Abstract
Diagnostic karyotype provides the framework for risk-stratification schemes in acute myeloid leukemia (AML); however, the prognostic significance of many rare recurring cytogenetic abnormalities remains uncertain. We studied the outcomes of 5876 patients (16-59 years of age) who were classified into 54 cytogenetic subgroups and treated in the Medical Research Council trials. In multivariable analysis, t(15;17)(q22;q21), t(8;21)(q22;q22), and inv(16)(p13q22)/t(16;16)(p13;q22) were the only abnormalities found to predict a relatively favorable prognosis (P < .001). In patients with t(15;17) treated with extended all-trans retinoic acid and anthracycline-based chemotherapy, additional cytogenetic changes did not have an impact on prognosis. Similarly, additional abnormalities did not have a significant adverse effect in t(8;21) AML; whereas in patients with inv(16), the presence of additional changes, particularly +22, predicted a better outcome (P = .004). In multivariable analyses, various abnormalities predicted a significantly poorer outcome, namely abn(3q) (excluding t(3;5)(q25;q34)), inv(3)(q21q26)/t(3;3)(q21;q26), add(5q)/del(5q), −5, −7, add(7q)/del(7q), t(6;11)(q27;q23), t(10;11)(p11∼13;q23), other t(11q23) (excluding t(9;11)(p21∼22;q23) and t(11;19)(q23;p13)), t(9;22)(q34;q11), −17, and abn(17p). Patients lacking the aforementioned favorable or adverse aberrations but with 4 or more unrelated abnormalities also exhibited a significantly poorer prognosis (designated “complex” karyotype group). These data allow more reliable prediction of outcome for patients with rarer abnormalities and may facilitate the development of consensus in reporting of karyotypic information in clinical trials involving younger adults with AML. This study is registered at http://www.isrctn.org as ISRCTN55678797 and ISRCTN17161961.
Introduction
Diagnostic karyotype is one of the most powerful independent prognostic indicators in acute myeloid leukemia (AML), which serves to identify biologically distinct subsets of disease and has been widely adopted to provide the framework for risk-adapted treatment approaches (reviewed in Grimwade,1 Mrózek et al,2 and Grimwade and Hills3 ). The authors of large multicenter studies4-10 have consistently reported that patients with acute promyelocytic leukemia (APL) with the t(15;17)(q22;q21)/PML-RARA treated with all-trans retinoic acid (ATRA) and anthracycline-based protocols and those with core binding factor (CBF) leukemias with t(8;21)(q22;q22)/RUNX1-RUNX1T1 or inv(16)(p13q22)/t(16;16)(p13;q22)/CBFB-MYH11 receiving intensive chemotherapy involving cytarabine at a range of doses are associated with a relatively favorable outcome, whereas those AML patients with abnormalities of 3q (abn(3q)), deletions of 5q (del(5q)), monosomies of chromosome 5 and/or 7 (-5/-7), or complex karyotype are associated with very poor prognoses.
However, there has been little consensus as to the outcome of cases with rare recurring cytogenetic abnormalities (ie, individual incidence < 2%), which together account for approximately 10% of AML and have variably been considered to predict an intermediate or adverse prognosis.4-10 Further sources of inconsistency between cytogenetic classification systems adopted by different trial groups relate to the prognostic impact of additional abnormalities in patients with favorable karyotype, particularly accompanying the t(8;21), the outcome of translocations involving the MLL locus at 11q23, and the level of cytogenetic complexity considered to confer adverse risk. Moreover, the authors of a recent study involving 1975 adults (ages 15-60 years) with AML11 suggested the existence of a novel adverse-risk group characterized by the presence of an autosomal monosomy in conjunction with at least one other autosomal monosomy or structural abnormality (denoted monosomal karyotype positive, MK+).
In the hierarchical Medical Research Council (MRC) cytogenetic classification system, which was developed more than a decade ago by the analysis of a cohort of 1612 children and younger adults (< 55 years) treated in the MRC AML10 trial, 3 cytogenetic risk groups were distinguished.4 Patients with t(15;17), t(8;21), and inv(16), irrespective of the presence of additional cytogenetic changes, were assigned to the “favorable- risk” group; patients lacking any of these aberrations and found to have abn(3q), del(5q), −5/−7, or complex karyotype (ie, 5 or more unrelated cytogenetic abnormalities) were defined as “adverse risk.” The remaining patients, that is, those with normal karyotype and other structural or numerical abnormalities, comprised the “intermediate-risk” group. In the original MRC study, infrequent abnormalities that were present in fewer than 20 patients were not considered individually and were assigned to the intermediate-risk group; however, it was recognized that there was likely to be considerable heterogeneity in clinical outcome according to the nature of these rare cytogenetic entities, of relevance in informing clinical management and the development of more appropriate risk-stratified treatment approaches for such patients. To begin to address this issue and with the aim of further refining cytogenetic classification of AML, which could ultimately facilitate comparison of clinical trial data from different groups, we considered the impact of karyotype on outcome in a much larger cohort of younger adult patients treated in the MRC trials.
Methods
Patients
The study cohort comprised 5876 AML cases with successful karyotype analysis enrolled in successive MRC trials conducted between May 1988 and January 2009: AML10 (1988-1995, n = 1238), AML12 (1995-2002, n = 2241), and AML15 (2002-2009, n = 2397), including 435 cases with secondary AML. The median age of the patients was 44 years (range, 16-59 years). AML was diagnosed and classified according to the French-American-British classification in the AML10 and AML12 trials; in AML15 the revised diagnostic criteria of the World Health Organization (WHO) classification12 were adopted. Sample collection and analyses were approved by the Multicenter Research Ethics Committee for Wales.
Therapy
All patients received intensive anthracycline and cytarabine (Ara-C)–based combination chemotherapy. Details of the AML10 treatment protocol have been published previously; in brief, patients were randomized to receive induction therapy with 2 courses of DAT (daunorubicin, Ara-C, 6-thioguanine: course 1, DAT 3 + 10; course 2, DAT 3 + 8) or ADE (Ara-C, daunorubicin, etoposide: course 1, ADE 10 + 3 + 5; course 2, ADE 8 + 3 + 5).13 The third and fourth courses of consolidation chemotherapy comprised MACE (m-amsacrine, Ara-C, etoposide) and MidAC (mitoxantrone, Ara-C), respectively. In this trial, researchers investigated the role of autologous and allogeneic bone marrow transplantation after this intensive therapy, as described.14,15
AML12 was determined on the basis of the marginally better induction regimen from AML 10 (ADE), and the standard treatment template became ADE, ADE, MACE, MidAC. AML12 investigated whether mitoxantrone might be superior to and less cardiotoxic than daunorubicin by comparing ADE with mitoxantrone, Ara-C, and etoposide (ie, MAE).16,17 AML10 risk group stratification was applied in AML12 for the delivery of risk-directed therapy,18 with allogeneic transplantation in first complete remission being restricted to standard and adverse risk patients and autologous transplantation was not a treatment option. After the randomization of 1658 patients to the ADE versus mitoxantrone, Ara-C, and etoposide, the induction schedule was changed to compare 2 dose levels of Ara-C (200 vs 100 mg/m2 given twice daily) in a DAT schedule with or without ATRA. All patients in AML12 were eligible for randomization to receive 4 versus 5 courses of treatment, where for standard- and adverse-risk adults the final course could be either chemotherapy or stem cell transplantation (allogeneic for patients with a sibling donor, autologous otherwise). The additional course of treatment was idarubicin 10 mg/m2 days 1 to 3, cytarabine 100 mg/m2 days 1 to 5 every 12 hours, and etoposide 100 mg/m2 days 1 to 5.
In AML15, adult patients who did not have APL were randomized to receive ADE (as given in AML10 and AML12), DA (course 1, DA 3 + 10: daunorubicin 50 mg/m2 days 1,3,5; Ara-C 100 mg/m2 days 1-10 every 12 hours; course 2, DA 3 + 8: daunorubicin 50 mg/m2 days 1,3,5; Ara-C 100 mg/m2 days 1-8 every 12 hours) or FLAG-Ida (course 1, fludarabine 30 mg/m2 days 2-6; Ara-C 2 g/m2 days 2-6; granulocyte colony-stimulating factor 263 μg days 1-7; idarubicin 8 mg/m2 days 4,5,6; course 2, idem). Patients also were randomized to receive gemtuzumab ozogamicin (3 mg protein/m2) or not on day 1 of course 1. After the recruitment of 1113 patients, the gemtuzumab ozogamicin randomization in induction was discontinued, and patients with FLT3 mutant AML were randomized to receive lestaurtinib or not after each of the first 4 treatment courses. Patients with standard- or poor-risk disease with an available donor were eligible for transplantation. Standard allograft was given as course 3; for patients older than 45 years of age, a miniallograft was recommended and given as course 4 after MACE consolidation. The remaining patients were randomized to standard MRC consolidation chemotherapy (MACE + MidAC) or high-dose cytarabine at doses of either 1.5 g/m2 or 3.0 g/m2 and were subrandomized to receive gemtuzumab ozogamicin (3 mg protein/m2) or not on day 1 of course 3 (excluding those patients receiving lestaurtinib). Patients also were randomized to receive a fifth course, that is, cytarabine at a dose of 1.5 g/m2; after the early results of AML12, this randomization was restricted to younger patients only (ie, aged < 45 years).
From January 1993, patients with APL were eligible for the MRC ATRA trial and were randomized to receive either short or extended courses of ATRA in combination with induction chemotherapy as per the AML 10/12 protocol.19 After the closure of the MRC ATRA trial (January 1997), patients with APL entered into AML12 routinely received an extended course of ATRA commenced simultaneously with induction chemotherapy and continued until achievement of morphologic remission (to a maximum of 60 days) followed by MRC combination chemotherapy. In AML15, the MRC treatment schedule used in AML12 was compared with a PETHEMA schedule. In addition, patients were randomized to receive gemtuzumab ozogamicin or not on day 1 of course 3, as described.20
Cytogenetics
Cytogenetic analysis was performed on metaphases from bone marrow aspirates taken at diagnosis with the use of standard procedures. This step was performed in the regional cytogenetics laboratories, whose satisfactory performance was monitored by a national external quality assurance scheme: United Kingdom National External Quality Assessment Service for Clinical Cytogenetics.21 Karyotypes were entered into the Leukaemia and Lymphoma Research (LLR) United Kingdom Cancer Cytogenetics Group (UKCCG) Karyotype Database of Acute Leukaemia,22 as well as the clinical trials database. Patients were classified as having an abnormal, normal, or failed cytogenetic result. A result was regarded as normal after analysis of 20 or more normal metaphases. Analysis of less than 20 normal metaphases was regarded as a failure. Karyotypes were not routinely analyzed centrally but were reviewed for accuracy in description of the structural and numerical, clonal chromosomal abnormalities, which were reported in accordance with the International System for Human Cytogenetic Nomenclature23 and classified according to the presence of the chromosomal abnormalities shown in Table 1. According to this scheme, abnormal karyotypes with more than 1 abnormality were classified into several relevant groups. Cases with none of these changes were classified as “other.” Karyotype complexity was defined by the number of unrelated abnormalities present from 1 to 5 or greater. A balanced translocation, for instance, t(8;21)(q22;q22), was defined as a single abnormality because the 2 events leading to it are related. Trisomy and monosomy were regarded as single abnormalities. Two abnormalities included the gain of 2 chromosomes, even if they were the same, or the gain of a derived chromosome. Unbalanced translocations leading to gain and loss of chromosomal material, for example, der(7)t(1;7)(q21;q22) also were counted as 2 abnormalities.
Frequency and demographics of chromosomal abnormalities
| Chromosome involved . | Description of abnormality . | Patients, no. (%)* . | Median age, y (range) . | P . | Secondary disease (% of those with abnormality) . | P . |
|---|---|---|---|---|---|---|
| — | Normal karyotype | 2432 (41) | 46 (16-59) | 135 (6) | ||
| 1 | Abnormality of 1p | 86 (2) | 43.5 (16-59) | .9 | 16 (19) | < .001 |
| t(1;22)(p13;q13) | 1 (< 0.5) | 37 | 0 | |||
| Abnormality of 1q | 84 (1) | 48 (16-59) | .09 | 12 (14) | .03 | |
| 3 | Monosomy 3 | 39 (1) | 51 (22-59) | .002 | 9 (23) | .002 |
| Abnormality of 3q | ||||||
| inv(3)(q21q26)/t(3;3)(q21;q26) | 69 (1) | 43 (18-59) | .7 | 11 (17) | .02 | |
| t(3;5)(q21∼25;q31∼35) | 26 (< .5) | 30.5 (16-58) | .005 | 3 (12) | .4 | |
| Other abnormality of 3q | 108 (2) | 46.5 (18-59) | .03 | 16 (15) | .008 | |
| 4 | Trisomy 4 | 70 (1) | 43 (18-59) | .7 | 5 (7) | > .999 |
| 5 | Abnormality of 5q | |||||
| Monosomy 5 | 129 (2) | 51 (18-59) | < .001 | 24 (19) | < .001 | |
| del(5q) | 146 (2) | 51 (16-59) | < .001 | 28 (19) | < .001 | |
| add(5q) | 60 (1) | 45.5 (18-59) | .4 | 9 (15) | .04 | |
| 6 | Trisomy 6 | 65 (1) | 49 (16-59) | .03 | 10 (15) | .03 |
| t(6;9)(p23;q34) | 42 (1) | 44 (19-59) | .5 | 2 (5) | .8 | |
| Abnormality of 6q, not t(6;11) | 79 (1) | 44 (16-59) | .9 | 9 (11) | .19 | |
| 7 | Monosomy 7 | 279 (5) | 47 (16-59) | < .001 | 54 (19) | < .001 |
| Abnormality of 7q | ||||||
| del(7q) | 145 (2) | 49 (16-59) | < .001 | 28 (19) | < .001 | |
| add(7q) | 68 (1) | 47 (16-59) | .02 | 9 (13) | .1 | |
| Abnormality of 7p | 81 (1) | 42 (16-59) | .5 | 17 (21) | < .001 | |
| 8 | Trisomy 8 | 547 (10) | 44 (16-59) | .7 | 57 (10) | .008 |
| t(8;21)(q22;q22) and variants | 421 (7) | 40 (16-59) | < .001 | 13 (3) | < .001 | |
| Abnormality of 8p11∼12 | 23 (< .5) | 32 (16-58) | .05 | 2 (9) | .7 | |
| 9 | Monosomy 9 | 25 (< .5) | 47 (17-57) | .4 | 3 (12) | .4 |
| t(9;22)(q34;q11) and variants | 47 (1) | 43 (22-58) | .7 | 1 (2) | .3 | |
| Deletion of 9q, including add(9q) | 133 (2) | 45 (16-59) | .7 | 10 (8) | .9 | |
| 11 | Trisomy 11 | 81 (1) | 51 (16-59) | < .001 | 7 (9) | .7 |
| All 11q23 | ||||||
| t(9;11)(p21∼22;q23) | 61 (1) | 38 (16-58) | < .001 | 6 (10) | .5 | |
| t(10;11)(p11∼14;q13∼23) | 34 (1) | 33.5 (16-59) | < .001 | 0 | .18 | |
| t(6;11)(q27;q23) | 24 (< .5) | 33 (17-57) | .001 | 1 (4) | > .999 | |
| t(11;19)(q23;p13) | 30 (1) | 35.5 (16-57) | .01 | 4 (13) | .3 | |
| Other 11q23 | 62 (1) | 38.5 (17-59) | .008 | 7 (11) | .2 | |
| Abnormality of 11q (not 11q23) | 117 (2) | 43 (16-59) | .9 | 9 (8) | .9 | |
| Abnormality of 11p13∼15 | 37 (1) | 40 (16-57) | .14 | 7 (19) | .007 | |
| 12 | Abnormality of 12p | |||||
| Monosomy 12 | 57 (1) | 52 (18-59) | < .001 | 11 (19) | .003 | |
| Other abnormality of 12p13 | 50 (1) | 46 (16-58) | .4 | 4 (8) | .8 | |
| Other abnormality of 12p, not 12p13 | 93 (2) | 45 (17-59) | .4 | 19 (20) | < .001 | |
| 13 | Trisomy 13 | 93 (2) | 50 (16-59) | < .001 | 12 (13) | .07 |
| Abnormality of 13q | ||||||
| Monosomy 13 | 73 (1) | 49 (16-59) | .01 | 14 (19) | < .001 | |
| Deletion of 13q | 27 (< .5) | 42 (20-59) | .17 | 2 (7) | > .999 | |
| 15 | t(15;17)(q22;q21) and variants | 788 (13) | 39 (16-59) | < .001 | 24 (3) | < .001 |
| Abnormality of 15q, not t(15;17) | 40 (1) | 42.5 (19-58) | .4 | 3 (8) | 1.0 | |
| 16 | inv(16)(p13q22)/t(16;16)(p13;q22) | 284 (5) | 38 (16-59) | < .001 | 12 (4) | .04 |
| Abnormality of 16q, not inv(16) | 91 (2) | 43 (16-59) | .8 | 12 (13) | .04 | |
| 17 | Monosomy 17 | 121 (2) | 51 (16-59) | < .001 | 15 (12) | .05 |
| Abnormality of 17p | 145 (2) | 46 (16-59) | .03 | 20 (14) | .006 | |
| 18 | Monosomy 18 | 97 (2) | 49 (18-59) | < .001 | 20 (21) | < .001 |
| 19 | Trisomy 19 | 58 (1) | 41 (16-59) | .15 | 4 (7) | > .999 |
| 20 | Monosomy 20 | 53 (1) | 49 (16-59) | .004 | 10 (19) | .005 |
| Abnormality of 20q | 48 (1) | 49 (20-59) | .04 | 4 (8) | .8 | |
| 21 | Trisomy 21 (acquired) | 148 (3) | 46 (17-59) | .13 | 21 (14) | .004 |
| Abnormality of 21q, not t(8;21) | 74 (1) | 49 (16-59) | .004 | 15 (20) | < .001 | |
| 22 | Trisomy 22 | 113 (2) | 42 (18-59) | .9 | 10 (9) | .6 |
| X | Loss of X | 109 (2) | 41 (16-59) | .16 | 9 (8) | .7 |
| Y | Loss of Y | 200 (3) | 42 (16-59) | .4 | 10 (5) | .2 |
| Other† | 139 (2) | 46 (16-59) | .05 | 14 (10) | .2 | |
| Level of karyotype complexity | ||||||
| 1 abnormality | 1830 (31) | 42 (16-59) | 131 (7) | |||
| 2 abnormalities | 786 (13) | 40 (16-59) | 70 (9) | |||
| 3 abnormalities | 275 (5) | 41 (17-59) | 17 (6) | |||
| 4 abnormalities | 123 (2) | 42 (16-59) | 14 (11) | |||
| 5 or more abnormalities | 430 (7) | 49 (16-59) | 68 (16) | |||
| P for trend | .09 | < .001 |
| Chromosome involved . | Description of abnormality . | Patients, no. (%)* . | Median age, y (range) . | P . | Secondary disease (% of those with abnormality) . | P . |
|---|---|---|---|---|---|---|
| — | Normal karyotype | 2432 (41) | 46 (16-59) | 135 (6) | ||
| 1 | Abnormality of 1p | 86 (2) | 43.5 (16-59) | .9 | 16 (19) | < .001 |
| t(1;22)(p13;q13) | 1 (< 0.5) | 37 | 0 | |||
| Abnormality of 1q | 84 (1) | 48 (16-59) | .09 | 12 (14) | .03 | |
| 3 | Monosomy 3 | 39 (1) | 51 (22-59) | .002 | 9 (23) | .002 |
| Abnormality of 3q | ||||||
| inv(3)(q21q26)/t(3;3)(q21;q26) | 69 (1) | 43 (18-59) | .7 | 11 (17) | .02 | |
| t(3;5)(q21∼25;q31∼35) | 26 (< .5) | 30.5 (16-58) | .005 | 3 (12) | .4 | |
| Other abnormality of 3q | 108 (2) | 46.5 (18-59) | .03 | 16 (15) | .008 | |
| 4 | Trisomy 4 | 70 (1) | 43 (18-59) | .7 | 5 (7) | > .999 |
| 5 | Abnormality of 5q | |||||
| Monosomy 5 | 129 (2) | 51 (18-59) | < .001 | 24 (19) | < .001 | |
| del(5q) | 146 (2) | 51 (16-59) | < .001 | 28 (19) | < .001 | |
| add(5q) | 60 (1) | 45.5 (18-59) | .4 | 9 (15) | .04 | |
| 6 | Trisomy 6 | 65 (1) | 49 (16-59) | .03 | 10 (15) | .03 |
| t(6;9)(p23;q34) | 42 (1) | 44 (19-59) | .5 | 2 (5) | .8 | |
| Abnormality of 6q, not t(6;11) | 79 (1) | 44 (16-59) | .9 | 9 (11) | .19 | |
| 7 | Monosomy 7 | 279 (5) | 47 (16-59) | < .001 | 54 (19) | < .001 |
| Abnormality of 7q | ||||||
| del(7q) | 145 (2) | 49 (16-59) | < .001 | 28 (19) | < .001 | |
| add(7q) | 68 (1) | 47 (16-59) | .02 | 9 (13) | .1 | |
| Abnormality of 7p | 81 (1) | 42 (16-59) | .5 | 17 (21) | < .001 | |
| 8 | Trisomy 8 | 547 (10) | 44 (16-59) | .7 | 57 (10) | .008 |
| t(8;21)(q22;q22) and variants | 421 (7) | 40 (16-59) | < .001 | 13 (3) | < .001 | |
| Abnormality of 8p11∼12 | 23 (< .5) | 32 (16-58) | .05 | 2 (9) | .7 | |
| 9 | Monosomy 9 | 25 (< .5) | 47 (17-57) | .4 | 3 (12) | .4 |
| t(9;22)(q34;q11) and variants | 47 (1) | 43 (22-58) | .7 | 1 (2) | .3 | |
| Deletion of 9q, including add(9q) | 133 (2) | 45 (16-59) | .7 | 10 (8) | .9 | |
| 11 | Trisomy 11 | 81 (1) | 51 (16-59) | < .001 | 7 (9) | .7 |
| All 11q23 | ||||||
| t(9;11)(p21∼22;q23) | 61 (1) | 38 (16-58) | < .001 | 6 (10) | .5 | |
| t(10;11)(p11∼14;q13∼23) | 34 (1) | 33.5 (16-59) | < .001 | 0 | .18 | |
| t(6;11)(q27;q23) | 24 (< .5) | 33 (17-57) | .001 | 1 (4) | > .999 | |
| t(11;19)(q23;p13) | 30 (1) | 35.5 (16-57) | .01 | 4 (13) | .3 | |
| Other 11q23 | 62 (1) | 38.5 (17-59) | .008 | 7 (11) | .2 | |
| Abnormality of 11q (not 11q23) | 117 (2) | 43 (16-59) | .9 | 9 (8) | .9 | |
| Abnormality of 11p13∼15 | 37 (1) | 40 (16-57) | .14 | 7 (19) | .007 | |
| 12 | Abnormality of 12p | |||||
| Monosomy 12 | 57 (1) | 52 (18-59) | < .001 | 11 (19) | .003 | |
| Other abnormality of 12p13 | 50 (1) | 46 (16-58) | .4 | 4 (8) | .8 | |
| Other abnormality of 12p, not 12p13 | 93 (2) | 45 (17-59) | .4 | 19 (20) | < .001 | |
| 13 | Trisomy 13 | 93 (2) | 50 (16-59) | < .001 | 12 (13) | .07 |
| Abnormality of 13q | ||||||
| Monosomy 13 | 73 (1) | 49 (16-59) | .01 | 14 (19) | < .001 | |
| Deletion of 13q | 27 (< .5) | 42 (20-59) | .17 | 2 (7) | > .999 | |
| 15 | t(15;17)(q22;q21) and variants | 788 (13) | 39 (16-59) | < .001 | 24 (3) | < .001 |
| Abnormality of 15q, not t(15;17) | 40 (1) | 42.5 (19-58) | .4 | 3 (8) | 1.0 | |
| 16 | inv(16)(p13q22)/t(16;16)(p13;q22) | 284 (5) | 38 (16-59) | < .001 | 12 (4) | .04 |
| Abnormality of 16q, not inv(16) | 91 (2) | 43 (16-59) | .8 | 12 (13) | .04 | |
| 17 | Monosomy 17 | 121 (2) | 51 (16-59) | < .001 | 15 (12) | .05 |
| Abnormality of 17p | 145 (2) | 46 (16-59) | .03 | 20 (14) | .006 | |
| 18 | Monosomy 18 | 97 (2) | 49 (18-59) | < .001 | 20 (21) | < .001 |
| 19 | Trisomy 19 | 58 (1) | 41 (16-59) | .15 | 4 (7) | > .999 |
| 20 | Monosomy 20 | 53 (1) | 49 (16-59) | .004 | 10 (19) | .005 |
| Abnormality of 20q | 48 (1) | 49 (20-59) | .04 | 4 (8) | .8 | |
| 21 | Trisomy 21 (acquired) | 148 (3) | 46 (17-59) | .13 | 21 (14) | .004 |
| Abnormality of 21q, not t(8;21) | 74 (1) | 49 (16-59) | .004 | 15 (20) | < .001 | |
| 22 | Trisomy 22 | 113 (2) | 42 (18-59) | .9 | 10 (9) | .6 |
| X | Loss of X | 109 (2) | 41 (16-59) | .16 | 9 (8) | .7 |
| Y | Loss of Y | 200 (3) | 42 (16-59) | .4 | 10 (5) | .2 |
| Other† | 139 (2) | 46 (16-59) | .05 | 14 (10) | .2 | |
| Level of karyotype complexity | ||||||
| 1 abnormality | 1830 (31) | 42 (16-59) | 131 (7) | |||
| 2 abnormalities | 786 (13) | 40 (16-59) | 70 (9) | |||
| 3 abnormalities | 275 (5) | 41 (17-59) | 17 (6) | |||
| 4 abnormalities | 123 (2) | 42 (16-59) | 14 (11) | |||
| 5 or more abnormalities | 430 (7) | 49 (16-59) | 68 (16) | |||
| P for trend | .09 | < .001 |
Cases were categorized according to presence of the cytogenetic entities previously defined in a large cohort of pediatric AML patients treated in the MRC AML trials.21 According to this scheme, abnormal karyotypes with more than 1 abnormality were classified into several relevant groups. AML indicates acute myeloid leukemia; and MRC, Medical Research Council.
As percentage of cases with a successful cytogenetic result.
Other karyotypes, not classified into any listed group.
End points and statistics
Outcome data were analyzed in patients with recurring cytogenetic abnormalities occurring in at least 20 patients across the 3 trials. Patients were defined as having a complete response (CR/CR with incomplete blood count recovery) if they exhibited a normocellular bone marrow aspirate containing less than 5% leukemic blasts and showed evidence of normal maturation of other marrow elements. Remission failures were classified by the investigating clinician as caused by induction death (death related to treatment and/or hypoplasia within 30 days) or resistant disease (failure to eliminate disease, including partial remissions). Where clinician evaluation was not available, deaths within 30 days were deemed induction death and other failures resistant disease. The following definitions are also used: overall survival (OS) was the time from randomization to death. For remitters, relapse-free survival was the time from CR/CR with incomplete blood count recovery to first event (relapse or death in CR); cumulative incidence of relapse (CIR) is the cumulative probability of relapse with death in CR as competing risk. OS/relapse-free survival/CIR percentages are quoted at 10 years. Surviving patients were censored on October 26, 2008 (AML10,12), or January 1, 2009 (AML15), when follow-up was complete for 97% of patients (the small number of patients lost to follow-up are censored at the date they were last known to be alive). Median follow-up was 7.3 years (range, 0.1-20.5 years). There was some difference in outcome between patients with cytogenetic data and those with either failed samples or no sample (10-year OS: with cytogenetics 40% vs failed 46% vs no sample 34%); after adjustment for age, white blood cell count (WBC), secondary disease, and performance status, the difference remained significant (P < .001).
Demographics, remission rates, and reasons for failure to achieve CR were compared by the use of χ2 and Mantel-Haenszel tests. Kaplan-Meier life-tables were constructed for time to event and unstratified comparisons were made by use of the log-rank test. Outcomes of patients with particular abnormalities were compared with the normal karyotype group. Odds ratios with standard errors were calculated. All P values were 2-tailed. To allow for multiple testing, the level of significance was set at P less than .01, and 99% confidence intervals (99% CIs) were presented for effect sizes. Multivariate modeling was performed by the use of logistic and Cox regression analyses with a forward selection method. All multivariate analyses had as candidate variables the cytogenetic abnormalities listed in Table 1, after adjustment for other well-known prognostic variables, including age, WBC, type of AML (de novo/secondary), with performance status as defined by the WHO and clinical trial (AML10/AML12/ AML15) as covariates. P values were those for entry to the model by use of the deviance statistic; Wald confidence intervals were used. Throughout the analyses, odds ratios greater than 1 indicated a worse outcome for the abnormality under consideration.
Results
Distribution of cytogenetic abnormalities in younger adults with AML
Overall, 2432 of 5876 (41%) of patients had a normal karyotype; frequencies of the various cytogenetic abnormalities identified in the remaining patients are shown in Table 1. Together, recurrent balanced chromosomal abnormalities that are the cytogenetic hallmarks of genetically defined disease entities in the revised WHO classification24 were identified in 28% of cases, namely t(15;17)(q22;q21) (13%), t(8;21)(q22;q22) (7%), inv(16)(p13q22)/t(16;16)(p13;q22) (5%), t(6;9)(p23;q34) (1%), t(9;11)(p21∼22;q23) (1%), and inv(3)(q21q26)/t(3;3)(q21;q26) (1%). These abnormalities were confirmed to be mutually exclusive (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In patients lacking one of the aforementioned recurrent genetic abnormalities (n = 4211); 750 (18%) harbored particular cytogenetic abnormalities that have been collectively designated as “MDS related” in the 2008 WHO classification. “MDS-related” unbalanced abnormalities were present in 711 cases (−7/del(7q), n = 336; −5/del(5q), n = 258; i(17q)/t(17p), n = 104; −13/del(13q), n = 97; del(11q), n = 109; del(12p)/t(12p), n = 88; del(9q), n = 69; idic(X)(q13), n = 4) and balanced abnormalities were identified in 42 cases (t(11;16)(q23;p13), n = 2; t(3;21)(q26;q22), n = 9; t(1;3)(p36;q21), n = 2; t(2;11)(p21;q23), n = 1; t(5;12)(q33;p12), n = 2; t(5;7)(q33;q11), n = 1; t(3;5)(q25;q34), n = 25). There were significant differences in the distribution of cytogenetic abnormalities with respect to age, with balanced rearrangements t(3;5)(q21∼25;q31∼35), t(8;21)(q22;q22), t(15;17)(q22;q21), inv(16)(p13q22), and t(11q23) typically occurring in younger patients, whereas unbalanced abnormalities, including various monosomies, del(5q), del(7q), and trisomies of chromosome 11 and 13, were overrepresented in older patients (Table 1). We also undertook a systematic analysis to establish which abnormalities tend to coexist, revealing several significant associations (supplemental Table 1).
Impact of cytogenetic abnormalities on disease outcome
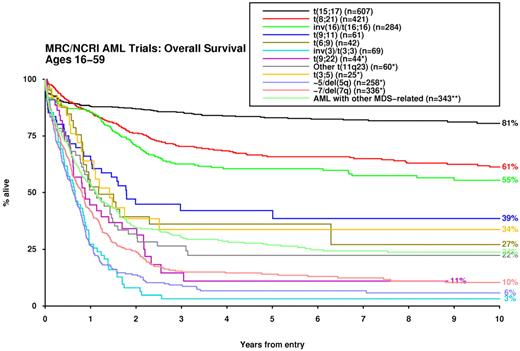
For analysis of patient outcomes, those with the t(15;17) who were not confirmed as receiving a long ATRA treatment regimen were excluded from all analyses (n = 181). In comparison with normal karyotype, a considerable number of cytogenetic abnormalities were predictive of disease outcome with respect to response to induction therapy, risk of relapse and OS, both in univariate analysis and after adjustment for age, WBC, secondary disease, and performance status (Table 2). Accordingly, significant differences in survival were observed among the cytogenetic entities specified in the 2008 WHO Classification (Figure 1).
Impact of cytogenetic abnormalities compared with normal karyotype on disease outcome
| Chromosome involved . | Description of abnormality . | CR* . | OS . | CIR . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rate, % . | Unadjusted OR (99% CI), P† . | Adjusted OR (99% CI), P† . | 10-y OS, % . | Unadjusted HR (99% CI), P‡ . | Adjusted HR (99% CI), P§ . | 10-y CIR . | Unadjusted HR (99% CI), P‡ . | Adjusted HR (99% CI), P§ . | ||
| — | Normal karyotype | 90 | 38 | 49 | ||||||
| 1 | Abnormality of 1p | 68 | 4.13 (2.21-7.71), < .001 | 5.51 (2.81-10.80), < .001 | 20 | 2.58 (1.64-4.05), < .001 | 2.20 (1.57-3.08), < .001 | 58 | 1.64 (0.93-2.91), .03 | 1.62 (1.01-2.60), .008 |
| Abnormality of 1q | 63 | 5.19 (2.82-9.55), < .001 | 5.78 (3.02-11.07), < .001 | 21 | 2.26 (1.45-3.54), < .001 | 1.88 (1.33-2.65), < .001 | 55 | 1.26 (0.72-2.19), .3 | 1.30 (0.78-2.15), .19 | |
| 3 | Monosomy 3 | 46 | 10.16 (4.36-23.65), < .001 | 12.11 (5.02-29.19), < .001 | 3 | 24.46 (10.01-59.76), < .001 | 4.20 (2.68-6.58), < .001 | 82 | 75.92 (17.15-336.0), < .001 | 5.17 (2.68-9.96), < .001 |
| Abnormality of 3q | ||||||||||
| inv(3)(q21q26)/t(3;3)(q21;q26) | 36 | 15.33 (7.86-29.88), < .001 | 19.80 (9.79-40.07), < .001 | 3 | 13.22 (7.14-24.48), < .001 | 4.07 (2.89-5.72), < .001 | 89 | 14.41 (4.64-44.74), < .001 | 4.04 (2.22-7.36), < .001 | |
| t(3;5)(q21∼25;q31∼35) | 96 | 0.36 (0.03-5.06), .3 | 0.44 (0.03-6.51), .4 | 34 | 1.38 (0.66-2.90), .3 | 1.41 (0.72-2.77), .18 | 52 | 1.46 (0.61-3.50), .3 | 1.53 (0.72-3.25), .14 | |
| Other abnormality of 3q | 59 | 6.08 (3.56-10.38), < .001 | 6.98 (3.97-12.25), < .001 | 11.3 | 5.20 (3.32-8.14), < .001 | 2.77 (2.08-3.68), < .001 | 71 | 5.21 (2.70-10.05), < .001 | 2.71 (1.79-4.09), < .001 | |
| 4 | Trisomy 4 | 87 | 1.31 (0.51-3.33), .5 | 1.55 (0.57-4.24), .3 | 16 | 1.15 (0.74-1.80), .4 | 1.23 (0.80-1.88), .2 | 54 | 1.09 (0.64-1.83), .7 | 1.13 (0.68-1.87), .5 |
| 5 | Abnormality of 5q | |||||||||
| Monosomy 5 | 57 | 6.56 (4.02-10.72), < .001 | 7.58 (4.49-12.80), < .001 | 0 | 20.31 (12.70-32.47), < .001 | 4.33 (3.35-5.61), < .001 | 75 | 16.98 (8.42-34.23), < .001 | 3.80 (2.59-5.57), < .001 | |
| del(5q) | 58 | 6.22 (3.90-9.93), < .001 | 7.28 (4.40-12.06), < .001 | 12 | 5.14 (3.47-7.62), < .001 | 2.90 (2.23-3.76), < .001 | 64 | 3.02 (1.75-5.19), < .001 | 2.19 (1.47-3.27), < .001 | |
| add(5q) | 53 | 7.62 (3.83-15.17), < .001 | 10.38 (4.96-21.72), < .001 | 10 | 10.49 (5.34-20.60), < .001 | 3.58 (2.42-5.30), < .001 | 64 | 6.05 (2.25-16.28), < .001 | 2.84 (1.53-5.26), < .001 | |
| 6 | Trisomy 6 | 78 | 2.44 (1.10-5.41), .004 | 2.52 (1.11-5.73), .003 | 21 | 2.34 (1.40-3.92), < .001 | 1.88 (1.28-2.76), < .001 | 62 | 2.12 (1.12-4.01), .005 | 1.75 (1.08-2.85), .003 |
| t(6;9)(p23;q34) | 88 | 1.18 (0.34-4.06), .7 | 1.65 (0.47-5.82), .3 | 27 | 1.47 (0.77-2.81), .14 | 1.55 (0.88-2.74), .04 | 62 | 1.70 (0.76-3.79), .10 | 1.61 (0.82-3.16), .06 | |
| Abnormality of 6q, not t(6;11) | 63 | 5.05 (2.70-9.44), < .001 | 6.15 (3.16-11.96), < .001 | 21 | 2.52 (1.58-4.02), < .001 | 2.09 (1.47-2.96), < .001 | 53 | 1.32 (0.73-2.38), .2 | 1.33 (0.78-2.26), .17 | |
| 7 | Monosomy 7 | 58 | 6.37 (4.46-9.11), < .001 | 7.63 (5.18-11.24), < .001 | 8 | 5.57 (4.20-7.38), < .001 | 3.03 (2.50-3.67), < .001 | 70 | 4.38 (2.92-6.55), < .001 | 2.60 (1.98-3.42), < .001 |
| Abnormality of 7q | ||||||||||
| del(7q) | 77 | 2.64 (1.54-4.52), < .001 | 2.76 (1.56-4.89), < .001 | 26 | 1.66 (1.20-2.31), < .001 | 1.51 (1.14-1.98), < .001 | 57 | 1.32 (0.88-1.98), .08 | 1.26 (0.87-1.81), .11 | |
| add(7q) | 68 | 4.17 (2.09-8.30), < .001 | 5.08 (2.43-10.61), < .001 | 30 | 2.25 (1.34-3.79), < .001 | 1.97 (1.33-2.94), < .001 | 33 | 0.96 (0.50-1.85), .9 | 1.01 (0.52-1.98), 1.0 | |
| Abnormality of 7p | 65 | 4.69 (2.50-8.79), < .001 | 6.06 (3.09-11.92), < .001 | 22 | 2.18 (1.39-3.43), < .001 | 1.97 (1.39-2.80), < .001 | 61 | 1.30 (0.75-2.25), .2 | 1.31 (0.79-2.17), .17 | |
| 8 | Trisomy 8 | 80 | 2.20 (1.58-3.07), < .001 | 2.64 (1.85-3.77), < .001 | 37 | 1.21 (1.01-1.44), .006 | 1.32 (1.12-1.57), < .001 | 46 | 0.99 (0.80-1.23), .9 | 1.08 (0.87-1.35), .3 |
| t(8;21)(q22;q22) and variants | 97 | 0.26 (0.12-0.56), < .001 | 0.36 (0.16-0.81), < .001 | 61 | .58 (0.49-0.70), < .001 | 0.60 (0.47-0.75), < .001 | 27 | 0.54 (0.44-0.67), < .001 | 0.51 (0.39-0.68), < .001 | |
| Abnormality of 8p11∼12 | 91 | 0.83 (0.12-5.62), 0.8 | 1.32 (0.19-9.26), .7 | 50 | 0.91 (0.43-1.92), .8 | 1.22 (0.56-2.68), .5 | 49 | 1.08 (0.46-2.53), .8 | 1.31 (0.57-2.98), .4 | |
| 9 | Monosomy 9 | 68 | 4.10 (1.34-12.53), .001 | 5.01 (1.57-15.99), < .001 | 8 | 4.04 (1.69-9.67), < .001 | 2.55 (1.42-4.57), < .001 | 63 | 3.00 (0.92-9.78), .03 | 2.13 (0.89-5.06), .02 |
| t(9;22)(q34;q11) and variants | 72 | 3.43 (1.45-8.12), < .001 | 2.80 (1.08-7.27), .004 | 14 | 2.83 (1.53-5.23), < .001 | 1.91 (1.22-3.00), < .001 | 65 | 3.95 (1.59-9.83), < .001 | 2.32 (1.27-4.23), < .001 | |
| Deletion of 9q, including add(9q) | 86 | 1.39 (0.71-2.73), .2 | 1.82 (0.90-3.68), .03 | 47 | 0.80 (0.58-1.09), .05 | 0.83 (0.58-1.19), .19 | 35 | 0.66 (0.46-0.94), .001 | 0.60 (0.38-0.96), .005 | |
| 11 | Trisomy 11 | 75 | 2.86 (1.44-5.67), < .001 | 3.25 (1.57-6.71), < .001 | 13 | 2.26 (1.44-3.55), < .001 | 1.84 (1.30-2.61), < .001 | 71 | 2.71 (1.52-4.84), < .001 | 2.01 (1.31-3.08), < .001 |
| All 11q23 | ||||||||||
| t(9;11)(p21∼22;q23) | 84 | 1.71 (0.69-4.23), .13 | 2.13 (0.82-5.51), .03 | 39 | 1.24 (0.75-2.04), .3 | 1.36 (0.86-2.17), .08 | 44 | 1.00 (0.56-1.79), 1.0 | 1.04 (0.57-1.89), .9 | |
| t(10;11)(p11∼14;q13∼23) | 85 | 1.50 (0.43-5.29), .4 | 2.58 (0.70-9.53), .05 | 12 | 3.57 (1.71-7.45), < .001 | 3.29 (1.99-5.45), < .001 | 71 | 4.94 (1.95-12.55), < .001 | 3.39 (1.87-6.15), < .001 | |
| t(6;11)(q27;q23) | 96 | 0.40 (0.03-5.54), .4 | 0.63 (0.04-9.04), .6 | 9 | 2.80 (1.23-6.38), .004 | 2.56 (1.42-4.61), < .001 | 76 | 5.10 (1.79-14.52), < .001 | 2.97 (1.53-5.74), < .001 | |
| t(11;19)(q23;p13) | 97 | 0.30 (0.02-4.15), .2 | 0.45 (0.03-6.36), .4 | 49 | 0.89 (0.46-1.71), .7 | 1.11 (0.55-2.22), .7 | 44 | 1.03 (0.48-2.19), .9 | 1.21 (0.57-2.56), .5 | |
| Other 11q23 | 75 | 3.03 (1.41-6.53), < .001 | 3.87 (1.70-8.82), < .001 | 21 | 2.55 (1.52-4.27), < .001 | 2.07 (1.41-3.04), < .001 | 65 | 2.40 (1.26-4.59), .002 | 1.84 (1.12-3.03), .001 | |
| Abnormality of 11q (not 11q23) | 69 | 3.92 (2.27-6.76), < .001 | 4.86 (2.72-8.68), < .001 | 20 | 2.33 (1.59-3.42), < .001 | 2.13 (1.59-2.86), < .001 | 62 | 2.02 (1.23-3.32), < .001 | 1.90 (1.28-2.82), < .001 | |
| Abnormality of 11p13∼15 | 73 | 3.23 (1.23-8.50), .002 | 4.58 (1.66-12.64), < .001 | 26 | 1.61 (0.86-3.01), .06 | 1.48 (0.87-2.52), .06 | 53 | 1.08 (0.51-2.27), .8 | 1.02 (0.48-2.15), 1.0 | |
| 12 | Abnormality of 12p | |||||||||
| Monosomy 12 | 59 | 6.07 (2.95-12.48), < .001 | 6.54 (3.05-14.00), < .001 | 6 | 11.79 (5.98-23.28), < .001 | 3.67 (2.51-5.37), < .001 | 84 | 21.17 (7.65-58.60), < .001 | 4.48 (2.65-7.58), < .001 | |
| Other abnormality of 12p13 | 63 | 5.06 (2.31-11.06), < .001 | 6.00 (2.62-13.73), < .001 | 14 | 4.18 (2.21-7.90), < .001 | 2.58 (1.69-3.92), < .001 | 48 | 1.44 (0.64-3.22), .3 | 1.31 (0.64-2.69), .3 | |
| Other abnormality of 12p, not 12p13 | 57 | 6.57 (3.73-11.58), < .001 | 8.45 (4.57-15.62), < .001 | 17 | 4.12 (2.56-6.65), < .001 | 2.75 (1.98-3.82), < .001 | 53 | 1.78 (0.94-3.37), .03 | 1.61 (0.93-2.80), .02 | |
| 13 | Trisomy 13 | 70 | 3.75 (2.05-6.89), < .001 | 3.62 (1.90-6.90), < .001 | 9 | 2.54 (1.64-3.93), < .001 | 1.86 (1.34-2.57), < .001 | 72 | 1.82 (1.05-3.14), .009 | 1.56 (1.00-2.42), .008 |
| Abnormality of 13q | ||||||||||
| Monosomy 13 | 60 | 5.87 (3.09-11.16), < .001 | 7.07 (3.57-14.02), < .001 | 8 | 9.02 (5.06-16.07), < .001 | 3.48 (2.48-4.88), < .001 | 67 | 5.88 (2.65-13.06), < .001 | 2.90 (1.76-4.75), < .001 | |
| Deletion of 13q | 85 | 1.51 (0.37-6.18), .4 | 2.10 (0.49-9.02), .18 | 31 | 1.25 (0.64-2.46), .4 | 1.46 (0.78-2.75), .12 | 61 | 1.45 (0.65-3.26), .2 | 1.61 (0.80-3.24), .07 | |
| 15 | t(15;17)(q22;q21) and variants | 93 | 0.67 (0.43-1.04), .02 | 1.11 (0.69-1.76), .6 | 81 | 0.40 (0.34-0.47), < .001 | 0.30 (0.23-0.39), < .001 | 13 | 0.34 (0.29-0.41), < .001 | 0.19 (0.13-0.27), < .001 |
| Abnormality of 15q, not t(15;17) | 78 | 2.53 (0.94-6.81), .02 | 3.36 (1.15-9.83), .002 | 46 | 0.96 (0.53-1.72), .9 | 1.10 (0.60-2.03), .7 | 45 | 0.91 (0.44-1.85), .8 | 1.07 (0.50-2.25), .8 | |
| 16 | inv(16)(p13q22)/t(16;16)(p13;q22) | 92 | 0.81 (0.46-1.44), .3 | 0.88 (0.48-1.62), .6 | 55 | 0.66 (0.53-0.82), < .001 | 0.64 (0.49-0.84), < .001 | 46 | 0.86 (0.67-1.10), .10 | 0.85 (0.65-1.12), .12 |
| Abnormality of 16q, not inv(16) | 78 | 2.45 (1.25-4.82), < .001 | 2.85 (1.41-5.79), < .001 | 31 | 1.66 (1.10-2.51), .003 | 1.60 (1.13-2.26), < .001 | 58 | 1.44 (0.87-2.40), .07 | 1.36 (0.87-2.13), .08 | |
| 17 | Monosomy 17 | 56 | 6.76 (4.07-11.21), < .001 | 8.20 (4.78-14.09), < .001 | 3 | 15.22 (9.37-24.72), < .001 | 3.96 (3.02-5.19), < .001 | 80 | 13.68 (6.68-28.01), < .001 | 3.62 (2.43-5.41), < .001 |
| Abnormality of 17p | 68 | 4.08 (2.47-6.73), < .001 | 4.91 (2.86-8.41), < .001 | 25 | 2.50 (1.75-3.59), < .001 | 2.11 (1.60-2.78), < .001 | 56 | 1.84 (1.16-2.92), .002 | 1.75 (1.20-2.56), < .001 | |
| 18 | Monosomy 18 | 61 | 5.46 (3.10-9.63), < .001 | 6.04 (3.29-11.06), < .001 | 4 | 12.40 (7.28-21.11), < .001 | 3.57 (2.64-4.84), < .001 | 78 | 15.76 (7.21-34.42), < .001 | 3.61 (2.35-5.56), < .001 |
| 19 | Trisomy 19 | 81 | 2.04 (0.85-4.91), .04 | 2.28 (0.91-5.70), .02 | 12 | 3.11 (1.81-5.34), < .001 | 2.35 (1.61-3.42), < .001 | 74 | 3.97 (2.00-7.86), < .001 | 2.54 (1.62-3.99), < .001 |
| 20 | Monosomy 20 | 67 | 4.35 (1.99-9.54), < .001 | 4.96 (2.19-11.21), < .001 | 6 | 6.65 (3.43-12.89), < .001 | 2.98 (1.99-4.45), < .001 | 88 | 8.77 (3.68-20.89), < .001 | 3.10 (1.88-5.14), < .001 |
| Abnormality of 20q | 66 | 4.49 (2.00-10.12), < .001 | 4.93 (2.05-11.86), < .001 | 16 | 3.59 (1.86-6.94), < .001 | 2.59 (1.65-4.06), < .001 | 71 | 3.81 (1.54-9.42), .001 | 2.62 (1.44-4.77), < .001 | |
| 21 | Trisomy 21 (acquired) | 74 | 3.01 (1.79-5.07), < .001 | 3.36 (1.92-5.88), < .001 | 21 | 1.88 (1.35-2.62), < .001 | 1.69 (1.28-2.21), < .001 | 66 | 1.94 (1.28-2.96), < .001 | 1.69 (1.20-2.37), < .001 |
| Abnormality of 21q, not t(8;21) | 71 | 3.59 (1.80-7.15), < .001 | 4.10 (1.98-8.50), < .001 | 17 | 2.37 (1.44-3.88), < .001 | 1.93 (1.34-2.80), < .001 | 61 | 1.89 (0.99-3.57), .02 | 1.66 (1.00-2.75), .009 | |
| 22 | Trisomy 22 | 84 | 1.67 (0.84-3.31), .05 | 2.04 (1.00-4.14), .009 | 47 | 0.85 (0.61-1.19), .2 | 0.90 (0.63-1.30), .5 | 42 | 0.81 (0.55-1.20), .16 | 0.81 (0.52-1.26), .2 |
| X | Loss of X | 87 | 1.28 (0.60-2.74), .4 | 1.89 (0.86-4.15), .03 | 56 | 0.70 (0.51-0.97), .003 | 0.75 (0.50-1.12), .06 | 23 | 0.54 (0.37-0.78), < .001 | 0.44 (0.24-0.79), < .001 |
| Y | Loss of Y | 89 | 1.08 (0.59-1.99), .7 | 1.47 (0.79-2.74), .11 | 51 | 0.75 (0.58-0.96), .002 | 0.79 (0.59-1.06), .04 | 33 | 0.65 (0.48-0.86), < .001 | 0.62 (0.42-0.90), < .001 |
| Level of karyotype complexity | ||||||||||
| 1 abnormality | 87 | per abnormality: OR 1.37 (1.30-1.46), < .001 | per abnormality: OR 1.42 (1.33-1.51), < .001 | 47 | per abnormality HR 1.17 (1.13-1.20), < .001 | per abnormality: HR 1.19 (1.15-1.22), < .001 | 42 | per abnormality: HR 1.09 (1.04-1.13), < .001 | per abnormality HR 1.11 (1.06-1.16), < .001 | |
| 2 abnormalities | 85 | 45 | 40 | |||||||
| 3 abnormalities | 83 | 48 | 35 | |||||||
| 4 abnormalities | 74 | 30 | 51 | |||||||
| 5 or more abnormalities | 62 | 10 | 69 | |||||||
| Chromosome involved . | Description of abnormality . | CR* . | OS . | CIR . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rate, % . | Unadjusted OR (99% CI), P† . | Adjusted OR (99% CI), P† . | 10-y OS, % . | Unadjusted HR (99% CI), P‡ . | Adjusted HR (99% CI), P§ . | 10-y CIR . | Unadjusted HR (99% CI), P‡ . | Adjusted HR (99% CI), P§ . | ||
| — | Normal karyotype | 90 | 38 | 49 | ||||||
| 1 | Abnormality of 1p | 68 | 4.13 (2.21-7.71), < .001 | 5.51 (2.81-10.80), < .001 | 20 | 2.58 (1.64-4.05), < .001 | 2.20 (1.57-3.08), < .001 | 58 | 1.64 (0.93-2.91), .03 | 1.62 (1.01-2.60), .008 |
| Abnormality of 1q | 63 | 5.19 (2.82-9.55), < .001 | 5.78 (3.02-11.07), < .001 | 21 | 2.26 (1.45-3.54), < .001 | 1.88 (1.33-2.65), < .001 | 55 | 1.26 (0.72-2.19), .3 | 1.30 (0.78-2.15), .19 | |
| 3 | Monosomy 3 | 46 | 10.16 (4.36-23.65), < .001 | 12.11 (5.02-29.19), < .001 | 3 | 24.46 (10.01-59.76), < .001 | 4.20 (2.68-6.58), < .001 | 82 | 75.92 (17.15-336.0), < .001 | 5.17 (2.68-9.96), < .001 |
| Abnormality of 3q | ||||||||||
| inv(3)(q21q26)/t(3;3)(q21;q26) | 36 | 15.33 (7.86-29.88), < .001 | 19.80 (9.79-40.07), < .001 | 3 | 13.22 (7.14-24.48), < .001 | 4.07 (2.89-5.72), < .001 | 89 | 14.41 (4.64-44.74), < .001 | 4.04 (2.22-7.36), < .001 | |
| t(3;5)(q21∼25;q31∼35) | 96 | 0.36 (0.03-5.06), .3 | 0.44 (0.03-6.51), .4 | 34 | 1.38 (0.66-2.90), .3 | 1.41 (0.72-2.77), .18 | 52 | 1.46 (0.61-3.50), .3 | 1.53 (0.72-3.25), .14 | |
| Other abnormality of 3q | 59 | 6.08 (3.56-10.38), < .001 | 6.98 (3.97-12.25), < .001 | 11.3 | 5.20 (3.32-8.14), < .001 | 2.77 (2.08-3.68), < .001 | 71 | 5.21 (2.70-10.05), < .001 | 2.71 (1.79-4.09), < .001 | |
| 4 | Trisomy 4 | 87 | 1.31 (0.51-3.33), .5 | 1.55 (0.57-4.24), .3 | 16 | 1.15 (0.74-1.80), .4 | 1.23 (0.80-1.88), .2 | 54 | 1.09 (0.64-1.83), .7 | 1.13 (0.68-1.87), .5 |
| 5 | Abnormality of 5q | |||||||||
| Monosomy 5 | 57 | 6.56 (4.02-10.72), < .001 | 7.58 (4.49-12.80), < .001 | 0 | 20.31 (12.70-32.47), < .001 | 4.33 (3.35-5.61), < .001 | 75 | 16.98 (8.42-34.23), < .001 | 3.80 (2.59-5.57), < .001 | |
| del(5q) | 58 | 6.22 (3.90-9.93), < .001 | 7.28 (4.40-12.06), < .001 | 12 | 5.14 (3.47-7.62), < .001 | 2.90 (2.23-3.76), < .001 | 64 | 3.02 (1.75-5.19), < .001 | 2.19 (1.47-3.27), < .001 | |
| add(5q) | 53 | 7.62 (3.83-15.17), < .001 | 10.38 (4.96-21.72), < .001 | 10 | 10.49 (5.34-20.60), < .001 | 3.58 (2.42-5.30), < .001 | 64 | 6.05 (2.25-16.28), < .001 | 2.84 (1.53-5.26), < .001 | |
| 6 | Trisomy 6 | 78 | 2.44 (1.10-5.41), .004 | 2.52 (1.11-5.73), .003 | 21 | 2.34 (1.40-3.92), < .001 | 1.88 (1.28-2.76), < .001 | 62 | 2.12 (1.12-4.01), .005 | 1.75 (1.08-2.85), .003 |
| t(6;9)(p23;q34) | 88 | 1.18 (0.34-4.06), .7 | 1.65 (0.47-5.82), .3 | 27 | 1.47 (0.77-2.81), .14 | 1.55 (0.88-2.74), .04 | 62 | 1.70 (0.76-3.79), .10 | 1.61 (0.82-3.16), .06 | |
| Abnormality of 6q, not t(6;11) | 63 | 5.05 (2.70-9.44), < .001 | 6.15 (3.16-11.96), < .001 | 21 | 2.52 (1.58-4.02), < .001 | 2.09 (1.47-2.96), < .001 | 53 | 1.32 (0.73-2.38), .2 | 1.33 (0.78-2.26), .17 | |
| 7 | Monosomy 7 | 58 | 6.37 (4.46-9.11), < .001 | 7.63 (5.18-11.24), < .001 | 8 | 5.57 (4.20-7.38), < .001 | 3.03 (2.50-3.67), < .001 | 70 | 4.38 (2.92-6.55), < .001 | 2.60 (1.98-3.42), < .001 |
| Abnormality of 7q | ||||||||||
| del(7q) | 77 | 2.64 (1.54-4.52), < .001 | 2.76 (1.56-4.89), < .001 | 26 | 1.66 (1.20-2.31), < .001 | 1.51 (1.14-1.98), < .001 | 57 | 1.32 (0.88-1.98), .08 | 1.26 (0.87-1.81), .11 | |
| add(7q) | 68 | 4.17 (2.09-8.30), < .001 | 5.08 (2.43-10.61), < .001 | 30 | 2.25 (1.34-3.79), < .001 | 1.97 (1.33-2.94), < .001 | 33 | 0.96 (0.50-1.85), .9 | 1.01 (0.52-1.98), 1.0 | |
| Abnormality of 7p | 65 | 4.69 (2.50-8.79), < .001 | 6.06 (3.09-11.92), < .001 | 22 | 2.18 (1.39-3.43), < .001 | 1.97 (1.39-2.80), < .001 | 61 | 1.30 (0.75-2.25), .2 | 1.31 (0.79-2.17), .17 | |
| 8 | Trisomy 8 | 80 | 2.20 (1.58-3.07), < .001 | 2.64 (1.85-3.77), < .001 | 37 | 1.21 (1.01-1.44), .006 | 1.32 (1.12-1.57), < .001 | 46 | 0.99 (0.80-1.23), .9 | 1.08 (0.87-1.35), .3 |
| t(8;21)(q22;q22) and variants | 97 | 0.26 (0.12-0.56), < .001 | 0.36 (0.16-0.81), < .001 | 61 | .58 (0.49-0.70), < .001 | 0.60 (0.47-0.75), < .001 | 27 | 0.54 (0.44-0.67), < .001 | 0.51 (0.39-0.68), < .001 | |
| Abnormality of 8p11∼12 | 91 | 0.83 (0.12-5.62), 0.8 | 1.32 (0.19-9.26), .7 | 50 | 0.91 (0.43-1.92), .8 | 1.22 (0.56-2.68), .5 | 49 | 1.08 (0.46-2.53), .8 | 1.31 (0.57-2.98), .4 | |
| 9 | Monosomy 9 | 68 | 4.10 (1.34-12.53), .001 | 5.01 (1.57-15.99), < .001 | 8 | 4.04 (1.69-9.67), < .001 | 2.55 (1.42-4.57), < .001 | 63 | 3.00 (0.92-9.78), .03 | 2.13 (0.89-5.06), .02 |
| t(9;22)(q34;q11) and variants | 72 | 3.43 (1.45-8.12), < .001 | 2.80 (1.08-7.27), .004 | 14 | 2.83 (1.53-5.23), < .001 | 1.91 (1.22-3.00), < .001 | 65 | 3.95 (1.59-9.83), < .001 | 2.32 (1.27-4.23), < .001 | |
| Deletion of 9q, including add(9q) | 86 | 1.39 (0.71-2.73), .2 | 1.82 (0.90-3.68), .03 | 47 | 0.80 (0.58-1.09), .05 | 0.83 (0.58-1.19), .19 | 35 | 0.66 (0.46-0.94), .001 | 0.60 (0.38-0.96), .005 | |
| 11 | Trisomy 11 | 75 | 2.86 (1.44-5.67), < .001 | 3.25 (1.57-6.71), < .001 | 13 | 2.26 (1.44-3.55), < .001 | 1.84 (1.30-2.61), < .001 | 71 | 2.71 (1.52-4.84), < .001 | 2.01 (1.31-3.08), < .001 |
| All 11q23 | ||||||||||
| t(9;11)(p21∼22;q23) | 84 | 1.71 (0.69-4.23), .13 | 2.13 (0.82-5.51), .03 | 39 | 1.24 (0.75-2.04), .3 | 1.36 (0.86-2.17), .08 | 44 | 1.00 (0.56-1.79), 1.0 | 1.04 (0.57-1.89), .9 | |
| t(10;11)(p11∼14;q13∼23) | 85 | 1.50 (0.43-5.29), .4 | 2.58 (0.70-9.53), .05 | 12 | 3.57 (1.71-7.45), < .001 | 3.29 (1.99-5.45), < .001 | 71 | 4.94 (1.95-12.55), < .001 | 3.39 (1.87-6.15), < .001 | |
| t(6;11)(q27;q23) | 96 | 0.40 (0.03-5.54), .4 | 0.63 (0.04-9.04), .6 | 9 | 2.80 (1.23-6.38), .004 | 2.56 (1.42-4.61), < .001 | 76 | 5.10 (1.79-14.52), < .001 | 2.97 (1.53-5.74), < .001 | |
| t(11;19)(q23;p13) | 97 | 0.30 (0.02-4.15), .2 | 0.45 (0.03-6.36), .4 | 49 | 0.89 (0.46-1.71), .7 | 1.11 (0.55-2.22), .7 | 44 | 1.03 (0.48-2.19), .9 | 1.21 (0.57-2.56), .5 | |
| Other 11q23 | 75 | 3.03 (1.41-6.53), < .001 | 3.87 (1.70-8.82), < .001 | 21 | 2.55 (1.52-4.27), < .001 | 2.07 (1.41-3.04), < .001 | 65 | 2.40 (1.26-4.59), .002 | 1.84 (1.12-3.03), .001 | |
| Abnormality of 11q (not 11q23) | 69 | 3.92 (2.27-6.76), < .001 | 4.86 (2.72-8.68), < .001 | 20 | 2.33 (1.59-3.42), < .001 | 2.13 (1.59-2.86), < .001 | 62 | 2.02 (1.23-3.32), < .001 | 1.90 (1.28-2.82), < .001 | |
| Abnormality of 11p13∼15 | 73 | 3.23 (1.23-8.50), .002 | 4.58 (1.66-12.64), < .001 | 26 | 1.61 (0.86-3.01), .06 | 1.48 (0.87-2.52), .06 | 53 | 1.08 (0.51-2.27), .8 | 1.02 (0.48-2.15), 1.0 | |
| 12 | Abnormality of 12p | |||||||||
| Monosomy 12 | 59 | 6.07 (2.95-12.48), < .001 | 6.54 (3.05-14.00), < .001 | 6 | 11.79 (5.98-23.28), < .001 | 3.67 (2.51-5.37), < .001 | 84 | 21.17 (7.65-58.60), < .001 | 4.48 (2.65-7.58), < .001 | |
| Other abnormality of 12p13 | 63 | 5.06 (2.31-11.06), < .001 | 6.00 (2.62-13.73), < .001 | 14 | 4.18 (2.21-7.90), < .001 | 2.58 (1.69-3.92), < .001 | 48 | 1.44 (0.64-3.22), .3 | 1.31 (0.64-2.69), .3 | |
| Other abnormality of 12p, not 12p13 | 57 | 6.57 (3.73-11.58), < .001 | 8.45 (4.57-15.62), < .001 | 17 | 4.12 (2.56-6.65), < .001 | 2.75 (1.98-3.82), < .001 | 53 | 1.78 (0.94-3.37), .03 | 1.61 (0.93-2.80), .02 | |
| 13 | Trisomy 13 | 70 | 3.75 (2.05-6.89), < .001 | 3.62 (1.90-6.90), < .001 | 9 | 2.54 (1.64-3.93), < .001 | 1.86 (1.34-2.57), < .001 | 72 | 1.82 (1.05-3.14), .009 | 1.56 (1.00-2.42), .008 |
| Abnormality of 13q | ||||||||||
| Monosomy 13 | 60 | 5.87 (3.09-11.16), < .001 | 7.07 (3.57-14.02), < .001 | 8 | 9.02 (5.06-16.07), < .001 | 3.48 (2.48-4.88), < .001 | 67 | 5.88 (2.65-13.06), < .001 | 2.90 (1.76-4.75), < .001 | |
| Deletion of 13q | 85 | 1.51 (0.37-6.18), .4 | 2.10 (0.49-9.02), .18 | 31 | 1.25 (0.64-2.46), .4 | 1.46 (0.78-2.75), .12 | 61 | 1.45 (0.65-3.26), .2 | 1.61 (0.80-3.24), .07 | |
| 15 | t(15;17)(q22;q21) and variants | 93 | 0.67 (0.43-1.04), .02 | 1.11 (0.69-1.76), .6 | 81 | 0.40 (0.34-0.47), < .001 | 0.30 (0.23-0.39), < .001 | 13 | 0.34 (0.29-0.41), < .001 | 0.19 (0.13-0.27), < .001 |
| Abnormality of 15q, not t(15;17) | 78 | 2.53 (0.94-6.81), .02 | 3.36 (1.15-9.83), .002 | 46 | 0.96 (0.53-1.72), .9 | 1.10 (0.60-2.03), .7 | 45 | 0.91 (0.44-1.85), .8 | 1.07 (0.50-2.25), .8 | |
| 16 | inv(16)(p13q22)/t(16;16)(p13;q22) | 92 | 0.81 (0.46-1.44), .3 | 0.88 (0.48-1.62), .6 | 55 | 0.66 (0.53-0.82), < .001 | 0.64 (0.49-0.84), < .001 | 46 | 0.86 (0.67-1.10), .10 | 0.85 (0.65-1.12), .12 |
| Abnormality of 16q, not inv(16) | 78 | 2.45 (1.25-4.82), < .001 | 2.85 (1.41-5.79), < .001 | 31 | 1.66 (1.10-2.51), .003 | 1.60 (1.13-2.26), < .001 | 58 | 1.44 (0.87-2.40), .07 | 1.36 (0.87-2.13), .08 | |
| 17 | Monosomy 17 | 56 | 6.76 (4.07-11.21), < .001 | 8.20 (4.78-14.09), < .001 | 3 | 15.22 (9.37-24.72), < .001 | 3.96 (3.02-5.19), < .001 | 80 | 13.68 (6.68-28.01), < .001 | 3.62 (2.43-5.41), < .001 |
| Abnormality of 17p | 68 | 4.08 (2.47-6.73), < .001 | 4.91 (2.86-8.41), < .001 | 25 | 2.50 (1.75-3.59), < .001 | 2.11 (1.60-2.78), < .001 | 56 | 1.84 (1.16-2.92), .002 | 1.75 (1.20-2.56), < .001 | |
| 18 | Monosomy 18 | 61 | 5.46 (3.10-9.63), < .001 | 6.04 (3.29-11.06), < .001 | 4 | 12.40 (7.28-21.11), < .001 | 3.57 (2.64-4.84), < .001 | 78 | 15.76 (7.21-34.42), < .001 | 3.61 (2.35-5.56), < .001 |
| 19 | Trisomy 19 | 81 | 2.04 (0.85-4.91), .04 | 2.28 (0.91-5.70), .02 | 12 | 3.11 (1.81-5.34), < .001 | 2.35 (1.61-3.42), < .001 | 74 | 3.97 (2.00-7.86), < .001 | 2.54 (1.62-3.99), < .001 |
| 20 | Monosomy 20 | 67 | 4.35 (1.99-9.54), < .001 | 4.96 (2.19-11.21), < .001 | 6 | 6.65 (3.43-12.89), < .001 | 2.98 (1.99-4.45), < .001 | 88 | 8.77 (3.68-20.89), < .001 | 3.10 (1.88-5.14), < .001 |
| Abnormality of 20q | 66 | 4.49 (2.00-10.12), < .001 | 4.93 (2.05-11.86), < .001 | 16 | 3.59 (1.86-6.94), < .001 | 2.59 (1.65-4.06), < .001 | 71 | 3.81 (1.54-9.42), .001 | 2.62 (1.44-4.77), < .001 | |
| 21 | Trisomy 21 (acquired) | 74 | 3.01 (1.79-5.07), < .001 | 3.36 (1.92-5.88), < .001 | 21 | 1.88 (1.35-2.62), < .001 | 1.69 (1.28-2.21), < .001 | 66 | 1.94 (1.28-2.96), < .001 | 1.69 (1.20-2.37), < .001 |
| Abnormality of 21q, not t(8;21) | 71 | 3.59 (1.80-7.15), < .001 | 4.10 (1.98-8.50), < .001 | 17 | 2.37 (1.44-3.88), < .001 | 1.93 (1.34-2.80), < .001 | 61 | 1.89 (0.99-3.57), .02 | 1.66 (1.00-2.75), .009 | |
| 22 | Trisomy 22 | 84 | 1.67 (0.84-3.31), .05 | 2.04 (1.00-4.14), .009 | 47 | 0.85 (0.61-1.19), .2 | 0.90 (0.63-1.30), .5 | 42 | 0.81 (0.55-1.20), .16 | 0.81 (0.52-1.26), .2 |
| X | Loss of X | 87 | 1.28 (0.60-2.74), .4 | 1.89 (0.86-4.15), .03 | 56 | 0.70 (0.51-0.97), .003 | 0.75 (0.50-1.12), .06 | 23 | 0.54 (0.37-0.78), < .001 | 0.44 (0.24-0.79), < .001 |
| Y | Loss of Y | 89 | 1.08 (0.59-1.99), .7 | 1.47 (0.79-2.74), .11 | 51 | 0.75 (0.58-0.96), .002 | 0.79 (0.59-1.06), .04 | 33 | 0.65 (0.48-0.86), < .001 | 0.62 (0.42-0.90), < .001 |
| Level of karyotype complexity | ||||||||||
| 1 abnormality | 87 | per abnormality: OR 1.37 (1.30-1.46), < .001 | per abnormality: OR 1.42 (1.33-1.51), < .001 | 47 | per abnormality HR 1.17 (1.13-1.20), < .001 | per abnormality: HR 1.19 (1.15-1.22), < .001 | 42 | per abnormality: HR 1.09 (1.04-1.13), < .001 | per abnormality HR 1.11 (1.06-1.16), < .001 | |
| 2 abnormalities | 85 | 45 | 40 | |||||||
| 3 abnormalities | 83 | 48 | 35 | |||||||
| 4 abnormalities | 74 | 30 | 51 | |||||||
| 5 or more abnormalities | 62 | 10 | 69 | |||||||
All odds ratios/hazard ratios are given compared with normal karyotype. CI indicates confidence interval; CIR, cumulative incidence of relapse; CR, complete remission; HR, hazard ratio; OR, odds ratio; and OS, overall survival.
Remission rates include CR with incomplete count recovery (CRi).
Method of analysis was logistic regression.
Method of analysis was log-rank test except for complexity (Cox regression).
Method of analysis was Cox regression.
Impact of cytogenetic entities recognized in 2008 WHO classification24 on survival. *Excluding patients with t(15;17), t(8;21), inv(16), t(9;11), t(6;9), inv(3)/t(3;3). **Excluding patients with any other abnormalities listed previously.
Impact of cytogenetic entities recognized in 2008 WHO classification24 on survival. *Excluding patients with t(15;17), t(8;21), inv(16), t(9;11), t(6;9), inv(3)/t(3;3). **Excluding patients with any other abnormalities listed previously.
