To the editor:
We read with interest the report in Blood by Lausten-Thomsen et al.1 In 2002, we published that 6 of 567 unselected cord bloods (∼ 1%) had ETV6-RUNX1 (also known as TEL-AML1)–positive cells at a frequency of 10−3 to 10−4.2 This was in accord with the evidence that pediatric acute lymphoblastic leukemia (ALL) is frequently initiated before birth via gene fusion3 and indicated that preleukemic clones may be generated during fetal development at a substantially higher frequency (∼ 100 times) than manifests as overt leukemia (ETV6-RUNX1+ ALL) during childhood.
Lausten-Thomsen et al screened 1417 cord blood samples for ETV6-RUNX1 fusion expression. Fourteen (∼ 1%) registered positive on first polymerase chain reaction (PCR) screen on cDNA prepared from fresh, unfrozen cord blood but none were confirmed on rescreening stored mRNA or RNA derived from frozen, immunosorted cells. They suggest that our positive results (and their own “first-round” results) could have been false-positives with reverse transcription–PCR (RT-PCR) contamination. We think this is very unlikely in Mori et al.2
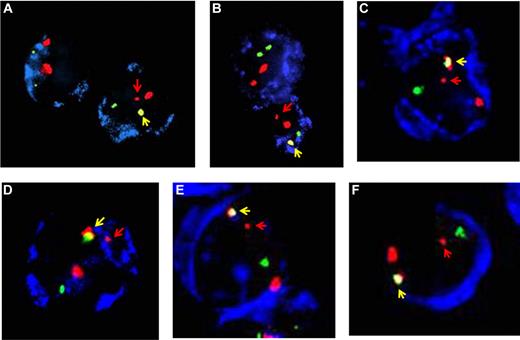
First, we used 4 separate laboratories for extracting RNA, making cDNA, running PCR arrays, and handling our positive leukemia samples or cell lines. Second, we performed parallel quantitative PCR (Q-PCR; TaqMan) screens on all samples and the results were entirely concordant between RT-PCR and Q-PCR. Third, we confirmed positivity (by RT-PCR and Q-PCR) on a separate sample of cells retrieved from the frozen whole cord blood unit. Fourth, and critically, we validated positive samples by fluorescence in situ hybridization (FISH). This latter result cannot be attributed to contamination. Lausten-Thomsen et al also suggest that our FISH data are unconvincing. They say that the image we provided (Figure 5C in Mori et al) is compromised by overlapping signals or subjective interpretation. On this they are plainly wrong. The FISH scoring was very stringent in only including those cells in which there was 1 large and 1 small RUNX1 signal which were well separated. As further examples, we are including 5 other “preleukemic” cells from cord blood that we screened in the same study (Figure 1).2 The designation of these cells as having a ETV6-RUNX1 fusion signal plus a small truncated RUNX1 (= remnant of translocation) plus larger noninvolved RUNX1 (chromosome 21) is unambiguous. This was the case for all 35 cells scored as positive. We also draw to the attention of Lausten-Thomsen et al and readers of Blood that all ETV6-RUNX1+ cells in cord blood were exclusively CD19 and/or CD10 positive (as in ETV6-RUNX1+ ALL itself). In the one cord blood positive for AML1-ETO, the FISH-positive cells were myeloid lineage. FISH screening was also concordant with Q-PCR with levels of ETV6-RUNX1+ cells recorded in both tests as 10−3 to 10−4 in samples scored as positive.
FISH images of cells in newborn cord blood with ETV6-RUNX1 gene fusion. (A) Original image from Mori et al2 (Figure 5C). (B-F) Additional images from other ETV6-RUNX1+ cord blood screened in Mori et al.2 Blue indicates CD19 or CD10; red, RUNX1; green, ETV6; yellow (arrowed), ETV6-RUNX1 fusion; and red arrow, residual (small) RUNX1 signal.
FISH images of cells in newborn cord blood with ETV6-RUNX1 gene fusion. (A) Original image from Mori et al2 (Figure 5C). (B-F) Additional images from other ETV6-RUNX1+ cord blood screened in Mori et al.2 Blue indicates CD19 or CD10; red, RUNX1; green, ETV6; yellow (arrowed), ETV6-RUNX1 fusion; and red arrow, residual (small) RUNX1 signal.
Lausten-Thomsen et al further suggest that our control cord blood sample (ETV6-RUNX1 negative by both RT-PCR and Q-PCR) for FISH was also “positive.” In that control (Table 1 in Mori et al2 ), one CD10-positive cell was scored as ETV6-RUNX1+ of 3199 cells screened giving a crude frequency of < 0.03, some 10-fold less than in our designated “positive” cord samples. Little can be said on the significance of a single cell, though it would have been helpful to have screened by FISH more cord blood that was negative by RT-PCR/Q-PCR. Collectively, the FISH data cannot be explained by artefacts.
If our original data2 were entirely correct, as we firmly believe, then the negative results of Lausten-Thomsen et al do require an explanation. We have none to offer except to point out that in their first round of PCR screening on freshly extracted mRNA, they did in fact find 14 of 1417 positive, that is, as in Mori et al,2 ∼ 1% positives. They failed to confirm that any of these 14 samples were positive with frozen mRNA or on mRNA extracted from frozen cells. Perhaps the authors should ask whether, in their frozen material, ETV6-RUNX1 mRNA stability is robust and whether their assay with stored cord blood cells or mRNA really does have the sensitivity implied by their model experiment with ETV6-RUNX1+ leukemic cells (Figure 1 in Lausten-Thomsen et al1 ).
Another group has independently produced a similar result to ours with ∼ 1%-2% ETV6-RUNXI–positive samples.4 Nevertheless, it would be helpful if others with access to a large series of cord blood samples would carry out another, carefully controlled, independent screen for ETV6-RUNX1 and perhaps other leukemic fusion genes.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Mel Greaves, Section of Haemato-Oncology, The Institute of Cancer Research, Brookes Lawley Bldg, 15 Cotswold Rd, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: mel.greaves@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal