels of 10−3-10−4 is almost exclusively supported by the study by Mori et al,12 whereas the present study (the largest to date) indicates far lower levels (< 10−5, ie, below the threshold of positivity set by Mori et al12 ). Equally significant is the absence of positive findings in parallel or subsequent PCR runs, including CD19+ cell–enriched subpopulations, suggesting that our initial observations of ETV6-RUNX1–positive samples reflect very infrequent and low-level contamination20 or very low levels (< 10−5) of ETV6-RUNX1–positive cells in a higher number of newborns, thus with random RT-PCR postivity of the rarely occurring ETV6-RUNX1 transcript in a cord blood sample. The diverging results of the present study and the previous paper by Mori et al12 may reflect differences between the studied populations (this paper having studied a predominately white Danish population), or more likely, relate to differences in the applied methods of screening and confirmation.
First, we used fresh UCB samples to eliminate potential artifacts from the freezing and thawing process when using stored cells. Of relevance for the confirmation analyses, the prefreezing protocols were similar in the present study and that of Mori et al.12
Second, we chose mRNA over total RNA since (1) mRNA is generally the preferred choice when maximum sensitivity is required,21 (2) RNA-to-cDNA conversion may be negatively affected by nonspecific or background RNA present in the RT reaction,22 (3) mRNA serves as the initial template in the initial RT-dependent RNA-to-cDNA conversion of the fusion gene, and (4) previous findings indicated a slightly superior sensitivity of the former in detecting RUNX1-ETV6 transcripts at low levels.16
Third, ETV6-RUNX1–fusion gene sequence–specific primers were used in the initial RNA-to-cDNA conversion, as opposed to nonspecific20 random hexamers. The latter may overestimate mRNA copy numbers23 and has been described as the least reliable cDNA priming method.24 As the postsorting qRT-PCR analyses aimed at confirming first round results, random hexamers were used because overestimation would be an advantage when dealing with cell levels ≤ 10−5.
Fourth, we used 7 times as much mRNA-derived cDNA as Mori et al in each first-round PCR screening, which increases the chance of finding rare events in this study: mRNA was purified from 2.5 × 106 cells, approximately 45% hereof was used for RNA-to-cDNA conversion, and 16.7% of the cDNA was added to each of the 3 PCR wells. Mori et al started with approximately 106 cells, the proportion thereof used for RNA-to-cDNA conversion was unreported, and 1/40 μL was used for qRT-PCR.
Fifth, we chose not to use the nested PCR technique applied by Mori et al,12 as it is nonquantitative and involves an “open-tube” step with an increased risk of contamination of the laboratory environment. It seems unlikely that the lack of positive findings at cell-levels of 10−3-10−4 can be explained by the applied screening method as: (1) the chosen assay consistently demonstrated ETV6-RUNX1–positive cells at levels ≥ 10−5 in validation experiments,16,17 (2) qRT-PCR generally is the most sensitive technique for detection of rare mRNA targets,20 and (3) previous findings12 indicate that qRT-PCR is equal or probably superior to nested PCR when exploring ETV6-RUNX1 cDNA.
In addition, studies of infrequent cells and low copy number mRNA are vulnerable to contamination, especially when studying identical targets. Due to the lack of confirmation in repeated PCR runs, we cannot entirely exclude the possibility of low-level contamination in the screening round. A background of contaminating ETV6-RUNX1 signals may not only overestimate the prevalence, but also increase the signal of true positive cases.
Assessment of PCR-related contamination is ideally done by confirming result at cellular levels, which can be done by at least 3 approaches: (1) increased qRT-PCR signal obtained by enrichment of the relevant subpopulations leading to higher numbers of the relevant cells (eg, CD19+ positive, in subsequent analyses), (2) sorting of subpopulations and subsequent demonstration of positive/negative mRNA/cDNA findings in the expected positive/negative subpopulations, and (3) fluorescent in situ hybridization (FISH).
The first approach demands relatively high amounts of cells, and our study is the only one to have attempted this. However, the positive results of the screening round could not be confirmed.
The second approach may, similar to the first screening round, be affected by random contamination, and can thus only be relied upon if the patterns of positive subpopulation are as expected in a reasonably high number of samples. Mori et al12 apparently attempted this in one sample.
The third approach depends on the level of investigated ETV6-RUNX1–positive cells. However, the very low levels found in this study preclude the use thereof. Since preleukemic cells are rare, FISH is technically demanding and is further burdened by subjective interpretations of the signals. Overlapping cells and/or cell fragments can be falsely interpreted as ETV6-RUNX1–positive signals. Thus, the FISH signal for the normal RUNX1 and the weak signal for the truncated RUNX1 should be clearly separated by at least the size of one normal signal
In the Mori et al12 study, (1) approximately 22 000 hematopoietic cells were studied with 35 cells (in 8 subpopulations) being classified as ETV6-RUNX1–positive, (2) the only mentioned control sample also demonstrated one cell interpreted as being positive, and (3) the only positive FISH illustration in the paper of an UCB sample12 demonstrates a cell classified as ETV6-RUNX1–positive, but rests (in addition to the fusion signal) on 2 close RUNX1 signals that are not incontrovertibly separate and could represent an uneven normal fluorescent RUNX1 signal.
In conclusion, our study indicates that the frequency and/or levels of t(12;21)-positive cells in healthy neonates is considerably lower than previously reported, and more importantly below the levels that allow detection of clone-specific ETV6-RUNX1 translocations in Guthrie cards in the majority of children who later developed t(12;21)-positive ALL, since Guthrie cards rarely contain more than 105 cells. This supports that future screening of cord blood could be an option to identify children at risk of developing t(12;21)-positive leukemia.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michael Timm for assistance in the flow cytometric analysis.
This study received financial support from the Danish Cancer Society (grant no. DP06136), and The Danish Childhood Cancer Foundation. K.S. holds the Danish Childhood Cancer Foundation professorship in Paediatric Oncology.
Authorship
Contribution: U.L.-T. designed the study, preformed all laboratory analyses, and wrote the manuscript; H.O.M. assisted in the laboratory analyses; T.R.V. assisted in collecting the samples and preparing the manuscript; H.H. designed the study; J.N. assisted with the flow cytometric analyses; and K.S. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kjeld Schmiegelow, Department of Paediatrics, Section 5704, The University Hospital Rigshospitalet, Blegdamsvej 9, 2100 Copenhagen, Denmark; e-mail: Kjeld.schmiegelow@rh.regionh.dk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal