Certain oncology trials showed worse clinical outcomes in the erythropoiesis-stimulating agent (ESA) arm. A potential explanation was that ESA-activated erythropoietin (Epo) receptors (EpoRs) promoted tumor cell growth. Although there were supportive data from preclinical studies, those findings often used invalidated reagents and methodologies and were in conflict with other studies. Here, we further investigate the expression and function of EpoR in tumor cell lines. EpoR mRNA levels in 209 human cell lines representing 16 tumor types were low compared with ESA-responsive positive controls. EpoR protein production was evaluated in a subset of 66 cell lines using a novel anti-EpoR antibody. EpoR+ control cells had an estimated 10 000 to 100 000 EpoR dimers/cell. In contrast, 54 of 61 lines had EpoR protein levels lower than 100 dimers/cell. Cell lines with the highest EpoR protein levels (400-3200 dimers/cell) were studied further, and, although one line, NCI-H661, bound detectable levels of [125I]–recombinant human Epo (rHuEpo), none showed evidence of ESA-induced EpoR activation. There was no increased phosphorylation of STAT5, AKT, ERK, or S6RP with rHuEpo. In addition, EpoR knockdown with siRNAs did not affect viability in 2 cell lines previously reported to express functional EpoR (A2780 and SK-OV-3). These results conflict with the hypothesis that EpoR is functionally expressed in tumors.
Introduction
The primary biologic function of erythropoietin (Epo) is to bind to erythropoietin receptor (EpoR) on erythroid progenitor cells, which stimulates signaling pathways that lead to proliferation, survival, and differentiation of these cells. Reports of EpoR expression in tumor cell lines of nonhematopoietic cell origin have caused concern that erythropoiesis-stimulating agents (ESAs) could also have proliferative and/or survival effects on tumor cells.1,–3 Adding to this concern are certain clinical trials that reported an association with recombinant human Epo (rHuEpo) treatment and possible tumor progression in patients with head and neck, breast, cervical, and ovarian cancers.4,5
Many of the reports of high levels of EpoR were based on the detection of EpoR mRNA by nonquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) methodologies or by Western blot analysis and/or immunohistochemical (IHC) staining with anti-EpoR antibodies. Conclusions that EpoR was functionally expressed in tumor cell lines were based on EpoR knockdown,6,7 signaling,6,8,9 or proliferation studies.1,2 However, multiple methodologic issues with those studies exist. The EpoR mRNA studies frequently lacked positive controls, and the anti-EpoR antibodies used were recently shown to be nonspecific.10,,–13 Moreover, in direct conflict with those reports, in many studies there was no effect of ESAs on intracellular signaling, growth, or survival of tumor cells in vitro or in vivo.1,–3,10,13,,,–17 In addition, responses similar to that reported for ESAs were reproduced with media changes or addition of vehicle lacking rHuEpo,18 suggesting that factors other than rHuEpo19 may be responsible for the observed effects. We therefore sought to re-examine the hypothesis of functional EpoR expression using a panel of human tumor cell lines.
Methods
Cell lines and culture conditions
Cell lines were obtained from a variety of sources, including ATCC, European Collection of Cell Cultures, and German Collection of Microorganisms and Cell Culture (DMSZ). The EpoR+ control leukemia line UT-7/Epo, an Epo-dependent megakaryoblastic leukemia line,20 was a gift from Dr Norio Komatsu (Jichi Medical School, Minamikawachi, Japan). A subline of UT-7/Epo was prepared by selection in medium containing granulocyte-macrophage colony-stimulating factor (GM-CSF) and lacking rHuEpo (UT-7/GM-CSF cells). OCIM-1 cells, an Epo-independent erythroleukemia line,21,22 were a gift from Dr Virginia Broudy (University of Washington). All cell lines were grown in supplier-recommended media, and positive control lines were maintained in Iscove modified Dulbecco medium (IMDM) plus 10% fetal bovine serum (FBS) and essential growth factors: 1 U/mL rHuEpo (Amgen) or 10 ng/mL human recombinant GM-CSF (Invitrogen). Erythroid progenitor cells were generated from CD34+ peripheral blood cells mobilized with granulocyte colony-stimulating factor (G-CSF; ALLCELLS) cultured for 6 days in StemSpan SFEM (StemCell Technologies) containing 10 ng/mL IL-3, 10 ng/mL IL-6, 50 ng/mL stem cell factor (SCF; R&D Systems), and 5 U/mL rHuEpo. These cells were primarily erythroid, considered to express physiologically relevant amounts of EpoR,23,24 and were Epo-responsive (increased STAT5 phosphorylation with ESA addition [data not shown]).
Hypoxia treatments of the cell lines were carried out at 1% O2, 5% CO2, and 94% N2 in an In vivo2 Hypoxia Workstation (Ruskinn Life Sciences). For hypoxia experiments, cells were seeded in duplicate flasks and incubated for 24 hours in hypoxia (1% O2) or kept in a standard ambient O2 incubator. Analyses were based on viable cell counts using a Vi-cell XR cell viability analyzer (Beckman Coulter) as viability was reduced in some cell lines after hypoxia, but viability was always greater than 60%.
Quantification of mRNA levels by RT-PCR
RNA was extracted from up to 1 × 107 cells using 1 mL RNeasy Mini Kit and homogenized with the QIAGEN Shredder (QIAGEN). Total RNA (10 μg) was treated with DNAse (Ambion), followed by inactivation per the manufacturer's protocol. cDNA was generated using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR (Q-RT-PCR; Taqman) was performed on test samples using primers to the housekeeping gene human β-actin (ACTB) and 3 different EpoR transcript regions. Probes were labeled with FAM (5′), VIC (5′), and TAMRA (3′).
Primers and probes were as follows: ACTB primers, CCT GGC ACC CAG CAC AA, GCC GAT CCA CAC GGA GTA CT, and probe ATC AAG ATC ATT GCT CCT CCT GAG CG; EPOR exon 3 primers, CTT CGT GCC CCT AGA GTT GC, TGA TGT GGA TGA CAC GGT GAT, and probe TC ACA GCA GCC TCC GGC GCT; EPOR exons 6 and 7 primers, ACC GCC GGG CTC TGA A, TTC AAA CTC GCT CTC TGG GC, and probe AGA AGA TCT GGC CTG GCA TCC CG; and EPOR exon 8 primers, TGC CAG CTT TGA GTA CAC TAT CCT, GCT CAG GGC ACA GTG TCC AT, and probe CCC AGC TCC CAG CTC TTG CGT C.
PCR reaction was as follows: 10 ng cDNA, Taqman 2× Universal PCR Master Mix (Applied Biosystems), 300nM primers, and 200nM probe. The RT-PCR amplification program was (1) activation at 50°C for 2 minutes; (2) denaturation at 95°C for 10 minutes; and (3) amplification for 40 cycles at 95°C for 15 seconds and 60°C for 1 minute with fluorescence capture at each step (ABI PRISM 7900HT Sequence Detection Systems; Applied Biosystems). Levels of EPOR transcripts were normalized to ACTB. The 2−ΔCT was calculated for each observed value where ΔCT was the difference in the observed CT values between the gene of interest and ACTB. The 2−ΔCT values from replicate data were then averaged. A human universal cDNA reference (20 ng) was used to merge and compare data across experiments.
Quantification of mRNA levels by bDNA signal amplification assay
RNA expression levels were determined using the QuantiGene branched DNA (bDNA) assay (Panomics Inc) following the manufacturer's instructions for the singleplex QG1.0 reagent system. The assay is based on sequential hybridization of single-stranded DNA probe sets that capture and detect target RNAs. The probe sets used in this study detect RNA species as follows: EPOR nucleotides 647 to 1660, NM_000121 (GenBank accession number, http://www.ncbi.nlm.nih.gov/Genbank/); cyclophilin B (CYCLOB) nucleotides 69 to 629, NM_000942; vascular endothelial growth factor (VEGF) nucleotides 978 to 1680, NM_001025366; BCL2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3) nucleotides 178 to 683, NM_004052; and 18s ribosomal RNA (18s rRNA) nucleotides 767 to 1232 NR_003286. The bDNA assay was performed on lysates from cells grown for 24 hours. A total of 3 independent samples were made for every cell line, and assays were run at least twice for each sample. RNA expression levels were normalized to the amount of 18s rRNA.