Significance of additional cytogenetic abnormalities in CBF leukemias
To address previous inconsistencies in the risk group assignment of t(8;21)–associated CBF leukemia on the basis of the presence of particular additional abnormalities (eg, deletions of the long arm of chromosome 9 (del(9q))25 and karyotype complexity,26 we investigated a cohort of 421 patients. No significant difference in OS was observed according to whether the t(8;21) was accompanied by del(9q) or the presence of additional abnormalities, compared with those with t(8;21) alone (supplemental Figure 1a-b). Loss of an X chromosome had no impact on outcome, although loss of the Y chromosome in male subjects was associated with a trend (P = .04) for better OS (supplemental Figure 1c-d). In patients with inv(16)/t(16;16), the presence of any additional abnormality was associated with a significantly better outcome, with patients with an additional chromosome 22 having a particularly favorable prognosis (supplemental Figure 2a-b). However, it should be noted that those patients with an additional chromosome 22 had a significantly lower presenting WBC compared with those with inv(16)/t(16;16) alone (median, 18.9; range, 1.6-224.2 vs median, 44.6; range, 1.5-370.0; P < .001).
Significance of additional cytogenetic abnormalities in t(15;17) and other aberrations specified in the 2008 WHO classification
Among the cohort of 607 patients with t(15;17) treated with extended ATRA and anthracycline-based chemotherapy, the presence of additional abnormalities had no significant impact on outcome, with all subsets showing a relatively favorable outcome. This included patients in whom the t(15;17) was accompanied by abnormalities of 17p or karyotypic changes that in their own right would have been considered adverse according to the original MRC classification (supplemental Figure 3a-c).4 We also considered the impact of additional cytogenetic abnormalities in patients with the other recurrent balanced rearrangements recognized by the WHO classification. These did not influence outcome in patients with t(9;11)(p21∼22;q23) (supplemental Figure 4). Patients with inv(3)/t(3;3) had a dismal prognosis irrespective of the presence of monosomy 7 (supplemental Figure 5). The t(6;9)(p23;q34) typically occurred as the sole cytogenetic abnormality and showed a trend to poorer outcome (Table 2), with a 10-year survival of 27% (P = .04 in adjusted analyses).
Refining cytogenetic-risk group classification in younger adults with AML
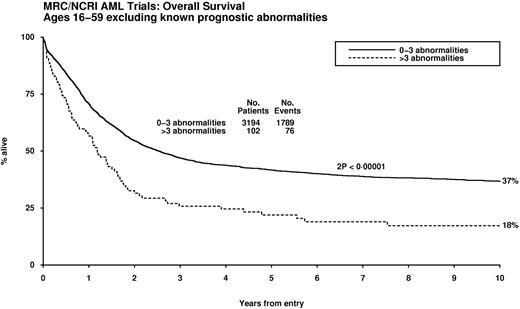
Multivariable analyses were conducted to identify karyotypic abnormalities with independent prognostic significance, taking into account age, presenting WBC, performance status, and type of AML (de novo/secondary). The t(15;17), t(8;21), and inv(16)/t(16;16) were the only abnormalities that were predictive of significantly better outcome (P < .001). After exclusion of this favorable prognostic group, Cox regression analyses revealed various abnormalities that were independently predictive of a significantly poorer outcome (Table 3). The impact of karyotype complexity on outcome in patients without favorable features was investigated after adjustment for the abnormalities with independent prognostic significance in multivariable analysis (ie, any of the aberrations specified in Table 3). Patients with4 or more unrelated abnormalities exhibited a significantly poorer prognosis (hazard ratio [HR] 1.58, 95% CI 1.29-1.94, P < .001), and this was seen in the effect of complexity on patients lacking any of the prognostically significant abnormalities (ie, t(15;17), t,(8;21) inv(16)/t(16;16), and those in Table 3; Figure 2).
Cytogenetic entities predicting significantly poorer overall survival in multivariable analysis
| Factor . | HR . | 99% CI . | P to enter model . |
|---|---|---|---|
| Individual abnormalities | |||
| Age, per year | 1.018 | 1.013-1.023 | |
| WBC, per unit increase | 1.003 | 1.002-1.003 | |
| Secondary disease | 1.54 | 1.31-1.82 | |
| Performance status | 1.15 | 1.09-1.21 | |
| −5 | 1.82 | 1.34-2.48 | < .001 |
| del(5q)/add(5q) | 1.73 | 1.37-2.19 | < .001 |
| inv(3) | 2.52 | 1.76-3.62 | < .001 |
| abn(3q) | 1.85 | 1.38-2.48 | < .001 |
| −7 | 1.51 | 1.22-1.88 | < .001 |
| t(10;11) | 2.62 | 1.59-4.29 | < .001 |
| +8 | 1.33 | 1.12-1.57 | < .001 |
| abn(17p) | 1.63 | 1.21-2.20 | < .001 |
| −17 | 1.58 | 1.15-2.17 | < .001 |
| t(6;11) | 2.25 | 1.26-4.03 | < .001 |
| add(7q)/del(7q) | 1.34 | 1.05-1.72 | .003 |
| t(11q23)* | 1.55 | 1.06-2.28 | .003 |
| t(9;22) | 1.64 | 1.04-2.56 | .004 |
| Additional effect of complexity in above model | |||
| > 3 abnormalities | 1.58 | 1.29-1.93 | < .001 |
| Factor . | HR . | 99% CI . | P to enter model . |
|---|---|---|---|
| Individual abnormalities | |||
| Age, per year | 1.018 | 1.013-1.023 | |
| WBC, per unit increase | 1.003 | 1.002-1.003 | |
| Secondary disease | 1.54 | 1.31-1.82 | |
| Performance status | 1.15 | 1.09-1.21 | |
| −5 | 1.82 | 1.34-2.48 | < .001 |
| del(5q)/add(5q) | 1.73 | 1.37-2.19 | < .001 |
| inv(3) | 2.52 | 1.76-3.62 | < .001 |
| abn(3q) | 1.85 | 1.38-2.48 | < .001 |
| −7 | 1.51 | 1.22-1.88 | < .001 |
| t(10;11) | 2.62 | 1.59-4.29 | < .001 |
| +8 | 1.33 | 1.12-1.57 | < .001 |
| abn(17p) | 1.63 | 1.21-2.20 | < .001 |
| −17 | 1.58 | 1.15-2.17 | < .001 |
| t(6;11) | 2.25 | 1.26-4.03 | < .001 |
| add(7q)/del(7q) | 1.34 | 1.05-1.72 | .003 |
| t(11q23)* | 1.55 | 1.06-2.28 | .003 |
| t(9;22) | 1.64 | 1.04-2.56 | .004 |
| Additional effect of complexity in above model | |||
| > 3 abnormalities | 1.58 | 1.29-1.93 | < .001 |
CI indicates confidence interval; HR, hazard ratio; and WBC, white blood cell.
Excluding t(9;11)(p21∼22;q23) and t(11;19)(q23;p13).
Impact of karyotype complexity on survival in patients lacking cytogenetic abnormalities that confer relatively favorable or adverse prognoses in multivariable analysis.
Impact of karyotype complexity on survival in patients lacking cytogenetic abnormalities that confer relatively favorable or adverse prognoses in multivariable analysis.
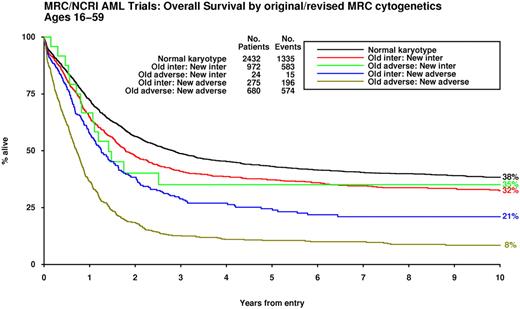
Results of the multivariable analysis conducted in this large study cohort were used to further refine the original MRC cytogenetic classification (Table 4). Although statistically significant, presence of the +8 abnormality did not lead to poor outcomes and was therefore not included in the revised definition (supplemental Figure 6). Application of the revised classification scheme led to reassignment of 299 cases (ie, 15% of the 1951 with abnormal karyotype, excluding those with t(15;17) and CBF leukemia); 275 cases moved from intermediate to adverse and conversely 24 (ie, those with t(3;5) without additional adverse features) were transferred from the adverse- to the intermediate-risk group (Figure 3, supplemental Figure 7a-b).
Revised MRC prognostic classification based on multivariable analyses
| Cytogenetic abnormality . | Comments . |
|---|---|
| Favorable | |
| t(15;17)(q22;q21) | |
| t(8;21)(q22;q22) | Irrespective of additional cytogenetic abnormalities* |
| inv(16)(p13q22)/t(16;16)(p13;q22) | |
| Intermediate | |
| Entities not classified as favorable or adverse | |
| Adverse | |
| abn(3q) [excluding t(3;5)(q21∼25;q31∼35)], | |
| inv(3)(q21q26)/t(3;3)(q21;q26), | |
| add(5q), del(5q), −5, | |
| −7, add(7q)/del(7q), | Excluding cases with favorable karyotype† |
| t(6;11)(q27;q23), | |
| t(10;11)(p11∼13;q23), | |
| t(11q23) [excluding t(9;11)(p21∼22;q23) and t(11;19)(q23;p13)] | |
| t(9;22)(q34;q11), | |
| −17/abn(17p), | |
| Complex (≥ 4 unrelated abnormalities) |
| Cytogenetic abnormality . | Comments . |
|---|---|
| Favorable | |
| t(15;17)(q22;q21) | |
| t(8;21)(q22;q22) | Irrespective of additional cytogenetic abnormalities* |
| inv(16)(p13q22)/t(16;16)(p13;q22) | |
| Intermediate | |
| Entities not classified as favorable or adverse | |
| Adverse | |
| abn(3q) [excluding t(3;5)(q21∼25;q31∼35)], | |
| inv(3)(q21q26)/t(3;3)(q21;q26), | |
| add(5q), del(5q), −5, | |
| −7, add(7q)/del(7q), | Excluding cases with favorable karyotype† |
| t(6;11)(q27;q23), | |
| t(10;11)(p11∼13;q23), | |
| t(11q23) [excluding t(9;11)(p21∼22;q23) and t(11;19)(q23;p13)] | |
| t(9;22)(q34;q11), | |
| −17/abn(17p), | |
| Complex (≥ 4 unrelated abnormalities) |
All favorable-risk abnormalities.
All adverse-risk abnormalities.
Outcome of patients according to original and refined MRC cytogenetic classification. The patients previously assigned to the “adverse-risk” group and reclassified as “intermediate-risk” all had t(3;5)(q21∼25;q31∼35).
Outcome of patients according to original and refined MRC cytogenetic classification. The patients previously assigned to the “adverse-risk” group and reclassified as “intermediate-risk” all had t(3;5)(q21∼25;q31∼35).
Impact of NPM1 and FLT3-ITD mutation status in patients with refined intermediate-risk cytogenetic abnormalities
Molecular genetics are increasingly being used to risk-stratify AML in conjunction with conventional cytogenetics, with the authors of many previous studies focusing on the prognostic impact of molecular markers in AML with normal karyotype.3,27 We wished to investigate the impact of nucleophosmin (NPM1) and Fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD) mutations on outcome in the cohort of patients with cytogenetic abnormalities defined as intermediate risk according to the refined MRC classification (Table 4). Genotyping information was available from 215 AML patients from this group28 ; in accordance with the findings of previous studies in normal karyotype AML (reviewed in Grimwade and Hills3 and Mrózek et al27 ), the presence of FLT3-ITD with wild-type NPM1 predicted a poor prognosis, whereas NPM1 mutation in the absence of FLT3-ITD was associated with a reduced risk of relapse with improved OS (supplemental Figure 8a-b). Sample sizes were too small to address the prognostic impact of CCAAT enhancer binding protein alpha (CEBPA) mutations in this group, or of NPM1 and CEBPA mutation status within the cohort of patients with adverse-risk cytogenetic abnormalities.
Relationship between cytogenetic-risk groups and monosomal karyotype
Finally we considered the distribution of cases with monosomal karyotype (designated MK+) as defined by Breems et al11 within the original and revised MRC cytogenetic classification systems. Most cases with monosomal karyotype (318/338; 94%) fell within the original MRC adverse-risk group (accounting for 45% of this group), with only 20 cases assigned to the intermediate-risk group, and were confirmed to have a very poor prognosis (supplemental Figure 9a). Application of the revised MRC classification (Table 4) led to reassignment of a further 13 MK+ cases to the adverse-risk group. Although the outcome of MK+ cases was noted to be particularly poor (5% OS at 10 years), the OS of patients with adverse karyotype as defined in the revised MRC classification and lacking a monosomal karyotype was also extremely low (OS 16%; supplemental Figure 9b).