Western blot analysis
Western blots of whole-cell lysates from a known number of cells were prepared and probed with a rabbit monoclonal antibody specific to EpoR (A82) as described previously.23 A strip of the membrane was cut off below the 28-kDa marker to stain with anti–cyclophilin B (CycloB), used as a loading control. A82 was used at 0.1 μg/mL (Amgen), and anti-CycloB antibodies were rabbit polyclonals (Abcam) used at 1:20 000. Secondary antibodies were horseradish peroxidase–linked anti–rabbit IgGs (HRP-IgG) used at 10 ng/mL for anti-EpoR (The Jackson Laboratory) and 1:50 000 for anti-CycloB blots. EpoR blots were developed with ECL-Plus, CycloB blots were developed with ECL, and both were exposed on hyperfilm ECL (GE Healthcare). In hypoxia experiments, anti-EpoR blots were stripped with Restore (Thermo Scientific) and reprobed with a mouse monoclonal antibody to BNIP3 (Sigma-Aldrich) and secondary anti–mouse HRP-IgG (GE Healthcare). In siRNA knockdown experiments, EpoR westerns were loaded with 25 to 50 μg of protein from cell lysates and probed with 0.2 μg/mL A82 and 0.1 to 0.4 μg/mL of the secondary HRP-IgG antibodies. The blots were stripped and reprobed with anti-CycloB antibodies.
Estimation of EpoR protein levels using semiquantitative Western blots
Human EpoR extracellular domain (ECD; amino acids 1-225; 25 kDa) was expressed in Chinese hamster ovary (CHO) cells. Protein was purified from culture medium as described,25 with absorbance (A280) protein concentration determination. A 2-fold serial dilution of cell lysates or a 2-fold serial dilution of EpoR-ECD mixed together with EpoR− 769-P cell lysate was generated and subjected to A82 immunoblot analysis. Band intensities (25-kDa EpoR-ECD and 59-kDa full-length EpoR in cell lysates) were compared visually on the same immunoblot.
[125I]-rHuEpo cell-surface binding
[125I]-rHuEpo binding studies were performed with intact cells as described.14 This method only detects Epo receptors on the cell surface because cytochalasin B and sodium azide were included to inhibit receptor internalization. Specific binding was nonspecific binding subtracted from total binding. Results from 2 to 3 experiments (where n = 3-5 for each cell line) are expressed as percentage of control (POC) cells (UT-7/Epo). An unpaired 2-tailed t test assessed statistical significance of difference (P < .05).
FACS analysis of the phosphorylation state of signal transduction proteins after rHuEpo addition
Cell lines were serum- and rHuEpo-starved overnight. Cells were then stimulated for 5 and 30 minutes with vehicle (rHuEpo formulation buffer), a 5-fold serial dilution of rHuEpo (300-0.02 U/mL) or an EGF/HGF/IGF-1 cocktail (EGF 100 ng/mL [Roche], HGF 500 ng/mL [R&D Systems], and IGF-1 500 ng/mL [R&D Systems]). Treated cells were fixed in 4% paraformaldehyde (10 minutes at 37°C), washed once with ice-cold Ca2+/Mg2+-free phosphate-buffered saline (PBS; Invitrogen) and permeabilized in ice-cold 90% (vol/vol) methanol (30 minutes on ice).
Samples were stained for 1 hour at room temperature with fluorochrome-conjugated antibodies that are specific for the phosphorylated forms of AKT, ERK1/2 (both Alexa Fluor 647 [Cell Signaling Technology]), S6 Ribosomal Protein (S6RP; Alexa Fluor 488 [Cell Signaling Technology]), and STAT5 (Alexa Fluor 488 [BD Biosciences]) run on a fluorescence-activated cell sorter (FACS) instrument (LSRII; BD Biosciences) and analyzed using FACSDiva software (BD Biosciences). The specificity of phosphospecific antibodies was confirmed in cell lines using growth factor stimulations. Antibodies were selected that cross-reacted with a single band of the predicted molecular weight under stimulation conditions that are known to lead to activation of the targeted phosphoprotein. Results are reported as fold change compared with vehicle treatment alone. Experiments were performed 3 times for each cell line.
siRNA transfection and screening
Small interfering RNAs (siRNAs) were transfected into cells using Lipofectamine RNAiMAX transfection reagent (Invitrogen). SiRNA and transfection reagent diluted in media were delivered to 384-well assay plates and after 20 minutes of room temperature incubation, cells were added with or without growth factor. After 4 days, cells viability was determined with Cell Titer Glo (Promega), and luminescence was measured on a luminometer according to the manufacturer's instructions. The final siRNA concentrations used per well were 10nM (A2780, NCI-H1299, SK-OV-3) and 30nM (UT-7/Epo, UT-7/GM-CSF).
A total of 8 each of EPOR, GM-CSF receptor α chain and β chain (CSFRA and CSFRB), Janus kinase 2 (JAK2), and 9 polo-like kinase-1 (PLK1) siRNAs were tested in the UT-7 lines, and EPOR, JAK2, and PLK1 siRNAs were tested in the other 3 cell lines. To determine normal viability, a library of approximately 10 000 single siRNAs was also tested and processed through Screener (Genedata). Normalized, corrected viability measurements for the siRNAs for each gene were compared with the remainder of the siRNAs in a Wilcoxon rank-sum test, and P values were corrected for multiple hypotheses testing by the Benjamini-Hochberg method as implemented in the statistical package in R (R Foundation for Statistical Computing). A82 Western blots as described in “Western blot analysis” were made from lysates of cells 4 days after transfection to assess the knockdown efficiency of the EpoR siRNAs. Quantification of band intensity was performed using ImageJ software (National Institutes of Health).
Results
EpoR mRNA expression levels
To assess the significance of EPOR mRNA expression, EpoR+ (UT-7/Epo, OCIM-1, HEL92.1.7, and K562) and EpoR− control (769-P and COLO677) cells were used. The EpoR+ controls have all been reported to bind [125I]-rHuEpo,20,–22,26 but only UT-7/Epo cells are dependent on Epo for growth. Also included were Epo-responsive erythroid progenitor cells differentiated in vitro from peripheral blood CD34+ cells. The negative control lines showed neither [125I]-rHuEpo surface binding nor EpoR protein according to A82 Western immunoblotting.23
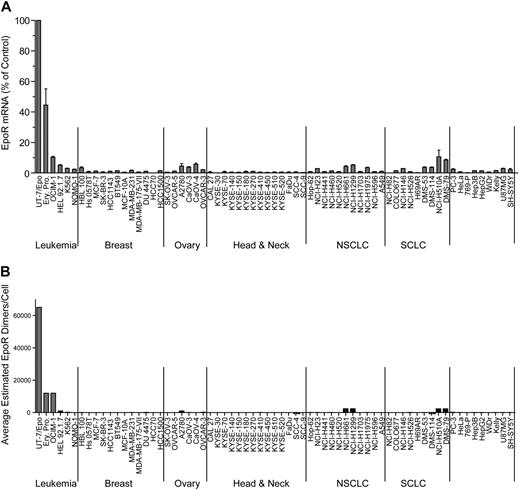
EPOR mRNA was measured in a panel of 209 cell lines by 2 methods: 175 by Q-RT-PCR and 66 by bDNA assay with 32 cell lines measured by both methods to allow cross-comparison of the methodologies (Table 1). The cell lines were selected because of published reports of high EpoR levels and/or because they were derived from cancers where an association between disease progression and rHuEpo treatment was reported: breast (n = 24), ovary (n = 14), lung (n = 31), and head and neck (including esophagus; n = 15). Although there was a difference in the absolute levels of mRNA with different methods, overall, the results with both methods were comparable, with the same rank order of expression levels for the positive and negative controls. Since the EPOR mRNA levels were highest in UT-7/Epo cells, results from other cells were expressed as a POC of these. The erythroid progenitor cells had somewhat lower levels of EPOR mRNA (POC = 37%-52% by bDNA; 6% by Q-RT-PCR), followed by OCIM-1 cells (POC = 10% by bDNA; 4% by Q-RT-PCR). None of the 175 tumor cell lines assayed by Q-RT-PCR had EPOR mRNA levels greater than a POC of 2%; 11 cell lines had a POC of 0.8% to 2%, with the remainder having a POC of 0.01% to 0.7% (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). For the set of cell lines assayed by bDNA, 57 of the nonhematopoietic tumor cell lines had a POC of less than 5%, with the majority having a POC less than 1%. Two nonhematopoietic cell lines measured by bDNA (NCI-H510A and DMS-79) had mRNA levels comparable with that of OCIM-1 cells (10% and 9%, respectively). The same 2 lines were also among the highest when measured by Q-RT-PCR (POC = 1% and 0.9%, respectively). Typical results from a subset of 66 cell lines are shown in Figure 1A. Based on these results, low-level EPOR mRNA expression was common among tumor cell lines.
The 209 cell lines examined for EpoR mRNA
| Tissue . | Cell lines . |
|---|---|
| Blood (n = 20) | UT-7/Epo+ control*†, OCIM-1+ control*†, NOMO-1†, HEL92.1.7†, K562*†, HL-60, HL60/MX-1, HL-60/MX-2, CCRF-CEM, CEM/C1, MOLT-4, RPMI-8226, SR, BJAB, Daudi, THP-1, U-937, Jurkat, CloneE6-1, T1 (174×CEM.T1), U266B1 |
| Breast (n = 24) | HBL 100†, Hs 0578T†, MCF-7*†, MCF-7ADR, MCF-10A†, SK-BR-3*†, HCC1143†, ZR-75-1, MDA-MB-231*†, MDA-MB-175-VII†, MDA-MB-468, MDA-MB-435s, MDA-MB-134, MDA-MB-157, MDA-MB-453, HS-578T, T-47D, HCC70†, HCC1500†, DU4475*†, alaB, BT-20, BT474, BT549*† |
| Head and neck (n = 15) | CAL 2†, KYSE-30†, KYSE-70†, KYSE-140†, KYSE-150†, KYSE-180†, KYSE-270†, KYSE-410†, KYSE-450†, KYSE-510†, KYSE-520†, FaDu*†, SCC-4†, SCC-9†, KB-3-1 |
| Ovary (n = 14) | SK-OV-3*†, IGR-OV1, CaOV-1, CaOV-3*†, CaOV-4*†, OVCAR, OVCAR-4, OVCAR-3*†, OVCAR-5*†, A2780*, ES-2, OV-90, PA-1, PTX-10 |
| Non–small cell lung (n = 31) | Hop-62*†, Hop-92, NCI-H226, NCI-H522, NCI-H292, NCI-H358, NCI-H23*†, NCI-H441†, NCI-H460*†, NCI-H520*†, NCI-H661†, NCI-H1299*†, NCI-H1703†, NCI-H1395, NCI-H1650, NCI-H2170, NCI-H1975*†, NCI-H596†, A549*†, A-427, BEN, EKVX, 9812, Colo-699, DV-90, EPLC-272H, HCC-15, LCLC97-TM1, SKLC-13, SK-LU-1, SK-MES-PD |
| Small cell lung (n = 12) | NCI-H82*†, NCI-H69, NCI-H146†, NCI-H526*†, NCI-H510A*†, NCI-H128, NCI-H211, COLO677*†, DMS-79*†, DMS-53†, DMS-114†, H69AR† |
| CNS (n = 9) | U-251, Kelly*†, SF-268, SF-295, SF-539, SNB-75, SH-SY5Y†, U-118MG, U87MG*† |
| Colorectal (n = 27) | HCC2998, KM12, NCI-H716, DLD-1, WiDR*†, HCT-15, HCT-116, HT-29, SW-620, CaCo-2, C2BBe1, Colo-203, Colo-205, Colo-206, Colo-320DM, Colo-320HSR, Colo-678, HCT-8, Hs-255.T, LOVO, LS174t, LS-180, SK-CO-1, SW-48, SW-403, SW-1116, T-84 |
| Kidney (n = 10) | 769-P negative control*†, RXF 393, TK-10, UO-31, 786-O, A-498, ACHN, CAKI-1, SN-12C, G-401 |
| Skin (n = 10) | LOXIMVI, UACC-62, UACC-257, MALME-3M, M14, SK-MEL-2, SK-MEL-5, SK-MEL-28, A-431, WM-115 |
| Liver (n = 8) | Hep-3B*†, Hep-G2*†, PLC/PRF/5, SK-Hep 1, SNU-182, SNU-398, SNU-449, SNU-475 |
| Bladder (n = 7) | 5637, BC-3C, HT-1376, J82, SW-780, T24, TCCSUP |
| Other (n = 22) | PC-3*†, HeLa†, ME-180, NCCIT, A-673, RD, JEG-3, SNU-1, NTERA-2, MES-SA, MES-SA/MX-2, AsPC-1, BxPC-3, CAPAN-1, CAPAN-2, Mia-PaCa-2, PANC-1, A-253, 143B, MG-63, SaOS-2, DU-145 |
| Tissue . | Cell lines . |
|---|---|
| Blood (n = 20) | UT-7/Epo+ control*†, OCIM-1+ control*†, NOMO-1†, HEL92.1.7†, K562*†, HL-60, HL60/MX-1, HL-60/MX-2, CCRF-CEM, CEM/C1, MOLT-4, RPMI-8226, SR, BJAB, Daudi, THP-1, U-937, Jurkat, CloneE6-1, T1 (174×CEM.T1), U266B1 |
| Breast (n = 24) | HBL 100†, Hs 0578T†, MCF-7*†, MCF-7ADR, MCF-10A†, SK-BR-3*†, HCC1143†, ZR-75-1, MDA-MB-231*†, MDA-MB-175-VII†, MDA-MB-468, MDA-MB-435s, MDA-MB-134, MDA-MB-157, MDA-MB-453, HS-578T, T-47D, HCC70†, HCC1500†, DU4475*†, alaB, BT-20, BT474, BT549*† |
| Head and neck (n = 15) | CAL 2†, KYSE-30†, KYSE-70†, KYSE-140†, KYSE-150†, KYSE-180†, KYSE-270†, KYSE-410†, KYSE-450†, KYSE-510†, KYSE-520†, FaDu*†, SCC-4†, SCC-9†, KB-3-1 |
| Ovary (n = 14) | SK-OV-3*†, IGR-OV1, CaOV-1, CaOV-3*†, CaOV-4*†, OVCAR, OVCAR-4, OVCAR-3*†, OVCAR-5*†, A2780*, ES-2, OV-90, PA-1, PTX-10 |
| Non–small cell lung (n = 31) | Hop-62*†, Hop-92, NCI-H226, NCI-H522, NCI-H292, NCI-H358, NCI-H23*†, NCI-H441†, NCI-H460*†, NCI-H520*†, NCI-H661†, NCI-H1299*†, NCI-H1703†, NCI-H1395, NCI-H1650, NCI-H2170, NCI-H1975*†, NCI-H596†, A549*†, A-427, BEN, EKVX, 9812, Colo-699, DV-90, EPLC-272H, HCC-15, LCLC97-TM1, SKLC-13, SK-LU-1, SK-MES-PD |
| Small cell lung (n = 12) | NCI-H82*†, NCI-H69, NCI-H146†, NCI-H526*†, NCI-H510A*†, NCI-H128, NCI-H211, COLO677*†, DMS-79*†, DMS-53†, DMS-114†, H69AR† |
| CNS (n = 9) | U-251, Kelly*†, SF-268, SF-295, SF-539, SNB-75, SH-SY5Y†, U-118MG, U87MG*† |
| Colorectal (n = 27) | HCC2998, KM12, NCI-H716, DLD-1, WiDR*†, HCT-15, HCT-116, HT-29, SW-620, CaCo-2, C2BBe1, Colo-203, Colo-205, Colo-206, Colo-320DM, Colo-320HSR, Colo-678, HCT-8, Hs-255.T, LOVO, LS174t, LS-180, SK-CO-1, SW-48, SW-403, SW-1116, T-84 |
| Kidney (n = 10) | 769-P negative control*†, RXF 393, TK-10, UO-31, 786-O, A-498, ACHN, CAKI-1, SN-12C, G-401 |
| Skin (n = 10) | LOXIMVI, UACC-62, UACC-257, MALME-3M, M14, SK-MEL-2, SK-MEL-5, SK-MEL-28, A-431, WM-115 |
| Liver (n = 8) | Hep-3B*†, Hep-G2*†, PLC/PRF/5, SK-Hep 1, SNU-182, SNU-398, SNU-449, SNU-475 |
| Bladder (n = 7) | 5637, BC-3C, HT-1376, J82, SW-780, T24, TCCSUP |
| Other (n = 22) | PC-3*†, HeLa†, ME-180, NCCIT, A-673, RD, JEG-3, SNU-1, NTERA-2, MES-SA, MES-SA/MX-2, AsPC-1, BxPC-3, CAPAN-1, CAPAN-2, Mia-PaCa-2, PANC-1, A-253, 143B, MG-63, SaOS-2, DU-145 |
CNS indicates central nervous system.
32 cell lines analyzed by both q-RT-PCR and bDNA method.
66 cell lines analyzed for EpoR protein.
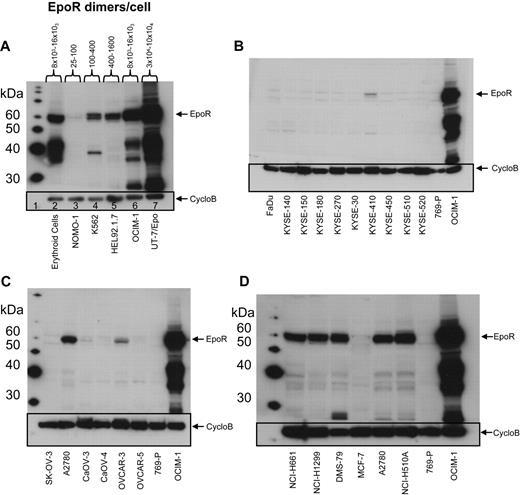
EpoR mRNA and protein levels for representative tumor cell lines. (A) EPOR mRNA levels were determined by bDNA assay from cell lysates (n = 3 from each culture). Expression of EPOR was normalized to the level of 18S rRNA and is expressed as a percentage of control (POC; 100% = EPOR mRNA levels in UT-7/Epo cells). (B) EpoR protein in lysates from the indicated cell lines: 2-fold serial dilutions of purified EpoR extracellular domain protein (EpoR ECD amino acid 1-225) and cell lysates were subjected to SDS-PAGE and Western blotting with detection of EpoR by mAb A82. Direct comparison of band intensities of the 59-kDa EpoR protein with those of the purified EpoR ECD was used to estimate EpoR dimers/cell. NSCLC indicates non–small cell lung carcinoma; SCLC, small cell lung carcinoma; and Ery. Pro., erythroid progenitor cells.
EpoR mRNA and protein levels for representative tumor cell lines. (A) EPOR mRNA levels were determined by bDNA assay from cell lysates (n = 3 from each culture). Expression of EPOR was normalized to the level of 18S rRNA and is expressed as a percentage of control (POC; 100% = EPOR mRNA levels in UT-7/Epo cells). (B) EpoR protein in lysates from the indicated cell lines: 2-fold serial dilutions of purified EpoR extracellular domain protein (EpoR ECD amino acid 1-225) and cell lysates were subjected to SDS-PAGE and Western blotting with detection of EpoR by mAb A82. Direct comparison of band intensities of the 59-kDa EpoR protein with those of the purified EpoR ECD was used to estimate EpoR dimers/cell. NSCLC indicates non–small cell lung carcinoma; SCLC, small cell lung carcinoma; and Ery. Pro., erythroid progenitor cells.
EpoR protein levels
EpoR protein levels were examined using Western blots with anti-EpoR antibody A82 in the same 66 cell lines examined for EPOR mRNA by the bDNA assay. A82 detected full-length 59-kDa EpoR protein in the positive control cell lines UT-7/Epo and OCIM-1 and in the erythroid progenitor cells (Figure 2A), but not in the negative control cell line 769-P (Figure 2B-D). In addition to the 59-kDa EpoR protein, A82 also detected a ladder of smaller proteins. The 36-kDa and 42-kDa bands were shown by mass spectroscopy analysis to be fragments of EpoR that contained the A82 epitope, amino acids 5 to 11 of EpoR.23
EpoR Western blots of representative cell lines. EpoR Western blot of cell lysates detected by mAb A82. Cyclophilin B (CycloB) is the loading control. Position of the 59-kDa full-length EpoR is indicated (arrow). 769-P is a negative control, and OCIM-1 is a positive control for EpoR. (A) Hematopoietic cells, including erythroid progenitor cells differentiated in vitro from peripheral blood CD34+ cells (lane 2) and 5 leukemia cell lines (8 × 104 cells/lane; lanes 3-7). EpoR amount estimated as in supplemental Figure 2A. (B) Head and neck cancer cell lines (1.6 × 105 cells/lane). (C) Ovarian cancer cell lines (1.6 × 105 cells/lane). (D) The 5 cell lines with the highest amount of EpoR among the solid tumor lines (Figure 1B) and MCF-7 (1.6 × 105 cells/lane). Data used to estimate EpoR levels for 2 of the higher expressing lines (DMS-79 and NCI-H1299) can be seen in panel 2 of supplemental Figure 2A.
EpoR Western blots of representative cell lines. EpoR Western blot of cell lysates detected by mAb A82. Cyclophilin B (CycloB) is the loading control. Position of the 59-kDa full-length EpoR is indicated (arrow). 769-P is a negative control, and OCIM-1 is a positive control for EpoR. (A) Hematopoietic cells, including erythroid progenitor cells differentiated in vitro from peripheral blood CD34+ cells (lane 2) and 5 leukemia cell lines (8 × 104 cells/lane; lanes 3-7). EpoR amount estimated as in supplemental Figure 2A. (B) Head and neck cancer cell lines (1.6 × 105 cells/lane). (C) Ovarian cancer cell lines (1.6 × 105 cells/lane). (D) The 5 cell lines with the highest amount of EpoR among the solid tumor lines (Figure 1B) and MCF-7 (1.6 × 105 cells/lane). Data used to estimate EpoR levels for 2 of the higher expressing lines (DMS-79 and NCI-H1299) can be seen in panel 2 of supplemental Figure 2A.
Semiquantitative A82 Western blotting was used to estimate EpoR protein levels in each cell line. The functional form of EpoR is a homodimer27,28 ; thus, EpoR levels were divided by 2 and are reported as dimers per cell. Because of the sensitivity of A82, it was possible to detect as little as 150 to 300 femtograms (fg) of EpoR (equivalent to 2-4 × 106 EpoR dimers); thus, lysates from 160 000 cells loaded on the gel would equate to 12 to 25 dimers/cell. EpoR levels in UT-7/Epo cells ranged from 30 000 to 100 000 dimers/cell. EpoR protein levels in erythroid progenitor cells (supplemental Figure 2A panel 1), and OCIM-1 cells were similar (Figure 2A) and ranged from 8000 to 16 000 EpoR dimers/cell.
This semiquantitative EpoR Western immunoblotting was performed on the 66 cell lines. To simplify analysis, the cell lines were arbitrarily organized into 5 groups based on the EpoR protein levels estimated (excluding the positive control UT-7/Epo, OCIM-1, HEL92.1.7, and K562 hematopoietic cell lines). Accordingly, 28 cell lines had undetectable EpoR (< 25 dimers/cell), 27 had 25 to 100 dimers/cell, 2 had 100 to 400 dimers/cell, 1 had 400 to 1600 dimers/cell, and 4 had an estimated 1600 to 3200 dimers/cell (examples for each group are shown in supplemental Figure 2B). Whereas there were some examples in which there was a correlation between levels of EpoR mRNA and protein (compare Figure 1A with 1B), there were many examples in which this was not the case. However the positive controls UT-7/Epo, OCIM-1, and the erythroid progenitor cells showed the highest EpoR protein levels, negative controls showed no detectable EpoR protein, and the 8 cell lines with the highest levels of EpoR protein were among the 11 cell lines expressing the highest mRNA levels. A total of 4 representative Western blots summarizing EpoR protein results are shown in Figure 2. Head and neck tumor lines as a group (Figure 2B) had the lowest EpoR protein expression of any tumor type (comparable with that of negative control 769-P cells). Only one line, KYSE-410, had detectable EpoR (25-100 dimers/cell). Ovarian tumor lines (Figure 2C) had EpoR levels ranging from undetectable for SK-OV-3 to 25 to 100 dimers/cell in Caov-3, Caov-4, and OVCAR-5; to 100 to 400 dimers/cell in OVCAR-3; and 400 to 1600 dimers/cell in A2780 cells. Some lung cancer lines produced higher levels of EpoR protein, with 4 expressing 1600 to 3200 EpoR dimers/cell. However, 7 had undetectable levels, and 9 expressed 25 to 100 dimers/cell. Breast tumor lines expressed low EpoR protein levels, with 7 having undetectable levels and 5 with 25 to 100 EpoR dimers/cell. The 5 solid tumor lines that showed the higher EpoR protein levels (approximately 400-3200 dimers/cell; Figure 2D) were from lung (NCI-H661, NCI-H1299, DMS-79, NCI-H510A) and ovary (A2780). MCF-7 cells had low EpoR protein levels but were included in this and subsequent studies because of published claims of high EpoR expression and Epo responsiveness.1,2 EPOR mRNA and protein levels are summarized in Table 2 for the higher-expressing lines. Those lines with the higher levels of EpoR protein had a POC of 1.5% to 3.7%, representing 8.3% to 20% that of erythroid progenitor cells. Overall, the EpoR protein levels estimated in the diverse human tumor cell lines did not reach the levels found in positive control cells known to respond to Epo.
EpoR mRNA and protein level comparison for positive and negative control cells, the 5 highest EpoR protein-producing cell lines, and the MCF-7 cell line
| Tumor cell line . | Tissue . | EpoR mRNA, % of control . | EpoR protein, dimers/cell . |
|---|---|---|---|
| UT-7/Epo | Megakaryoblastic leukemia | 100 | 30 000-100 000 |
| Erythroid progenitor cells | Differentiated CD34+ PBMCs | 37-52 | 8000-16 000 |
| OCIM-1 | Erythroleukemia | 10.3 | 8000-16 000 |
| NCI-H510A | Lung, small cell | 10.6 | 1600-3200 |
| DMS-79 | Lung, small cell | 8.6 | 1600-3200 |
| NCI-H1299 | Lung, non–small cell | 5.2 | 1600-3200 |
| NCI-H661 | Lung, non–small cell | 4.2 | 1600-3200 |
| A2780 | Ovary | 4.5 | 400-1600 |
| COL0677 | Colon | 1.4 | Undetectable (< 25) |
| 769-P | Kidney | 0.2 | Undetectable (< 25) |
| MCF-7 | Breast | 1.2 | 25-100 |
| Tumor cell line . | Tissue . | EpoR mRNA, % of control . | EpoR protein, dimers/cell . |
|---|---|---|---|
| UT-7/Epo | Megakaryoblastic leukemia | 100 | 30 000-100 000 |
| Erythroid progenitor cells | Differentiated CD34+ PBMCs | 37-52 | 8000-16 000 |
| OCIM-1 | Erythroleukemia | 10.3 | 8000-16 000 |
| NCI-H510A | Lung, small cell | 10.6 | 1600-3200 |
| DMS-79 | Lung, small cell | 8.6 | 1600-3200 |
| NCI-H1299 | Lung, non–small cell | 5.2 | 1600-3200 |
| NCI-H661 | Lung, non–small cell | 4.2 | 1600-3200 |
| A2780 | Ovary | 4.5 | 400-1600 |
| COL0677 | Colon | 1.4 | Undetectable (< 25) |
| 769-P | Kidney | 0.2 | Undetectable (< 25) |
| MCF-7 | Breast | 1.2 | 25-100 |
PBMCs indicates peripheral blood mononuclear cells.
Effect of hypoxia on EpoR expression
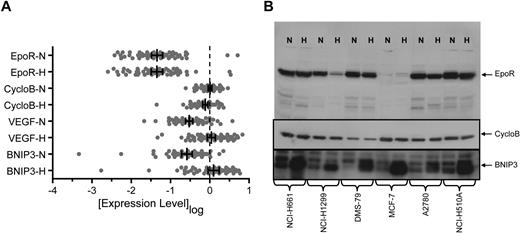
It has been proposed that EpoR was increased in response to the hypoxic environment of a tumor.29 To address this, we compared EPOR mRNA levels after 24 hours growth in normoxia or hypoxia (1% O2). Transcript levels of VEGF and BNIP3, known hypoxia-regulated genes,30,–32 and CYCLOB, a non–hypoxia-regulated gene, were included as positive and negative controls. CYCLOB showed a median of −0.083-fold (P ≤ .001; Wilcoxon sign-rank) and a mean of −0.094-fold mRNA decrease in hypoxia. One standard deviation around the mean (1.9-fold) was used to define a significant change in the assay. In contrast to CYCLOB, growth in hypoxia significantly increased VEGF and BNIP3 mRNA levels (median increase, 5.4-fold and 3.9-fold, respectively; P < .001 for both; Wilcoxon sign-rank; Figure 3A). All but 2 of the 66 cell lines (KYSE510 and NCI-H460) showed greater than a 1.9-fold increase in either BNIP3 or VEGF mRNA in hypoxia with maximum observed increases of 43-fold and 18-fold, respectively (supplemental Figure 3).
EpoR mRNA and protein levels in normoxia versus hypoxia in cell lines. (A) Expression of EPOR, CYCLOB, VEGF, and BNIP3 mRNA was examined in the human tumor cell lines shown in Figure 1 following growth in normoxia (N) or hypoxia (H) using a quantitative bDNA assay. Each point represents the mean level of transcript for each cell line from duplicate experiments from 3 independent cultures. Vertical bars represent the mean, and error bars represent the 95% confidence interval. (B) EpoR, CycloB, and BNIP3 Western blots of cell lysates from N and H treated cells from the 5 cell lines that express higher levels of EpoR protein and MCF-7 (1.6 × 105 cells/lane). CYCLOB is the loading control and a non–hypoxia-regulated gene. BNIP3 is a known hypoxia-regulated gene.
EpoR mRNA and protein levels in normoxia versus hypoxia in cell lines. (A) Expression of EPOR, CYCLOB, VEGF, and BNIP3 mRNA was examined in the human tumor cell lines shown in Figure 1 following growth in normoxia (N) or hypoxia (H) using a quantitative bDNA assay. Each point represents the mean level of transcript for each cell line from duplicate experiments from 3 independent cultures. Vertical bars represent the mean, and error bars represent the 95% confidence interval. (B) EpoR, CycloB, and BNIP3 Western blots of cell lysates from N and H treated cells from the 5 cell lines that express higher levels of EpoR protein and MCF-7 (1.6 × 105 cells/lane). CYCLOB is the loading control and a non–hypoxia-regulated gene. BNIP3 is a known hypoxia-regulated gene.
EPOR mRNA expression was not significantly different in normoxia versus hypoxia (median increase, 1.1-fold; P > .999; Wilcoxon sign-rank) and was similar to results observed for CYCLOB. The distribution was balanced and characterized by 5 lines that exhibited an increase of EPOR mRNA greater than 1.9-fold in hypoxia (maximum increase, 3.6-fold) and 5 lines that exhibited a decrease greater than 1.9-fold (maximum decrease, 6.7-fold) following growth in hypoxia (supplemental Figure 3). A total of 4 of the 5 lines with the greatest increase in EPOR mRNA in hypoxia were retreated and reanalyzed, and 3 of the 4 remained above 2-fold after hypoxia; the fourth had mRNA below the level of detection.
The level of EpoR protein after hypoxia was also examined and showed a median fold-change of 1.0 (data not shown). For the 5 lines which exhibited a greater than 2-fold increase in EPOR mRNA in hypoxia, only one showed an increase in EpoR protein (NCI-H1975), which was estimated to be 8-fold but was still a low level of only 100 to 400 EpoR dimers/cell in hypoxia. A representative Western blot of MCF-7 and the 5 cell lines NCI-H661, NCI-H1299, DMS-79, A2780, and NCI-H510A showed no increase in EpoR protein with hypoxia (Figure 3B). The experiment as performed allowed detection of increases due to hypoxia since an anti-BNIP3 antibody detected increased BNIP3 levels on the same blot.
One solid tumor cell line showed detectable 125I-rHuEpo binding to intact cells
The EpoR protein studies allowed an estimate of the total amount of EpoR per cell but did not address the question of whether EpoR was present on the cell surface. Only a fraction (1%-10%) of the EpoR protein is transported to the surface.33,–35 Cell-surface binding studies with [125I]-rHuEpo to intact cells were therefore performed. This assay is able to detect as few as 100 surface EpoRs and makes no assumption about the nature of the Epo-binding site on the cell surface. The lines with the higher levels of total EpoR protein were examined. Ovarian cancer cell lines were also examined because of reports that they respond to Epo.8,9 Specific [125I]-rHuEpo binding was detected with UT-7/Epo (POC = 100%) and OCIM-1 cells (POC = 20%), with no binding detected in the negative controls (769-P and COLO677). The ovarian lines (SK-OV-3, A2780, CaOV-3, and OVCAR-5; data not shown) and MCF-7 cells (Figure 4) also showed no detectable [125I]-rHuEpo binding. A total of 4 of the 5 cell lines with the higher EpoR protein also showed no detectable specific binding of [125I]-rHuEpo. NCI-H661 cells showed an increase in binding above control (POC = 6%; Figure 4). Thus, among cell lines that produced the higher levels of EpoR protein (estimated 400-3200 dimers/cell), only one, NCI-H661, had levels of surface EpoR protein sufficient for detectable binding of [125I]-rHuEpo. We concluded even in the presence of detectable EpoR protein by Western blot, very little if any EpoR was detected on the cell surface.
Specific cell-surface binding of [125I]-rHuEpo was detected in 1 of 5 of the higher EpoR protein–producing cell lines. Specific binding of [125I]-rHuEpo to live intact cells (1 × 106 cells) is expressed as percentage of control UT-7/Epo cells and is the average of 2 to 3 experiments (N = 5 samples/cell for each experiment). The negative control was 769-P cells. Error bars represent SD. Note specific binding with NCI-H661 cells (P ≤ .001).
Specific cell-surface binding of [125I]-rHuEpo was detected in 1 of 5 of the higher EpoR protein–producing cell lines. Specific binding of [125I]-rHuEpo to live intact cells (1 × 106 cells) is expressed as percentage of control UT-7/Epo cells and is the average of 2 to 3 experiments (N = 5 samples/cell for each experiment). The negative control was 769-P cells. Error bars represent SD. Note specific binding with NCI-H661 cells (P ≤ .001).
Cell lines with the higher EpoR protein showed no detectable stimulation of phosphorylation of intracellular proteins by rHuEpo
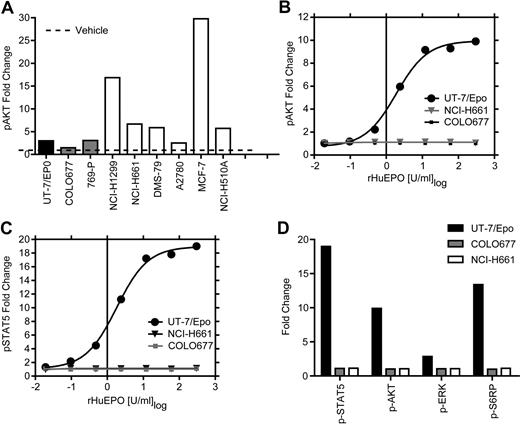
The presence of functional EpoR on the surface of cell lines that showed the higher levels of EpoR by Western blot was investigated using an additional strategy that makes no assumptions about the level or nature of the EpoR on cells. The signaling proteins STAT5, AKT, ERK, and S6RP are all known to be downstream of EpoR and phosphorylated in response to activation by rHuEpo. Levels of phosphorylated STAT5, AKT, ERK, and S6RP were measured in the 5 higher EpoR-expressing cell lines, including NCI-H661 cells, and in the MCF-7 cells. To heighten the sensitivity of the assay, cells were starved of serum and rHuEpo overnight before rHuEpo stimulation and analysis (Figure 5). Increased phosphorylation of AKT in tumor lines was seen after treatment with a positive control cocktail of growth factors (EGF/HGF/IGF-1; Figure 5A), indicating they were viable and responsive to cytokines. UT-7/Epo cells also demonstrated increased phosphorylation of all 4 proteins with rHuEpo (Figure 5B-D), as did differentiated CD34+ cells, which showed increased STAT5 phosphorylation (data not shown) after 5 or 30 minutes of addition in a dose-dependent manner. The negative control COLO677 cells had no detectable increase in any of the signaling proteins with rHuEpo treatment. In contrast to positive controls, there was no increase in multiple independent experiments in phosphorylation of AKT, ERK, S6RP, or STAT5 in NCI-H661 cells 5 minutes (Figure 5B-D) or after 30 minutes of treatment (data not shown) at any rHuEpo concentration up to 300 U/mL. Taken together, these results indicate that treatment with rHuEpo, even after overnight serum starvation, did not induce detectable intracellular signaling above the vehicle control under these conditions.
rHuEpo treatment of tumor cell lines cells did not lead to increased phosphorylation of signaling proteins. Tumor cell lines were starved of serum and rHuEpo overnight. Cells were stimulated for 5 minutes with vehicle (rHuEpo formulation buffer), a rHuEpo titration from 0.02 to 300 U/mL, or an EGF/HGF/IGF-1 growth factor cocktail (EGF 100 ng/mL, HGF 500 ng/mL, and IGF-1 500 ng/mL). UT-7/Epo cells were the positive control and COLO677 the negative control for rHuEpo treatment. Fixed and permeabilized cells were stained with fluorochrome-conjugated antibodies to the phosphorylated forms of the proteins, run on a FACS instrument, and analyzed as fold-change compared with vehicle treatment alone. Experiments were repeated 3 times with similar results. (A) Growth factor cocktail (EGF/HGF/IGF-1) 5-minute stimulation of 6 tumor cell lines analyzed for p-AKT. Note the stimulation of p-AKT in response to the growth factor cocktail in the tumor cell lines. (B) UT-7/Epo, NCI-H661, and COLO677 treated with increasing concentrations of rHuEpo and analyzed for p-AKT. (C) UT-7/Epo, NCI-H661, and COLO677 treated with increasing concentrations of rHuEpo and analyzed for p-STAT5. Note the lack of response in NCI-H661 cells with p-AKT and p-STAT5. (D) Effect of 5-minute stimulation of NCI-H661with 300 U/mL rHuEpo on phosphorylation of 4 signal transduction proteins. Similar results were seen after 30 minutes (data not shown).
rHuEpo treatment of tumor cell lines cells did not lead to increased phosphorylation of signaling proteins. Tumor cell lines were starved of serum and rHuEpo overnight. Cells were stimulated for 5 minutes with vehicle (rHuEpo formulation buffer), a rHuEpo titration from 0.02 to 300 U/mL, or an EGF/HGF/IGF-1 growth factor cocktail (EGF 100 ng/mL, HGF 500 ng/mL, and IGF-1 500 ng/mL). UT-7/Epo cells were the positive control and COLO677 the negative control for rHuEpo treatment. Fixed and permeabilized cells were stained with fluorochrome-conjugated antibodies to the phosphorylated forms of the proteins, run on a FACS instrument, and analyzed as fold-change compared with vehicle treatment alone. Experiments were repeated 3 times with similar results. (A) Growth factor cocktail (EGF/HGF/IGF-1) 5-minute stimulation of 6 tumor cell lines analyzed for p-AKT. Note the stimulation of p-AKT in response to the growth factor cocktail in the tumor cell lines. (B) UT-7/Epo, NCI-H661, and COLO677 treated with increasing concentrations of rHuEpo and analyzed for p-AKT. (C) UT-7/Epo, NCI-H661, and COLO677 treated with increasing concentrations of rHuEpo and analyzed for p-STAT5. Note the lack of response in NCI-H661 cells with p-AKT and p-STAT5. (D) Effect of 5-minute stimulation of NCI-H661with 300 U/mL rHuEpo on phosphorylation of 4 signal transduction proteins. Similar results were seen after 30 minutes (data not shown).
siRNA knockdown of EpoR did not affect viability of tumor cells
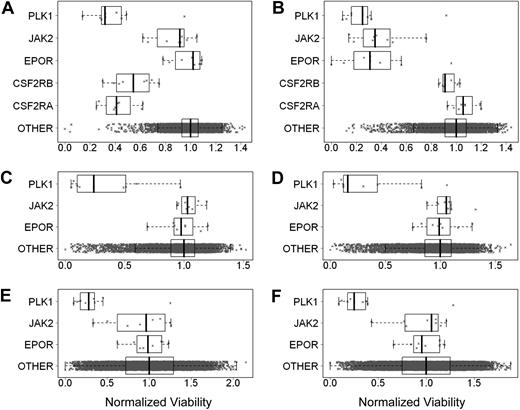
It was proposed that tumor cells may express EpoR and be dependent on autocrine Epo production.8,9 To test this hypothesis, EpoR siRNA knockdown experiments were performed in 3 cell lines selected because they either had higher EpoR expression, were reported to respond to rHuEpo, or because EpoR siRNA knockdown was reported to affect viability.7,–9 UT-7/Epo cells were positive control cells, and UT-7/GM-CSF cells, dependent on GM-CSF instead of rHuEpo, were used to detect nonspecific effects of the EPOR siRNAs on cell viability. Both cell lines were transfected with 8 different EPOR-targeting siRNAs, 8 different CSFRA-targeting and CSFRB-targeting siRNAs, 8 JAK2-targeting siRNAs (an essential gene in the EpoR signaling pathway), 9 PLK1-targeting siRNAs, a gene essential for viability (thus serving as an additional positive control), and approximately 10 000 siRNAs targeting genes other than EpoR as negative controls. At 4 days after transfection, cell viability was examined. As expected, EPOR siRNAs decreased viability of UT-7/Epo cells but not of UT-7/GM-CSF cells (Figure 6A-B). UT-7/GM-CSF cells had decreased viability after transfection with siRNAs to the GM-CSF receptor (Figure 6A), demonstrating the dependence of these cells on GM-CSF. Because the UT-7/Epo cells did not survive in the presence of EPOR siRNAs, the UT-7/GM-CSF cells served as a surrogate that allowed an assessment of the extent of EpoR knockdown seen with each of the EPOR siRNAs. Western blots of UT-7/GM-CSF lysates confirmed that EPOR siRNAs reduced EpoR protein, whereas random nontargeting or cyclophilin A (PPIA)–targeting siRNAs did not (Figure 7A). In addition, the extent of knockdown of EpoR in the UT-7/GM-CSF cells directly correlated with the magnitude of reduced viability of the UT-7/Epo cells (supplemental Figure 4A). In the UT-7/Epo cell line, JAK2 knockdown decreased cell viability more than 80% for the most effective siRNA. However, with UT-7/GM-CSF cells growing on GM-CSF, JAK2 siRNAs did not show a similar effect. UT-7/Epo and UT-7/GM-CSF cells did not survive in the presence of siRNAs targeting the cell-essential protein PLK1.
EpoR siRNA knockdown showed no effect on tumor cell line viability. Effect of knockdown of gene expression using siRNAs was assessed by determining cell viability. The cell viability for each siRNA was divided by the median viability of cells transfected with approximately 10 000 irrelevant siRNAs to give normalized viability where 1.0 represents no effect. Each point is a normalized value for an siRNA. The rectangle shows the interquartile range (IQR) from the 25th percentile to the 75th percentile. The whiskers go from the minimum value to the maximum value unless the distance from the minimum value to the first quartile is more than 1.5 times the IQR. In that case, the whisker extends out to the smallest value within 1.5 times the IQR from the first quartile. A similar rule is used for values larger than 1.5 times the IQR from the third quartile. The vertical bar is the mean of the siRNAs targeting specific genes. In each graph, the top plot represents siRNA for PLK1, the second plot JAK2, the third plot EPOR, and the fourth and fifth plots in panels A and B CSFRA and CSFRB, respectively; the fourth plot in panels C-F and the sixth plot in panels A and B is the approximately 10 000 siRNAs targeting other genes. (A) UT-7/GM-CSF grown with rHuGM-CSF (Benjamin-Hochberg corrected P > .999 for EPOR). (B) UT-7/Epo cells grown with rHuEpo (P < .002). (C) A2780 cells grown without rHuEpo (P > .999). (D) A2780 cells grown with rHuEpo (P > .999). (E) NCI-H1299 cells grown without rHuEpo (P = .99). (F) NCI-H1299 cells grown with rHuEpo (P = .98).
EpoR siRNA knockdown showed no effect on tumor cell line viability. Effect of knockdown of gene expression using siRNAs was assessed by determining cell viability. The cell viability for each siRNA was divided by the median viability of cells transfected with approximately 10 000 irrelevant siRNAs to give normalized viability where 1.0 represents no effect. Each point is a normalized value for an siRNA. The rectangle shows the interquartile range (IQR) from the 25th percentile to the 75th percentile. The whiskers go from the minimum value to the maximum value unless the distance from the minimum value to the first quartile is more than 1.5 times the IQR. In that case, the whisker extends out to the smallest value within 1.5 times the IQR from the first quartile. A similar rule is used for values larger than 1.5 times the IQR from the third quartile. The vertical bar is the mean of the siRNAs targeting specific genes. In each graph, the top plot represents siRNA for PLK1, the second plot JAK2, the third plot EPOR, and the fourth and fifth plots in panels A and B CSFRA and CSFRB, respectively; the fourth plot in panels C-F and the sixth plot in panels A and B is the approximately 10 000 siRNAs targeting other genes. (A) UT-7/GM-CSF grown with rHuGM-CSF (Benjamin-Hochberg corrected P > .999 for EPOR). (B) UT-7/Epo cells grown with rHuEpo (P < .002). (C) A2780 cells grown without rHuEpo (P > .999). (D) A2780 cells grown with rHuEpo (P > .999). (E) NCI-H1299 cells grown without rHuEpo (P = .99). (F) NCI-H1299 cells grown with rHuEpo (P = .98).
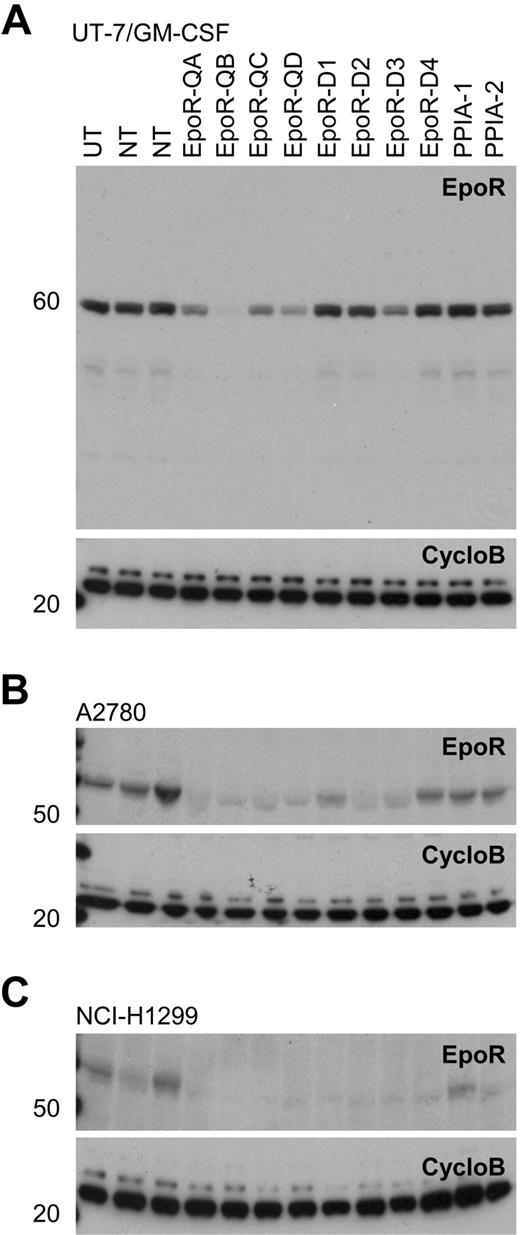
Western blots of EpoR knockdown for 8 different siRNAs. A82 Western blot of siRNA mediated knockdown of EPOR in transfected cells with 8 EPOR siRNAs, 2 nontargeting control siRNAs (NT), 2 cyclophilin A siRNAs (PPIA-1, PPIA-2), and an untransfected control (UT). CycloB is a loading control. Blots are representative of experiments performed on lysates from duplicate transfections on separate days. (A) UT-7/GM-CSF cells showing decreased EpoR protein in response to EPOR siRNA treatments. In the UT-7/Epo cells, there was a direct correlation between the degree of cell death observed (Figure 6) and the degree of EpoR knockdown with the 8 siRNAs (R2 = .6973; supplemental Figure 4). (B) A2780 cells and (C) NCI-H1299 cells both grown in rHuEpo show a decrease in EpoR protein in response to siRNA treatments.
Western blots of EpoR knockdown for 8 different siRNAs. A82 Western blot of siRNA mediated knockdown of EPOR in transfected cells with 8 EPOR siRNAs, 2 nontargeting control siRNAs (NT), 2 cyclophilin A siRNAs (PPIA-1, PPIA-2), and an untransfected control (UT). CycloB is a loading control. Blots are representative of experiments performed on lysates from duplicate transfections on separate days. (A) UT-7/GM-CSF cells showing decreased EpoR protein in response to EPOR siRNA treatments. In the UT-7/Epo cells, there was a direct correlation between the degree of cell death observed (Figure 6) and the degree of EpoR knockdown with the 8 siRNAs (R2 = .6973; supplemental Figure 4). (B) A2780 cells and (C) NCI-H1299 cells both grown in rHuEpo show a decrease in EpoR protein in response to siRNA treatments.
Viability of A2780, SK-OV-3, and NCI-H1299 cells was decreased with siRNAs to the positive control PLK1. Viability was not affected when cells were transfected with negative control siRNAs. There was no effect on viability when these cells were transfected with any of the EPOR siRNAs, and there was no effect on viability when cells were transfected with JAK2 siRNAs. Similar results were seen regardless of whether experiments were performed in the presence or absence of rHuEpo (Figure 6C-F; data not shown for SK-OV-3 cells). The lack of an effect was not due to a failure of the siRNAs to decrease EpoR protein, as the knockdown of EpoR in A2780 and NCI-H1299 was confirmed by Western blots (Figure 7B-C), and the decrease in EpoR protein was similar to that observed with UT-7/GM-CSF transfected with the same EPOR siRNAs. Taken together, these results suggest that in contrast to EPOR in UT-7/Epo cells and PLK1 in all the lines tested, knockdown of EPOR in A2780, SK-OV-3, and NCI-H1299 had no apparent effect on viability.
Discussion
This report describes the results of a survey of more than 200 human tumor cell lines. EPOR mRNA was detected in all cell lines, though at levels lower than rHuEpo-responsive control cell lines. Of these, 66 were selected for more detailed examination, and EpoR protein was detected in almost half of these. However, 90% of the cell lines had EpoR protein levels at a low level of 2% or less than that found in erythroid progenitor cells, and the remaining 10% at 20% or less. Cell lines with the highest level of EpoR were examined more thoroughly, and one line (NCI-H661) showed detectable surface EpoR. In that line (and the others examined), there was no evidence of activation of EpoR as assessed by rHuEpo-induced phosphorylation of intracellular proteins, although activation was evident in positive controls. It has been postulated that an inability to detect functional EpoR under standard culture conditions in ambient O2 is because EpoR levels are low in normoxia but are up-regulated by ischemia in nonvascularized solid tumors.29 This possibility is not supported by the results described here nor by other reports suggesting EpoR is not regulated by hypoxia.10,25,36 Hypoxia regulation, while proven for Epo the ligand, does not appear to be important for the receptor.
The lack of detectable functional EpoR in the tumor cell lines may have several explanations. One is that EpoR is inefficiently transported to the cell surface,33,–35 the causes of which are several-fold. The rate of assembly of functional EpoR, a homodimer,27,28 is concentration dependent.37,38 Thus, low-level protein production and inefficient surface translocation of EpoR may be a limiting factor in signal transduction for cells. In support of this possibility are examples in which a threshold level of EpoR must be reached before cells become demonstrably Epo responsive.39,,,,–44
Another possible explanation for the lack of functional EpoR in the cell lines is that EpoR expression alone is insufficient. Consistent with this notion, K562 and OCIM-1 cells do not respond to Epo despite detectable EpoR expression on the cell surface.21,26,45 Primary erythroid progenitor cells (CFUEs) have approximately 1000 EpoR surface receptors,40,46 a number sufficient for signaling; however, although OCIM-1 cells have a comparable amount of total and surface EpoR expression,21,47 they are not Epo-responsive.
A lack of a detectable effect of added Epo on various cell lines was noted despite detectable EpoR mRNA expression. It was suggested that the cell lines either expressed constitutively activated EpoR,7 or growth was supported by an Epo:EpoR autocrine loop.6,8 Alternatively, it is theoretically possible that autocrine Epo interacts with EpoR in an intracellular compartment, and/or that signaling occurs via different pathways. However, in experiments described here, EpoR siRNA knockdown performed in 2 of the higher EpoR-expressing lines, including A2780, one of the lines reported to exhibit an autocrine loop,8 did not lead to a loss of viability in the presence or absence of rHuEpo treatment. This suggested that neither Epo nor EpoR contributed to their survival.
Numerous in vitro and in vivo investigations of EpoR expression and function in tumor cell lines have been published to date, many of which are consistent with the results described here.1,10,13,14 In addition, there are many in vivo studies, the great majority of which demonstrate no tumor-stimulatory effect of ESAs. In fact, many demonstrate an enhanced tumor-inhibitory effect when combined with chemotherapy or radiotherapy (reviewed in Sinclair et al1 ). Although some of the contrary results may be explained by difficulties with reagents and experimental conditions, we cannot reconcile all the contrary data. However, the data presented here do not support a direct stimulatory effect of Epo on tumor cell growth. It is hoped that an ongoing pharmacovigilance program and future clinical trials will clarify the role of Epo in the outcome of patients who receive cancer therapy.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the members of the hematology group, especially Sean Yoder, who grew cell lines; Yun Lin for the bDNA assays; and Leigh Busse and Norma Rogers for the binding studies. Thanks to Miki Ben Jakov and Priya Mitty for help with the siRNA studies and especially to Elissa Swearingen. Thanks also to the Q-RT-PCR group, especially Kari Hale, Ivonne Archibeque, Samer Al-Assad and Michael Eschenberg. For the statistical analysis of the bDNA data, we thank Cheng Su.
Authorship
Contribution: S.S. cowrote the manuscript, designed EpoR expression and signaling experiments, interpreted data, and edited the manuscript; A.E. designed experiments and interpreted bDNA data; P.K. designed and interpreted siRNA data; I.McC. designed and interpreted signaling studies; J.R. performed signaling studies; A.S. designed and interpreted experiments and edited the manuscript; C.G.B. edited the manuscript; and S.E. designed and interpreted experiments and cowrote and edited the manuscript.
Conflict-of-interest disclosure: All authors are employees of and/or own stock in Amgen Inc, a manufacturer and marketer of erythropoiesis-stimulating agents.
Correspondence: Susan Swift, MS 29-1-A, 1 Amgen Center Dr, Thousand Oaks, CA 91320; e-mail: sswift@amgen.com.

![Figure 4. Specific cell-surface binding of [125I]-rHuEpo was detected in 1 of 5 of the higher EpoR protein–producing cell lines. Specific binding of [125I]-rHuEpo to live intact cells (1 × 106 cells) is expressed as percentage of control UT-7/Epo cells and is the average of 2 to 3 experiments (N = 5 samples/cell for each experiment). The negative control was 769-P cells. Error bars represent SD. Note specific binding with NCI-H661 cells (P ≤ .001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/21/10.1182_blood-2009-10-248674/5/m_zh89991050730004.jpeg?Expires=1768362123&Signature=Fe1g00m0l5V~lbfCHs2GnNQJJTwdp5QN2Zw1V-UX4iE7NbXl0KVwAV9rlGO9kFyghTQ6ETPGn6bbZg5OYvY8aj4Feb6wb4FmabtHTITaO479E1mW6ur9lleiWCOLMekFzUgROmxHNZhY4FEM~kVKqWyTWENbcHweyXAsUQ5DkvM4ZCQDmwDB7Qlkw7jJcf9aUo~-Iyi7KCc3rI4VZrJz2mfN9-zI9a~QDE-~2deh3K1vDv3NQtf89P8lKjceThKnXStyDccHCE1FKiUEyCGBys9A0geEHPF1aFa~V~99o~kbAtQezKQgwDs3NG4lI8tSaHF3qpMyd2QyMvnZhNB3dg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)