Discussion
Diagnostic karyotype is a major prognostic indicator in AML, which is widely used in conjunction with information on NPM1, FLT3, and CEBPA mutation status, particularly for cases with normal karyotype as the basis for directing risk-adapted treatment approaches.3,27 Nevertheless, informed clinical decision making in situations in which cytogenetic analysis shows rarer karyotypic abnormalities has been hampered by a lack of consensus regarding the likely outcome of such patients. Discrepancies in risk-group assignment of these cases according to commonly applied cytogenetic classification systems most likely reflect limitations imposed by small sample sizes that have rendered the outcome data unreliable. Further confounding factors include variations in inter- and intra-study treatment approach, as well as differences in the patient population, particularly with respect to age distribution.1
Despite these limitations, it would be helpful if greater standardization in risk stratification of AML could be achieved as a means of optimizing therapy and also for reporting outcome data, thereby enabling more reliable comparison of results from different international trial groups. Apart from the benefit of achieving greater consensus in cytogenetic classification, establishing the outcome associated with rarer cytogenetic abnormalities is important, particularly given the results of a recent meta-analysis that has suggested that a relapse risk in excess of 35% can provide a useful working threshold to identify patients in whom allogeneic transplantation may confer a survival benefit.10
To refine existing cytogenetic classification systems, we examined the prognostic significance of rarer abnormalities drawn from a large series of 5876 adult patients aged 16 to 59 years receiving comparable therapy. In multivariable analyses, t(15;17), t(8;21), and inv(16)/t(16;16) emerged as the only abnormalities conferring a relatively favorable prognosis. Among APL patients with the t(15;17), treated with standard ATRA and anthracycline-based protocols, the presence of additional cytogenetic abnormalities (irrespective of the nature or complexity) had no significant impact on prognosis (supplemental Figure 3), which is in accordance with data from large European APL Group and PETHEMA studies.29,30 This finding would suggest that an adverse impact on outcome from the presence of additional abnormalities reported previously31 may have been ameliorated by optimal ATRA and anthracycline-based therapy or reflect a chance effect associated with smaller sample size. On the basis of an analysis of 421 patients, we also found that particular additional cytogenetic abnormalities did not adversely affect outcome in t(8;21) CBF leukemia, in contrast with previous reports that suggested a negative impact for del(9q),25 complex karyotype,26 or loss of −Y chromosome in male subjects.32 Indeed, we noted a trend (P = .04) to more favorable survival in the latter group (supplemental Figure 1d). However, our data are in accordance with those of Cancer and Leukemia Group B33 and the German AML Intergroup32 with respect to the prognostic significance of additional abnormalities in patients with inv(16), showing that presence of +22 predicts a significantly better outcome (supplemental Figure 2b). The reasons for this difference remain to be established; however, results of a previous study34 have suggested that this cannot solely be accounted for by KIT mutation status. Patients with inv(16) as the sole abnormality were noted to have significantly greater WBC,32 and defining the mechanisms underlying this may provide insights into the greater risk of relapse.
Having excluded cases with favorable karyotype (ie, t(15;17), t(8;21) and inv(16)/t(16;16)), multivariable analyses, conducted on the enlarged MRC dataset, showed that several cytogenetic abnormalities were independent predictors of a poor prognosis (Table 3). These included t(3;3)/inv(3) del(5q)/-5, and -7, which were recognized as adverse-risk factors in the original MRC classification.4 However, several abnormalities that were too infrequent to be considered previously also were found to be independent predictors of poor outcome, including −17 and abnormalities of 17p, which are associated with loss of TP53,35 and t(9;22)(q34;q11), which has been associated with poor prognosis in a large case series,36 leading to the assignment of these entities to the adverse-risk group in several existing cytogenetic classification systems.5,8,9 Abnormalities of 3q and 5q also generally are considered as adverse prognostic indicators; however, we found that the outcome of the t(3;5), which is associated with formation of the NPM1-MLF1 fusion37 and is considered an MDS-related abnormality in the 2008 WHO classification,24 did not differ significantly from patients with normal karyotype, although we recognize that the number of cases with t(3;5) was relatively small and this should be confirmed in a larger patient cohort. Other WHO 2008-specified MDS-related abnormalities were associated with a poor prognosis, even when cases with -5/del(5q) and -7 were excluded (Figure 1).
The overall outcome for patients with t(11q23) was significantly worse than for those with normal karyotype (adjusted HR for survival 1.77, 95% CI 1.42-2.23, P < .001). However, the chromosomal partner was observed to have an important bearing upon prognosis. The t(9;11)(p21∼22;q23), which leads to the MLLT3-MLL fusion and which is now recognized as a distinct disease entity in the WHO classification,24 was found to have a relatively favorable outcome, in accordance with the majority of studies.38-41 A similar outcome was observed in patients with t(11;19)(q23;p13), although the involved fusion partner (ie, ELL or MLLT1 [ENL], located at 19p13.1 and 13.3, respectively)42 was not distinguished in this study. In multivariable analysis, cases with t(6;11)(q27;q23) and t(10;11)(p12;q23), involving MLLT4 (AF6) and MLLT10 (AF10) genes, respectively,42 predicted a very poor prognosis.
Interestingly, both of these abnormalities have been associated with a poor prognosis in previous studies,43-47 including a recent large international pediatric study considering 756 cases of AML with MLL translocations.47 In the latter study, the most favorable outcome was observed in cases with t(1;11)(q21;q23), although this abnormality was too infrequent in our series (n = 3) to consider its prognostic significance in adults. Our data are also in agreement with a large German study involving 180 adults with 11q23 translocations, reported by Krauter and colleagues,41 in which t(9;11) and t(6;11) were found to have a relatively favorable and adverse outcome, respectively, in multivariable analysis. In contrast to our data, t(11;19) and t(10;11) did not emerge as independent prognostic factors in the German study but were each identified in less than 20 cases.
Another disease entity recognized in the updated WHO classification is the t(6;9)/DEK-CAN,24 which was associated with a very poor prognosis in a large case series48 and is generally assigned to the adverse cytogenetic-risk group.1,2 The poor outcome may relate to a strong association with FLT3-ITD mutations.49 In the present study, there was some evidence of poorer survival in patients (n = 42) with the t(6;9)(p23;q34) compared with those with normal karyotype (27% vs 38%, adjusted HR 1.55 (95% 0.88-2.74, P = .04), but this effect was not sufficiently strong to emerge in multivariable analysis.
The level of karyotypic complexity that confers adverse prognosis provides a further source of inconsistency between cytogenetic classification schemes, with all groups with the exception of the MRC, adopting 3 or more (3+) abnormalities. Accordingly, the latest WHO classification has defined a complex karyotype as one with 3 or more unrelated abnormalities in the absence of t(15;17), t(8;21), inv(16)/t(16;16), or t(9;11), which, when present, denotes a case as “MDS-related” AML.24 On the basis of this definition, the outcome according to karyotype complexity was as follows: (2+: CIR 65%, OS 12%; 3+: CIR 67%, OS 10%; 4+: CIR 72%, OS 8%; 5+: CIR 74%, OS 6%; P < .001 for trend over number of abnormalities on CIR, OS). However, such a definition does not take into account the impact of cytogenetic entities that would confer adverse risk in their own right.
Because complex karyotype is widely considered as a predictor for very poor outcome and frequently is used as an indication for allogeneic transplantation or experimental treatment approaches, it is critical that the definition of this entity is robust. Therefore, we investigated the impact of karyotype complexity on outcome in patients with particular cytogenetic abnormalities that would, in their own right, have led to their assignment to the intermediate- or adverse-risk groups, respectively, disregarding the number of unrelated abnormalities. Level of karyotype complexity was observed to have little impact on outcome in patients already having at least one of the independent adverse-risk abnormalities identified on multivariable analysis, who, generally, had a very poor prognosis. Conversely, in patients lacking any of these independent adverse-risk abnormalities, t(15;17), CBF leukemia, or t(9;11), the presence of 4 or more unrelated changes was found to provide the most informative cutoff, predicting a significantly poorer prognosis even after adjustment for abnormalities known to be prognostic (HR 1.60, 95% CI 1.31-1.96, P < .001).
Analysis of this very large series of AML patients treated in the MRC trials with prolonged follow-up has allowed the prognostic significance of several rarer cytogenetic abnormalities to be established. This has achieved further refinement of the original hierarchical MRC cytogenetic classification scheme and reconciled several differences between existing classification systems. This study will hopefully provide impetus facilitating the development of consensus in the reporting of karyotype data, allowing more reliable comparison between clinical trials involving younger adults with AML. We have independently confirmed that the presence of a monosomal karyotype identifies a group of patients with very poor prognosis11 but note that the majority of such patients fall within the adverse-risk group as defined by the MRC. Importantly we have shown that a substantial proportion of patients cannot be reliably classified as having poor-risk AML based on the presence of a monosomal karyotype alone.
Our data lend support to the continued use of cytogenetic analysis as a component of the routine diagnostic work-up of AML to provide a framework for risk stratification, to be used in conjunction with screening for an increasing range of molecular markers—required not only to predict risk of relapse in those with normal karyotype but also to further dissect out groups of patients with differing prognoses who share particular cytogenetic abnormalities, or fall within the same cytogenetic-risk group. For example, we show that NPM1 and FLT3-ITD mutation status provide independent prognostic information in patients who would otherwise have been considered intermediate risk on the basis of the karyotypic abnormalities identified (supplemental Figure 8a-b), in accordance with a recent study by Haferlach and colleagues.50 Further refinement of risk groups may be achieved through molecular screening for other mutations, including CEBPA, WT1, RUNX1, MLL-PTD, and overexpression of genes such as EVI1 (reviewed in Grimwade and Hills3 ). A key and ongoing challenge is the integration of pretreatment parameters, including cytogenetics and an ever-expanding number of molecular markers, with early assessment of treatment response to develop robust algorithms that further refine risk stratification of AML to guide consolidation therapy.
Presented in part at the XXIV Symposium of the International Association for Comparative Research on Leukemia and Related Diseases (IACRLRD), Columbus, OH, October 15-16, 2009.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all the clinicians participating in the MRC/National Cancer Research Institute trials and members of laboratories of the United Kingdom Cancer Cytogenetics Group (UKCCG) and cytogenetic laboratories from Denmark and New Zealand (for a complete list, see the Supplemental Appendix). We are also grateful to members of the NCRI AML Working Group for helpful comments, particularly Anne Parker, Sahra Ali, Charles Craddock, Don Milligan, Lars Kjeldsen, Jamie Cavenagh, Paresh Vyas, Rosemary Gale, and Dominic Culligan.
D.G., C.J.H., and A.V.M. are grateful to Leukaemia and Lymphoma Research of Great Britain for funding; D.G., R.K.H., and A.K.B. also acknowledge support from the National Institute for Health Research and the European LeukemiaNet. The MRC trials databases have been supported by Kay Kendall Leukaemia Fund and Leukaemia and Lymphoma Research.
Authorship
Contribution: D.G., R.K.H., A.V.M., and C.J.H. designed the study and wrote the manuscript; R.K.H. performed statistical analyses; A.V.M. and C.J.H. classified the cytogenetic abnormalities and managed the cytogenetics database; H.W. and S.C. undertook cytogenetic analyses and coordinated data collection; K.W. contributed to the design of the MRC AML trials; A.H.G. and A.K.B. contributed to study design and were lead participants in the MRC AML trials; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
For a complete list of National Cancer Research Institute Adult Leukaemia Working Group participants, see the Supplemental Appendix.
Correspondence: David Grimwade, Cancer Genetics Laboratory, Department of Medical and Molecular Genetics, 8th Fl, Tower Wing, Guy's Hospital, Great Maze Pond, London SE1 9RT, United Kingdom; e-mail: david.grimwade@genetics.kcl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal