Abstract
The Additional sex combs like 1 (Asxl1) gene is 1 of 3 mammalian homologs of the Additional sex combs (Asx) gene of Drosophila. Asx is unusual because it is required to maintain both activation and silencing of Hox genes in flies and mice. Asxl proteins are characterized by an amino terminal homology domain, by interaction domains for nuclear receptors, and by a C-terminal plant homeodomain protein-protein interaction domain. A recent study of patients with myelodysplastic syndrome (MDS) and chronic myelomonocytic leukemia (CMML) revealed a high incidence of truncation mutations that would delete the PHD domain of ASXL1. Here, we show that Asxl1 is expressed in all hematopoietic cell fractions analyzed. Asxl1 knockout mice exhibit defects in frequency of differentiation of lymphoid and myeloid progenitors, but not in multipotent progenitors. We do not detect effects on hematopoietic stem cells, or in peripheral blood. Notably, we do not detect severe myelodysplastic phenotypes or leukemia in this loss-of-function model. We conclude that Asxl1 is needed for normal hematopoiesis. The mild phenotypes observed may be because other Asxl genes have redundant function with Asxl1, or alternatively, MDS or oncogenic phenotypes may result from gain-of-function Asxl mutations caused by genomic amplification, gene fusion, or truncation of Asxl1.
Introduction
Proteins of the Polycomb group (PcG) and trithorax group (trxG) ensure epigenetic maintenance of gene expression patterns through mitosis and faithful propagation of cell fates. PcG genes silence their targets, whereas trxG proteins maintain transcriptional activation. Although the precise mechanism of maintenance is unknown, trxG and PcG genes encode chromatin proteins required for histone modification or establishment or prevention of nucleosome remodeling, or those that enhance or prevent transcriptional elongation.1 The Enhancer of trithorax and Polycomb (ETP) genes encode proteins required for both maintenance of activation and silencing, as shown by simultaneous anterior and posterior transformations caused by failure to activate or repress Hox genes. The molecular basis of ETP function is unknown.2 Hematopoiesis is a dynamic process requiring coordination between genetic and epigenetic programs to regulate transitions between and maintenance of cell fates, which ultimately generates all blood lineages. Mammalian PcG and trxG genes display hematopoietic lineage- and differentiation stage–specific expression patterns, are required for normal and leukemic hematopoiesis, and show aberrant expression in leukemias and lymphomas.3-5 Mutations in vertebrate PcG and trxG genes can lead to oncogenic or tumor suppressor activity, depending on context.3-5
Additional sex combs like 1 (Asxl1) belongs to the ETP group. Asxl1 regulates Hox genes in axial patterning,6 and is 1 of 3 mammalian homologs of the Drosophila Asx gene.4,5,7-9 As shown in Figure 1A, all mammalian ASXL proteins have conserved sequence features: an amino-terminal ASX homology (ASXH) region, which contains 2 putative nuclear receptor coregulator binding (NR box) motifs, 3 other NR box motifs, and a carboxy-terminal plant homeodomain (PHD) domain.7,8
ASXL1 is a member of a repressive complex containing histone H1.2.9 Conversely, ASXL1 functions autonomously as a transcriptional activator in human cancer cell lines that are retinoic acid (RA) sensitive.10 ASXL1 enhances recruitment of RA receptors to their chromatin targets, and interacts with the activation domain core of RA receptors in the presence of the ligand RA to increase their activity. However, in other cancer cell lines that are RA resistant, ASXL1 is a corepressor of retinoic acid receptor (RAR) activity.10 These results support predictions of dual activator/repressor functions for mammalian ASXL proteins depending on cellular context.
ASXL1 is 1 of several fusion protein partners with PAX5 in B-cell precursor acute lymphoblastic leukemia patient samples.11 A recent study identified ASXL1 mutations in 13% of patients with myelodysplastic syndromes (MDSs) and 43% of patients with chronic myelomonocytic leukemia (CMML).12 These mutations cause truncation of the protein downstream of the ASXH domain with consequent loss of the PHD domain. These findings suggest that ASXL1 may function as a tumor suppressor in malignancies of the myeloid lineage by affecting stem or progenitor cell self-renewal or differentiation.12 Recent studies highlight the role of PHD domain mutations or translocations in leukemia,13 suggesting that the PHD domain of Asxl1 has an important functional role. In this study, we generated a loss-of-function mouse model of Asxl1 and show that Asxl1 is required for normal hematopoiesis, but is not required for function of stem cells and multipotent progenitors. Strikingly, these mice did not exhibit MDS or leukemia. These mild phenotypes of Asxl1 loss-of-function mutations may occur because Asxl genes have redundant function, or because ASXL1 mutations that lead to MDS and CMML are gain of function.
Methods
Gene targeting and generation of Asxl1tm1Bc mice
Genomic Asxl1 fragments from BAC clone no. gs12943 (Genome Systems) and vectors pMTL2214 and pPNT15 were used to prepare a replacement targeting vector designed to insert a PGK promoter–driven neomycin resistance cassette into exon 5 of Asxl1 at the XbaI site (amino acid 90), which is upstream of the conserved ASXH and PHD domains in Asxl1 (Figure 1A-B). The location of the 5′ (at the HindIII site) and 3′ (at the ClaI site) ends of the flanking genomic homology arms in the targeting vector are denoted in larger bold font in Figure 1B. This construct was linearized and electroporated into the E14 mouse embryonic stem cell line, and G418-resistant colonies were selected. All primer sequences are given in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Seven of 627 clones were correctly targeted as determined by genomic polymerase chain reaction (PCR) screening using external primer P1 specific for Asxl1 intron 4, and primer P4 within the neomycin gene (Figure 1B), which yields a 2161-bp PCR product. These were confirmed by Southern blotting. The neomycin transgene introduces an additional EcoR1 site so that a 3.9-kb fragment is detected when the 5′ external probe (0.5-kb EcoR1-HindIII fragment shown in Figure 1B) is hybridized to EcoRI digested genomic DNA, compared with the 7.3-kb fragment in wild type (Figure 1C). Two independent positive embryonic stem cell clones of normal karyotype were used to inject C57BL/6J blastocysts to generate 2 lines of chimeric mice that were mated to C57BL/6J mice. Chimeras and selected N1 offspring were genotyped by Southern blot and heterozygous mutant mice were backcrossed to C57BL/6J mice. Offspring from N2 or N3 generation Asxl1tm1Bc mutant heterozygous parents were used for all experiments reported here, except for the fetal liver competitive repopulation unit (CRU) assay, which used more than N8 generation mice. Adult mice used for experiments ranged in age from 6 to 58 weeks, and at least 1 wild-type littermate control was included for each Asxl1tm1Bc mutant sampled concurrently. Genotyping was routinely performed on DNA from adult tail tips, liver from newborns, or yolk sac from embryos, by PCR using primer P2 specific for Asxl1 intron 4, primer P3 specific for the PGK promoter, and common primer P5 specific for exon 5 downstream of the neomycin cassette. Fragments of 253 bp and 426 bp were indicative of the wild-type and targeted alleles, respectively (Figure 1D). All experiments with animals conformed to the regulations established by the Canadian Council on Animal Care. All protocols were approved by the University of British Columbia Animal Care Committee.
RT-PCR and sequencing of Asxl1tm1Bc mRNA
Mouse embryonic fibroblasts (MEFs) were prepared from homozygous mutant E12.5 to E14.5 Asxl1tm1Bc embryos, and immortalized by infection with a retroviral vector expressing Tbx2 as described.16 RNA was prepared from these MEFs, or from pooled neonate tissues, using Trizol (Invitrogen) and used as templates for cDNA synthesis with poly(dT) and random hexamer primers (Superscript III; Invitrogen). Neonate tissue cDNA was used as a template for PCR reactions under standard conditions with a forward primer from exon 1 and primer P5 or with the exon 1 primer and primer P4. The MEF cDNA was used as a template for PCR reactions with exon 4 and exon 6 primers carried out in the presence of 0.45 M betaine (Sigma). After gel purification, size-selected PCR products were sequenced by the Nucleic Acid Protein Service Unit at the University of British Columbia.
Gene expression analysis
Blood and FACS analyses
Peripheral blood of newborn or adult mice was obtained by cardiac puncture immediately after killing. White blood cells were prepared from peripheral blood after incubation in ammonium chloride (StemCell Technologies) to lyse erythrocytes. Cell suspensions of spleen and thymus were prepared using 40-μm nylon mesh strainers. Bone marrow was flushed from both femurs by irrigation of the excised bone using a 22-gauge × 1½ needle and 3-cc syringe and was brought to single-cell suspension by multiple passages through the syringe. Red blood cell and all nucleated cell counts were determined using a hemocytometer.
Single-cell suspensions at a density of 5 to 10 × 106 cells/mL were incubated on ice, protected from light, for 30 to 60 minutes, and were singly, doubly, or triply stained with specific monoclonal antibodies to the murine cell surface antigens: Gr-1 (Ly-6G) and Mac-1 (CD11b) monocyte and granulocyte markers; Ter-119 (Ly-76) erythroid lineage marker; CD4 (L3T4), CD8a (Ly-2), CD44 (Pgp-1, Ly-24), and CD25 (interleukin-2 [IL-2] receptor chain α) T-lymphoid lineage markers; B220 (CD45R), CD43 (Ly-48, leukosialin), surface immunoglobulin M (IgM; R6-60.2), and surface IgD B-lymphoid lineage markers; and Ly5.2 mouse strain–specific marker. The antibodies were conjugated to the fluorochromes fluorescein isothiocyanate (FITC) or phycoerythrin (PE) as indicated in the figures. All antibodies were from BD PharMingen, except for the anti-IgD antibody, which was from Southern Biotech Associates Inc. Cell sorting was performed on FACSCalibur, FACScan, or FACSort flow cytometers (Becton Dickinson). Dead cells were excluded from analysis using forward and side scatter and phosphatidyl inositol staining. Acquisition and analysis were performed with CELLQuest software (Becton Dickinson).
Clonogenic progenitor assays
To detect multipotential myeloid lineage clonogenic progenitors 2.5 × 104 cells from embryonic day (E) 18.5 fetal liver, newborn spleen, or adult bone marrow were plated in a 1.1-mL volume per Greiner Petri dish. Cells were plated in Methocult M3434 methylcellulose medium (StemCell Technologies) containing 10 ng/mL recombinant murine IL-3, 10 ng/mL recombinant human IL-6, 50 ng/mL recombinant murine steel factor, and 3 U/mL recombinant human erythropoietin. Myeloid and erythroid cell colonies were counted and scored after 10 to 12 days. To detect pre-B-lymphoid clonogenic progenitors, 1.0 × 105 cells from adult bone marrow were plated in a 1.1-mL volume per Greiner Petri dish in Methocult M3630 methylcellulose medium (StemCell Technologies) containing 10 ng/mL IL-7.18 The number of B-cell colonies was scored after 5 to 7 days. All cell culture was performed in a humidified incubator at 37°C at 5% CO2. All colonies were scored microscopically using standard phenotypic criteria. Final colony-forming unit (CFU) values represent the average of 2 replicates per sample.
Fetal thymic organ culture
Fetal thymus lobes were dissected from E14.5 embryos and cultured as described.19 Thymocytes were stained with monoclonal antibodies against CD4 and CD8, and analyzed by fluorescence-activated cell sorting (FACS) as described in “Blood and FACS analyses.”
Day-12 CFU-S and competitive repopulating unit assay
Methods used for the day-12 spleen colony-forming unit (CFU-S12) and competitive repopulation unit (CRU) assays are as previously described.20 Reconstitution was tested at 11 and 20 weeks after transplant by FACS analysis to detect Ly5.2 and B220, CD4 and CD8, or Gr1 and Mac1, from peripheral blood from the tail vein. Mice whose blood contained greater than 1% donor-derived (Ly5.2+) myeloid cells (Gr-1+ and/or Mac-1+), B cells (B220+), and T cells (CD4+ and/or CD8+) were considered to have been repopulated with transduced cells. CRU frequencies in the test fetal liver samples were calculated by applying Poisson statistics to the proportion of negative recipients at different dilutions using L-Calc (StemCell Technologies) software.
Statistical analyses
Comparison of means between groups was done using the Student t test. Differences with P values less than .05 were considered statistically significant.
Results
Generation of Asxl1tm1Bc mutant mice
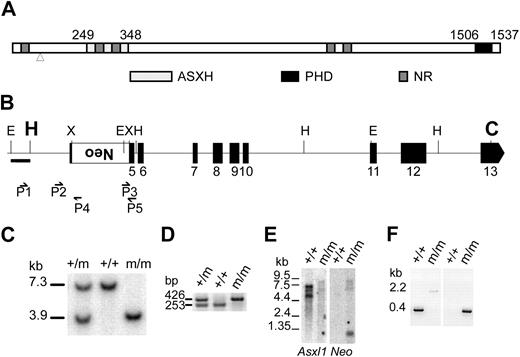
The Asxl1 gene was disrupted using targeted replacement mutagenesis by permanent insertion of a PGK promoter–driven neomycin-resistance (neo) gene in reverse orientation to the Asxl1 direction of transcription into the XbaI site of exon 5 (Figure 1B; see “Gene targeting and generation of Asxl1tm1Bc mice” for details and controls). This insertion interrupts the reading frame of Asxl1 by introducing several premature termination codons after the corresponding amino acid 90 position, upstream of the conserved ASXH domain (Figure 1A).8 To determine whether Asxl1 mRNAs containing the transgene were expressed, we performed Northern blots to show that all detectable transcripts in Asxl1tm1Bc/tm1Bc mutant embryos contain the transgene and to confirm neomycin expression (Figure 1E). It is likely these Asxl1 mutant transcripts are not transcribed due to nonsense-mediated decay, however any stable truncated protein generated from these mRNAs would lack the conserved ASXH and PHD domains and all NR box motifs except one at the extreme amino-terminus (Figure 1A),8 and likely would be nonfunctional.
Generation of Asxl1 mutant mice. (A) Schematic representation of the conserved ASXH and PHD domains and NR sequence motifs in ASXL1 homologs is shown.8 Numbers above the diagram show the first and last amino acids that define the ASXH and PHD domains. Mutations causing ASXL1 truncations found in MDS and CMML patients lie between amino acids 596 and 1457, all within exon 12.12 The arrowhead shows the site of insertion of the neomycin (neo) transgene at amino acid 90. (B) Diagram of part of the Asxl1 locus showing exons 5 to 13 indicated as black boxes. The PGKneo expression cassette (large white box) was inserted into the XbaI site in exon 5 by a replacement gene-targeting approach and was used for positive selection of clones. Position of relevant restriction sites (C indicates ClaI; E, EcoRI; H, HindIII; and X, XbaI), the location of external probe (bar below the E-H fragment shown at the left), and PCR primers (small arrows) are indicated. The location of the 5′ (HindIII site) and 3′ (ClaI site) ends of the flanking genomic homology arms in the targeting vector is denoted in larger bold font. (C) Southern blot analysis of genomic DNA isolated from newborn offspring of an Asxl1tm1Bc intercross after digestion with EcoRI and hybridized with an external probe shown in panel B. This probe detects a 7.3-kb fragment from the wild-type allele (+/+), and a 3.9-kb fragment from the targeted allele (m/m). (D) Multiple primer PCR analysis of liver genomic DNA of E18.5 embryos from an Asxl1tm1Bc intercross. Primers P2 and P5 amplify a 253-bp fragment of the wild-type allele, whereas primers P3 and P5 amplify a 426-bp fragment of the targeted allele. (E) Northern blot analysis of poly(A)+RNA from pooled tissue of neonate wild-type and Asxl1tm1Bc mice using probes for Asxl1 (left panel) and neomycin (right panel). (F) RT-PCR analysis of Asxl1tm1Bc mutant neonate tissue. RT-PCR of total RNA from pooled tissue of individual newborn Asxl1+/+ (+/+) and Asxl1tm1Bc/tm1Bc (m/m) mice. Primer from exon 1 and primer P5 (in exon 5) amplify an approximately 400-bp product in Asxl1+/+ samples, and a 2.2-kb product in Asxl1tm1Bc/tm1Bc (left panel). Primer from exon 1 and primer P4 (within neo) amplify a 300-bp product from the Asxl1tm1Bc/tm1Bc samples, whereas no amplification occurs in the Asxl1+/+ samples, as expected (right panel).
Generation of Asxl1 mutant mice. (A) Schematic representation of the conserved ASXH and PHD domains and NR sequence motifs in ASXL1 homologs is shown.8 Numbers above the diagram show the first and last amino acids that define the ASXH and PHD domains. Mutations causing ASXL1 truncations found in MDS and CMML patients lie between amino acids 596 and 1457, all within exon 12.12 The arrowhead shows the site of insertion of the neomycin (neo) transgene at amino acid 90. (B) Diagram of part of the Asxl1 locus showing exons 5 to 13 indicated as black boxes. The PGKneo expression cassette (large white box) was inserted into the XbaI site in exon 5 by a replacement gene-targeting approach and was used for positive selection of clones. Position of relevant restriction sites (C indicates ClaI; E, EcoRI; H, HindIII; and X, XbaI), the location of external probe (bar below the E-H fragment shown at the left), and PCR primers (small arrows) are indicated. The location of the 5′ (HindIII site) and 3′ (ClaI site) ends of the flanking genomic homology arms in the targeting vector is denoted in larger bold font. (C) Southern blot analysis of genomic DNA isolated from newborn offspring of an Asxl1tm1Bc intercross after digestion with EcoRI and hybridized with an external probe shown in panel B. This probe detects a 7.3-kb fragment from the wild-type allele (+/+), and a 3.9-kb fragment from the targeted allele (m/m). (D) Multiple primer PCR analysis of liver genomic DNA of E18.5 embryos from an Asxl1tm1Bc intercross. Primers P2 and P5 amplify a 253-bp fragment of the wild-type allele, whereas primers P3 and P5 amplify a 426-bp fragment of the targeted allele. (E) Northern blot analysis of poly(A)+RNA from pooled tissue of neonate wild-type and Asxl1tm1Bc mice using probes for Asxl1 (left panel) and neomycin (right panel). (F) RT-PCR analysis of Asxl1tm1Bc mutant neonate tissue. RT-PCR of total RNA from pooled tissue of individual newborn Asxl1+/+ (+/+) and Asxl1tm1Bc/tm1Bc (m/m) mice. Primer from exon 1 and primer P5 (in exon 5) amplify an approximately 400-bp product in Asxl1+/+ samples, and a 2.2-kb product in Asxl1tm1Bc/tm1Bc (left panel). Primer from exon 1 and primer P4 (within neo) amplify a 300-bp product from the Asxl1tm1Bc/tm1Bc samples, whereas no amplification occurs in the Asxl1+/+ samples, as expected (right panel).
We generated a polyclonal antibody against amino acids 811 to 929 of Asxl1, but we were unable to detect endogenous Asxl1 protein expression in Western blots of MEFs (data not shown). We therefore performed RT-PCR on Asxl1tm1Bc mutant tissues and embryonic fibroblast (MEF) lines, to confirm the predicted transcript structure, and to assess whether use of cryptic splice sites resulted in the generation of aberrant splice variants from the mutated Asxl1 allele that may be translated. PCR with the exon 1 primer and primer P5 amplifies an approximately 400-bp band in Asxl1+/+ samples, and a 2.2-kb band in Asxl1tm1Bc/tm1Bc samples consistent with read-through of the inserted PGKneo cassette (Figure 1F). The exon 1 primer and primer P4 amplify a band from the Asxl1tm1Bc/tm1Bc samples, confirming expected reverse orientation of the neo cassette, whereas no amplification occurs in the Asxl1+/+ samples (Figure 1F). We performed PCR with primers from exon 4 and exon 6 that flank the insertion site of the targeting cassette in exon 5. Sequencing of the product showed that an abnormal splicing event in Asxl1tm1Bc mutants can remove all of exon 5, including the targeting cassette, from a minority of the transcripts (supplemental Figure 1). This abnormally spliced Asxl1tm1Bc mRNA would have an altered reading frame with premature termination codons and would likely undergo nonsense-mediated decay. We therefore predict that the Asxl1tm1Bc mutant allele is a null allele. The Asxl1tm1Bc mutant mice exhibit partially penetrant perinatal lethality, and show developmental phenotypes consistent with Enhancer of trithorax and Polycomb (ETP) function, as described in detail elsewhere.6
Asxl1 is expressed ubiquitously in hematopoietic cells
To assess Asxl1 expression in hematopoietic cells, we used global cDNA amplification of hematopoietic cells fractionated by FACS into hematopoietic stem cells (HSCs) and primitive clonogenic, pluripotent progenitors and mature cells, followed by Southern blot analysis using gene-specific probes as described.17 Asxl1 is expressed ubiquitously in all progenitor-enriched and mature hematopoietic cell–enriched fractions analyzed in fetal liver (Figure 2A) and adult bone marrow (Figure 2B). Asxl1 expression is ubiquitous in mature myeloid cells (monocytes/macrophages [Mac-1+] and granulocytes [Gr-1+]), in lymphoid lineages, including FACS fractionated cells from adult bone marrow enriched for B-cell progenitors (cKit+B220+, B220+CD25+, B220+CD43+) or immature B cells (B220+IgM+), in mature spleen-derived B cells (IgD+IgM+; Figure 2A), and in all fractionated CD4 CD8 subpopulations representing the major stages of T-lymphoid cell differentiation obtained from adult thymus. In contrast, the expression of Bmi1 is highest in progenitor-enriched Sca+Lin− and Sca+Lin+ fractions, whereas expression was low to undetectable in all other fractions analyzed (Figure 2).
Asxl1 is ubiquitously expressed in hematopoietic cells. Southern blot analysis of Asxl1, Bmi1, and actin gene expression in total amplified cDNAs from FACS-sorted hematopoietic cell populations from fetal liver and adult bone marrow, spleen, and thymus. Total cDNAs were amplified using RT-PCR from 104 sorted cells, and each sample with or without reverse transcriptase (RT + or −) was sequentially hybridized to each probe. Results for a representative experiment are shown; duplicate experiments were performed. (A) Asxl1 is expressed in unfractionated E14.5 fetal liver (TFL), and the hematopoietic stem cell–depleted (Sca-1−Lin+ [S−L+]), low (Sca-1+Lin+ [S+L+]), and enriched (Sca-1+Lin− [S+L−]) subpopulations, whereas Bmi1 expression is predominant in the 2 Sca-1+ fractions. (B) Asxl1 is expressed in unfractionated adult bone marrow (TBM) and in all fractionated adult bone marrow, spleen, and thymus cells investigated, whereas Bmi1 is expressed predominantly within the progenitor-enriched Sca-1+ fractions and the Gr-1 low fraction of total bone marrow, with very low or undetectable expression in other fractions.
Asxl1 is ubiquitously expressed in hematopoietic cells. Southern blot analysis of Asxl1, Bmi1, and actin gene expression in total amplified cDNAs from FACS-sorted hematopoietic cell populations from fetal liver and adult bone marrow, spleen, and thymus. Total cDNAs were amplified using RT-PCR from 104 sorted cells, and each sample with or without reverse transcriptase (RT + or −) was sequentially hybridized to each probe. Results for a representative experiment are shown; duplicate experiments were performed. (A) Asxl1 is expressed in unfractionated E14.5 fetal liver (TFL), and the hematopoietic stem cell–depleted (Sca-1−Lin+ [S−L+]), low (Sca-1+Lin+ [S+L+]), and enriched (Sca-1+Lin− [S+L−]) subpopulations, whereas Bmi1 expression is predominant in the 2 Sca-1+ fractions. (B) Asxl1 is expressed in unfractionated adult bone marrow (TBM) and in all fractionated adult bone marrow, spleen, and thymus cells investigated, whereas Bmi1 is expressed predominantly within the progenitor-enriched Sca-1+ fractions and the Gr-1 low fraction of total bone marrow, with very low or undetectable expression in other fractions.
Effects of Asxl1tm1Bc mutation on T lymphopoiesis
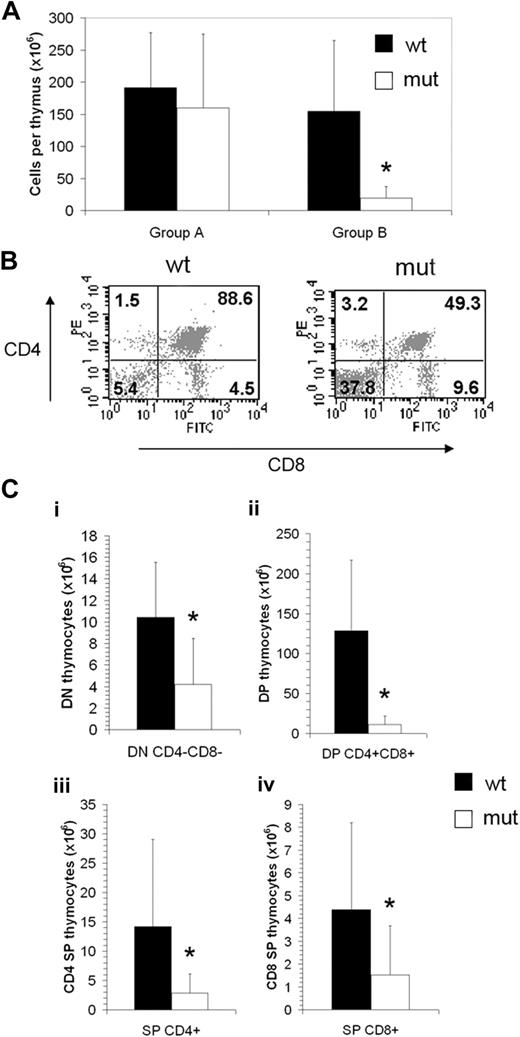
Thymus cellularity of Asxl1tm1Bc mutant mice older than 15 weeks was drastically decreased compared with Asxl1+/+ controls, and so we grouped mice according to age for further analysis (Figure 3A). There is a relative increase in the number of double-negative (CD4−CD8−) thymocytes in Asxl1tm1Bc mutant compared with control thymi (Figure 3B). When the overall reduction in thymus cellularity of older mutant Asxl1tm1Bc mice is taken into account, all CD4 CD8 subcompartments show a significant reduction in absolute cell number (Figure 3C). However, there were no significant differences in the absolute numbers of single-positive (CD4+ or CD8+) or double-positive (CD4+CD8+) cells in the peripheral blood (supplemental Figure 2).
Asxl1tm1Bc/tm1Bc mutant mice exhibit reduced thymopoiesis. (A) Thymus cellularity is reduced in older (older than 15 weeks; group B, n = 8 each genotype) compared with younger (15 weeks or younger; group A, n = 6 wt, 4 mutant) Asxl1tm1Bc/tm1Bc (mut) adult mice compared with wild type. (B) Flow cytometry of T lymphocytes from adult thymus expressing the lineage markers CD4 and CD8 shows a relative increase in the double-negative and both single-positive fractions, and a relative decrease in the double-positive fraction in Asxl1tm1Bc/tm1Bc thymus compared with wild type. (C) Absolute cell number per thymus is reduced in group B Asxl1tm1Bc/tm1Bc mice compared with wild type for all fractions: (i) double-negative CD4−CD8− (DN), (ii) double-positive CD4+CD8+ (DP), (iii) single-positive (SP) CD4+, and (iv) single-positive CD8+ (n = 8). *P < .05.
Asxl1tm1Bc/tm1Bc mutant mice exhibit reduced thymopoiesis. (A) Thymus cellularity is reduced in older (older than 15 weeks; group B, n = 8 each genotype) compared with younger (15 weeks or younger; group A, n = 6 wt, 4 mutant) Asxl1tm1Bc/tm1Bc (mut) adult mice compared with wild type. (B) Flow cytometry of T lymphocytes from adult thymus expressing the lineage markers CD4 and CD8 shows a relative increase in the double-negative and both single-positive fractions, and a relative decrease in the double-positive fraction in Asxl1tm1Bc/tm1Bc thymus compared with wild type. (C) Absolute cell number per thymus is reduced in group B Asxl1tm1Bc/tm1Bc mice compared with wild type for all fractions: (i) double-negative CD4−CD8− (DN), (ii) double-positive CD4+CD8+ (DP), (iii) single-positive (SP) CD4+, and (iv) single-positive CD8+ (n = 8). *P < .05.
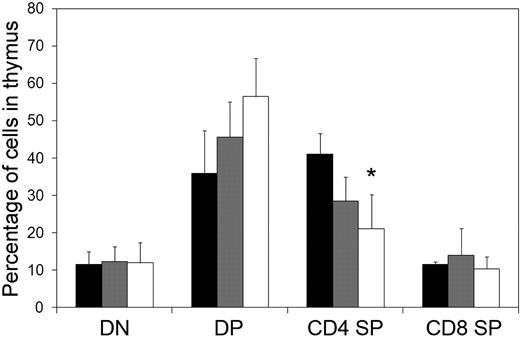
To determine whether defects in thymopoiesis could be detected earlier in development, we used the fetal thymic organ culture assay to measure relative proportions of CD4- and CD8-expressing T cells from E14.5 thymi. We detected no significant differences in the relative proportion of double-negative, double-positive, or CD8 single-positive populations. However there was a modest decrease in the proportion of CD4 single-positive cells from Asxl1tm1Bc/tm1Bc embryos compared with wild-type littermate-derived samples (Figure 4).
Flow cytometric analysis of T cells cultured from E14.5 thymus. Cells were analyzed using monoclonal antibodies against CD4 and CD8. Numbers indicate the percentage of cells within each subcompartment: double-negative CD4−CD8− (DN), double-positive CD4+CD8+ (DP), single-positive CD4 (CD4+SP), and single-positive CD8 (CD8+SP). *P < .05. ■ indicates wild type (n = 2); ▩, Asxl1+/tm1Bc (n = 6); and □, Asxl1tm1Bc/tm1Bc mutant (n = 7).
Flow cytometric analysis of T cells cultured from E14.5 thymus. Cells were analyzed using monoclonal antibodies against CD4 and CD8. Numbers indicate the percentage of cells within each subcompartment: double-negative CD4−CD8− (DN), double-positive CD4+CD8+ (DP), single-positive CD4 (CD4+SP), and single-positive CD8 (CD8+SP). *P < .05. ■ indicates wild type (n = 2); ▩, Asxl1+/tm1Bc (n = 6); and □, Asxl1tm1Bc/tm1Bc mutant (n = 7).
Effects of Asxl1tm1Bc mutation on B lymphopoiesis
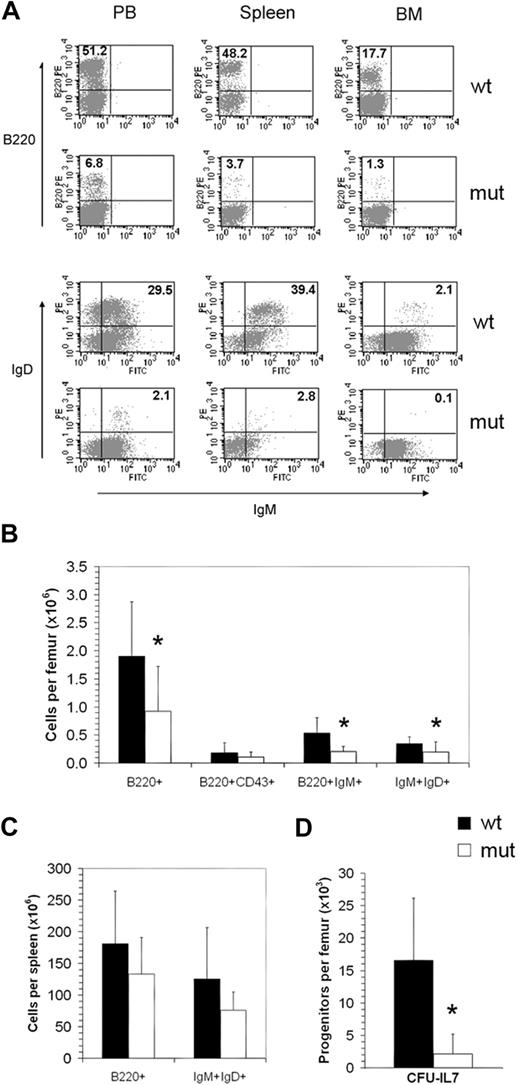
There was no effect of the Asxl1tm1Bc mutation on E18.5 fetal liver cellularity, or adult bone marrow cellularity, regardless of age (supplemental Figure 3). However, a subset of Asxl1tm1Bc mutant mice older than 15 weeks showed a relative decrease in B-lymphoid cell numbers within peripheral blood, spleen, and bone marrow, as illustrated by reduced expression of the pan-B-cell marker B220 and the mature B-cell markers IgM and IgD, compared with littermate wild-type controls (Figure 5A; severe case shown). Absolute numbers of B220+ cells, B220+IgM+ immature B cells, and IgM+IgD+ mature B cells were significantly reduced in bone marrow of mice older than 15 weeks (Figure 5B and supplemental Figure 3), whereas a modest though not statistically significant decrease was seen in bone marrow of mice younger than 15 weeks (supplemental Figure 3A) and in spleen (Figure 5C). However, no significant difference in absolute number of B220+ and IgM+IgD+ cells was seen in the peripheral blood (supplemental Figure 2), suggesting highly variable penetrance of the effect on B cells of Asxl1tm1Bc mice compared with wild-type controls. To determine whether there was a reduction in the number of pro-B precursor cells in Asxl1tm1Bc/tm1Bc bone marrow, we monitored coexpression of B220 and CD43 using flow cytometry. Overall, the differences between Asxl1tm1Bc/tm1Bc and Asxl1+/+ control samples were not statistically significant (Figure 5B). This suggests that the major defect in B-cell progression for most Asxl1tm1Bc/tm1Bc mice occurred after pro-B differentiation. To determine whether Asxl1tm1Bc mutation affected progression through the IL-7–responsive, pre-B progenitor stage, we analyzed bone marrow from adult Asxl1tm1Bc/tm1Bc mice (all older than 12 weeks) in a CFU–IL-7 methylcellulose colony assay. There were severe reductions in the frequency of CFU–IL-7 generated from the Asxl1tm1Bc/tm1Bc bone marrow samples compared with wild-type controls (Figure 5D), consistent with an impaired transition to the pre-B stage.
Asxl1tm1Bc/tm1Bc mutant mice exhibit reduced B-cell lymphopoiesis. (A) Flow cytometric profiles of adult (> 15 weeks old) peripheral blood (PB), spleen, and bone marrow (BM) show a relative decrease in B220- and IgM/IgD-positive cells in Asxl1tm1Bc/tm1Bc (mut) tissues compared with wild type (wt). (B) Absolute bone marrow cell numbers in adult femur expressing the markers B220; B220 and CD43; B220 and IgM; and IgM and IgD are significantly lower in Asxl1tm1Bc/tm1Bc mice compared with wild type (n = 8 each genotype except n = 4 for B220 and IgM). (C) Absolute cell numbers from > 15-week-old spleen expressing the markers B220, and IgM and IgD, are lower in Asxl1tm1Bc/tm1Bc mice compared with wild type (n = 8 each genotype). (D) In vitro colony formation of committed pre-B-lymphocyte progenitors (CFU–IL-7 assay) from adult bone marrow (> 15 weeks old) is significantly reduced in cultures of Asxl1tm1Bc/tm1Bc group B compared with wild type (n = 7 wt, 8 mutant). *P < .05.
Asxl1tm1Bc/tm1Bc mutant mice exhibit reduced B-cell lymphopoiesis. (A) Flow cytometric profiles of adult (> 15 weeks old) peripheral blood (PB), spleen, and bone marrow (BM) show a relative decrease in B220- and IgM/IgD-positive cells in Asxl1tm1Bc/tm1Bc (mut) tissues compared with wild type (wt). (B) Absolute bone marrow cell numbers in adult femur expressing the markers B220; B220 and CD43; B220 and IgM; and IgM and IgD are significantly lower in Asxl1tm1Bc/tm1Bc mice compared with wild type (n = 8 each genotype except n = 4 for B220 and IgM). (C) Absolute cell numbers from > 15-week-old spleen expressing the markers B220, and IgM and IgD, are lower in Asxl1tm1Bc/tm1Bc mice compared with wild type (n = 8 each genotype). (D) In vitro colony formation of committed pre-B-lymphocyte progenitors (CFU–IL-7 assay) from adult bone marrow (> 15 weeks old) is significantly reduced in cultures of Asxl1tm1Bc/tm1Bc group B compared with wild type (n = 7 wt, 8 mutant). *P < .05.
Effects of Asxl1tm1Bc mutation on erythropoiesis and myelopoiesis
We analyzed properties of hematopoietic cells from homozygous Asxl1tm1Bc mice at stages E18.5, P1, and adult (> P42). P1 and adult Asxl1tm1Bc homozygous null mutants showed no differences in peripheral blood red or white cell counts compared with Asxl1+/+, regardless of whether they were grouped according to age as described in the previous section (supplemental Figure 4). We did not observe any significant changes in platelets, erythrocyte, leukocyte, or thrombocyte morphology, by visual examination of Wright-Giemsa–stained blood smears or changes in cell morphology in cytospin preparations of spleen and bone marrow cells of Asxl1tm1Bc mutants compared with wild-type controls (data not shown).
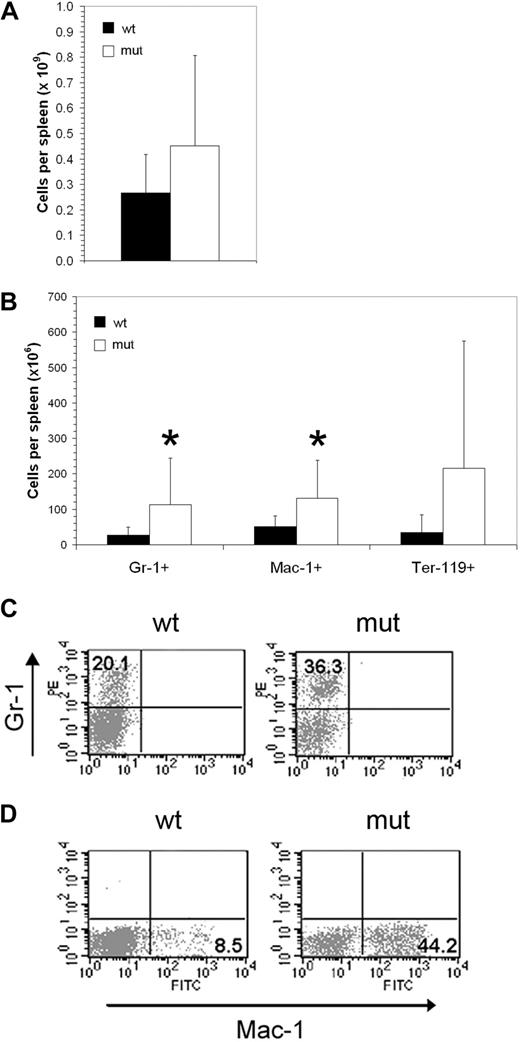
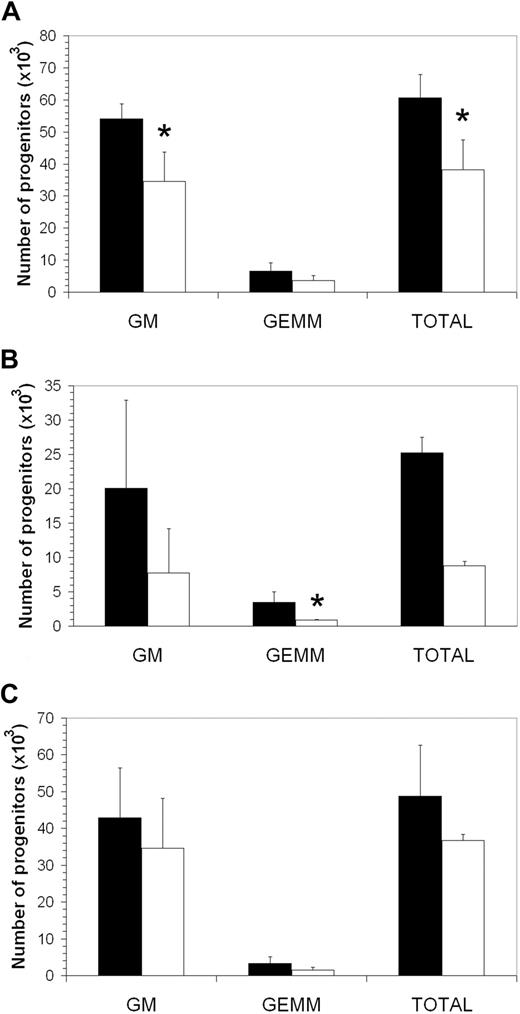
The Asxl1tm1Bc mutation had modest and variable effects on myelopoiesis. Consistent with increased splenic cellularity of a subset of adult mice (Figure 6A), a significant increase in Asxl1tm1Bc adult splenocytes expressing Gr-1 and Mac-1 was observed (Figure 6B-C). Sections of adult Asxl1tm1Bc showed normal morphology with prominent extramedullary hematopoiesis present within the red pulp regions of the enlarged Asxl1 mutant spleens (not shown). Cellularity and histology of P1 spleens was normal (not shown). We conducted in vitro colony assays of E18.5 fetal liver, P1 spleen, and adult bone marrow cells, grown in methylcellulose and supplemented with growth factors that support CFU-C myeloerythroid lineage multipotential and unipotential progenitor differentiation (Figure 7). Overall, there is a slight reduction in myeloerythroid colony-forming ability across the colony types scored: granulocyte-macrophage (GM), and granulocyte-erythrocyte-macrophage-megakaryocyte (GEMM). Statistically significant reductions in colony number were seen only for E18.5 fetal liver GM and total CFU-C myeloid, and newborn spleen GEMM (P < .05). No significant changes in the percentages of myeloid cells were observed in adult peripheral blood (supplemental Figure 2) or bone marrow (not shown).
Asxl1tm1Bc/tm1Bc mutant mice exhibit splenomegaly and increase in myeloid cell number. (A) Cellularity of Asxl1tm1Bc/tm1Bc (mut) adult spleens is increased compared with wild type (wt; n = 14 wt, 12 mutant). (B) In adult Asxl1tm1Bc/tm1Bc mice, the absolute numbers of myeloid (Gr1 or Mac1 positive) and erythroid (Ter119 positive) cells are increased in spleen (Gr1+ n = 13 wt, 11 mutant; Mac1+ n = 11; Ter119+ n = 10 wt, 7 mutant). (C) The proportion of cells bearing the myeloid lineage markers Gr1 (i) and Mac (ii) is increased in adult spleen of Asxl1tm1Bc/tm1Bc mutants.
Asxl1tm1Bc/tm1Bc mutant mice exhibit splenomegaly and increase in myeloid cell number. (A) Cellularity of Asxl1tm1Bc/tm1Bc (mut) adult spleens is increased compared with wild type (wt; n = 14 wt, 12 mutant). (B) In adult Asxl1tm1Bc/tm1Bc mice, the absolute numbers of myeloid (Gr1 or Mac1 positive) and erythroid (Ter119 positive) cells are increased in spleen (Gr1+ n = 13 wt, 11 mutant; Mac1+ n = 11; Ter119+ n = 10 wt, 7 mutant). (C) The proportion of cells bearing the myeloid lineage markers Gr1 (i) and Mac (ii) is increased in adult spleen of Asxl1tm1Bc/tm1Bc mutants.
Asxl1tm1Bc/tm1Bc mutant mice have fewer committed myeloerythroid progenitors. In vitro colony formation of committed myeloerythroid progenitors (CFU-GEMM assay) is reduced in Asxl1tm1Bc/tm1Bc mice compared with wild type in cultures of (A) E18.5 fetal liver (n = 2 wt, 3 mutant), (B) newborn spleen (n = 6 wt, 5 mutant), and (C) adult bone marrow (n = 9 wt, 6 mutant) *P < .05. ■ indicates wild type; □, Asxl1tm1Bc/tm1Bc mutant.
Asxl1tm1Bc/tm1Bc mutant mice have fewer committed myeloerythroid progenitors. In vitro colony formation of committed myeloerythroid progenitors (CFU-GEMM assay) is reduced in Asxl1tm1Bc/tm1Bc mice compared with wild type in cultures of (A) E18.5 fetal liver (n = 2 wt, 3 mutant), (B) newborn spleen (n = 6 wt, 5 mutant), and (C) adult bone marrow (n = 9 wt, 6 mutant) *P < .05. ■ indicates wild type; □, Asxl1tm1Bc/tm1Bc mutant.
Effects of Asxl1tm1Bc mutation on progenitor and hematopoietic stem cell activity
Because we observed Asxl1 expression in cell fractions enriched for hematopoietic progenitors and HSC in fetal liver and adult bone marrow (Figure 2), we assessed the functional requirement for Asxl1 in these cells using the day-12 spleen colony-forming unit (CFU-S12) assay.21 In 2 independent experiments, we found no differences in CFU-S12 colony numbers between Asxl1 mutant and control samples within either experiment (data not shown).
To assess whether the Asxl1tm1Bc mutation affected function of long-term repopulating HSCs, we carried out a standard competitive repopulation unit (CRU) assay in which various numbers of either wild-type or homozygous Asxl1tm1Bc/tm1Bc;Ly5.2+ cells derived from E17 fetal liver were injected into Ly5.1+ congenic irradiated hosts in the presence of helper Ly5.2+ wild-type cells. We observed no significant difference in CRU frequency, based on FACS immunophenotyping of peripheral blood, between recipients of Asxl1tm1Bc mutant and Asxl1+/+ cells in this assay, at 20 weeks after transplant (data not shown).
Discussion
We generated a knockout mouse model of the ETP group gene Asxl1 to assess the effect of loss of function of Asxl1 in hematopoiesis. The Asxl1tm1Bc mutation had consistent and profound effects on lymphopoiesis. The data suggest that the T-lymphopoiesis defect in the Asxl1tm1Bc mutants lies in the DN1 to DN4 precursor stage.18 Asxl1tm1Bc mice also exhibited striking defects in B lymphopoiesis at the progenitor level. The number of bone marrow pre-B progenitor cells detectable by the CFU–IL-7 in vitro assay is markedly lower in all samples from Asxl1tm1Bc/tm1Bc adult mice compared with wild-type controls, consistent with the lower absolute number of B220+ cells within the bone marrow of most adult Asxl1tm1Bc/tm1Bc mice. Of note, defective B-lymphoid development is seen in mice mutant for other PcG genes.19,22-27
Our data suggest that there is a progressive collapse of the lymphoid compartment in Asxl1tm1Bc mutant mice over time, because both T- and B-lymphoid defects were observed in mutant mice greater than 15 weeks of age but not in younger mice. However, as the experiments we conducted were not specifically designed to evaluate the timing of phenotypic changes, we cannot adequately address onset and progression of the defects in lymphopoiesis from our current study. This should be carefully assessed in future studies, which would ideally be conducted using a conditional mouse mutant model to specifically ablate Asxl1 in hematopoietic cells, and would also avoid problems associated with partial perinatal lethality of the constitutive Asxl1 null mice.
It is interesting to note that the reduced thymopoiesis observed in loss-of-function Hoxa9 mouse mutants ameliorates with age28 in contrast to the increase in severity of thymic defects observed in Asxl1 mutants. We report elsewhere6 that Asxl1 mutations cause misexpression of Hox genes in mesodermal derivatives during embryonic development. It may be that in the absence of Asxl1, aberrant expression of Hoxa9, or other Hox genes that have a known role in regulation of hematopoiesis, leads to the thymic defects seen in Asxl1 mutants.29 However not all hematopoietic defects seen in PcG mutants are a consequence of Hox gene misregulation, as evidence from Rae28 and eed mutant mice indicates that defects in fetal liver and adult hematopoiesis, respectively, are not mediated via Hox targets.30,31 Conversely, certain Hox genes do play a critical role in leukemogenesis involving the trithorax gene Mll, as Hoxa7 and Hoxa9 are required for myeloid transformation mediated by Mll chimeric fusion proteins,32 and are also likely regulated by Mll during normal hematopoiesis because down-regulation of Hoxa7, Hoxa9, and Hoxa10 was observed in E12.5 fetal liver of Mll homozygous mutant mice.33
Myeloerythroid lineage defects in Asxl1tm1Bc mutants are mild. This is consistent with observations that other PcG and trxG genes exhibit variable effects on myeloerythropoiesis and erythropoiesis.22,24,26,34-41 Bmi1, Phc1, and Mll1 mutations that affect myeloerythropoiesis may do so indirectly by negatively affecting HSC activity, resulting in reduced differentiation along all hematopoietic lineages,20,25,42-44 but this possibility is unlikely for Asxl1 as we did not observe effects on HSCs or multipotent progenitors.
Despite the observation of changes in cell frequency in liver, spleen, thymus, and bone marrow, mature hematopoietic cell types exhibiting normal morphology and number were observed within the peripheral blood of most adult Asxl1tm1Bc/tm1Bc mice. These results suggest that in the absence of Asxl1, homeostatic compensatory mechanisms normalize myeloid, erythroid, and lymphoid cells despite disturbances in progenitor cells, consistent with previous studies of Polycomb and trithorax group mutants.26,27,34-36,40
Many Polycomb and trithorax group mutants affect self-renewal of HSCs.34-37,42-45 However, Asxl1tm1Bc fetal liver reconstituted all blood lineages in irradiated adult mice up to 20 weeks in a competitive repopulation assay and the Asxl1tm1Bc mutation does not alter colony numbers in the CFU-S12 assay.21 Therefore Asxl1 is not required for HSC or multipotent progenitor activity. However, the in vitro clonogenic progenitor assays show a modest reduction in CFU-GEMM activity in Asxl1 mutants, indicating a possible role in common myeloid progenitor regulation. Serial transplant assays with Asxl1 mutant cells may reveal more progressive defects at the HSC level such as were seen in studies of the Polycomb group gene Phc1tm1Os mutant mice.20
The failure to observe lymphomas or leukemias in Asxl1tm1Bc/tm1Bcmutant mice monitored for up to 58 weeks suggests that loss of Asxl1 alone is insufficient to cause oncogenic transformation. In light of a recent study characterizing mutations in the human ASXL1 gene in bone marrow samples from patients with MDS and CMML,12 it is perhaps surprising that our investigations of loss-of-function Asxl1 mutant mice did not reveal more profound defects in HSC or progenitor function with respect to myelopoiesis, and that the mice did not develop myelodysplastic phenotypes other than splenomegaly. One explanation is that Asxl1, Asxl2, and perhaps Asxl3 have redundant functions in hematopoiesis. This idea is supported by observations that deletions of Asxl212 and an ASXL2-MOZ fusion (GenBank accession number AB08428146 ) are also associated with MDS. Furthermore, the chromosomal region at 2p23, the site of ASXL2, is amplified in a variety of tumor types, including B-cell leukemia and lymphomas.47
Another potential explanation for the mild phenotypes of loss-of-function Asxl1 mutations stems from the observation that mutations found in the MDS and CMML samples were all found in exon 12 and spanned the region from Arg596 to Ser1457 of the corresponding 1541 amino acid ASXL1 protein (Figure 1).12 These mutations would remove the C-terminal PHD domain but leave the N-terminal ASXH region containing 2 putative NR box motifs, and all putative nuclear localization signal sequences intact.7,8 These ASXL1 truncations may function as dominant negative mutations, leading to a more severe phenotype compared with the null mutation described here, which removes both the ASXH and the PHD domains. Loss-of-function ASXL1 mutations have not yet been associated with cancer to our knowledge. Interestingly, oncogenic effects of ASXL1 mutations are associated with gain-of-function ASXL1 mutations, consistent with overexpression of ASXL1 in cancer cell lines,7 with amplification of region 20q11.21 that contains the ASXL1 locus in a wide range of human tumors,48-51 and with the PAX5-ASXL1 fusions detected previously in acute lymphoblastic leukemia patients.11
ASXL1 functions as a nuclear receptor coregulator in human cancer cell lines.10 In transfected HeLa cells stably expressing a FLAG-ASXL1 construct, there was a striking increase in RA-dependent RAR-induced transcription, whereas in cells expressing a truncated form of ASXL1 lacking the PHD domain, there was only a slight enhancement of RAR activity, suggesting that the PHD domain mediates nuclear receptor activity.10 RA is a well-established therapeutic agent for acute promyelocytic leukemia.52 Perhaps aberrant RA receptor signaling in cells with truncation of the PHD domain of ASXL1 may contribute to development of dysplasia if RA cytotoxicity is enhanced in cells overexpressing ASXL1 truncations.10
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rewa Grewal, Carolyn Bateman, Pol Gomez, and Gosta Berg for technical assistance.
This research was supported by grants from the Canadian Institutes of Health Research to H.W.B., from the Terry Fox Foundation to R.K.H., and from the National Institutes of Health (RO1-CA-0078815) and a SCOR grant from the Leukemia & Lymphoma Society of America to J.L.H. C.L.F. was supported by a Medical Research Council of Canada studentship. C.D.H. was supported by a Canadian Institutes of Health Research New Investigator award, and is a Michael Smith Foundation for Health Research scholar.
National Institutes of Health
Authorship
Contribution: C.L.F., N.P., C.D.H., H.O., R.K.H., and H.W.B. designed experiments; N.P. analyzed Asxl1 expression; C.L.F. constructed the Asxl1 targeting vector; C.D.H. and C. Bodner performed embryonic stem cell tissue culture; C.L.F. performed genotyping assays; C.L.F. with assistance from C.D.H. and C. Bodner performed FACS immunophenotyping, in vitro functional assays, and general hematopoietic analysis in Asxl1 mutants; H.O. performed the thymic organ culture experiments; C. Brookes carried out the competitive repopulation assays; J.L.H. carried out pathologic examinations; C.L.F., R.K.H., and H.W.B. wrote the manuscript; and C.D.H., N.P., H.O., and J.L.H. suggested improvements to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hugh W. Brock, Department of Zoology, University of British Columbia, 2350 Health Sciences Mall, Vancouver, V6T 1Z3, BC Canada; e-mail: brock@zoology.ubc.ca.

![Figure 2. Asxl1 is ubiquitously expressed in hematopoietic cells. Southern blot analysis of Asxl1, Bmi1, and actin gene expression in total amplified cDNAs from FACS-sorted hematopoietic cell populations from fetal liver and adult bone marrow, spleen, and thymus. Total cDNAs were amplified using RT-PCR from 104 sorted cells, and each sample with or without reverse transcriptase (RT + or −) was sequentially hybridized to each probe. Results for a representative experiment are shown; duplicate experiments were performed. (A) Asxl1 is expressed in unfractionated E14.5 fetal liver (TFL), and the hematopoietic stem cell–depleted (Sca-1−Lin+ [S−L+]), low (Sca-1+Lin+ [S+L+]), and enriched (Sca-1+Lin− [S+L−]) subpopulations, whereas Bmi1 expression is predominant in the 2 Sca-1+ fractions. (B) Asxl1 is expressed in unfractionated adult bone marrow (TBM) and in all fractionated adult bone marrow, spleen, and thymus cells investigated, whereas Bmi1 is expressed predominantly within the progenitor-enriched Sca-1+ fractions and the Gr-1 low fraction of total bone marrow, with very low or undetectable expression in other fractions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/1/10.1182_blood-2009-07-230698/4/m_zh89990946250002.jpeg?Expires=1769924501&Signature=b7bas3u21kbqQ7TH28XyUeYXGthxdoripfGy3xdBAWK7na2X5lY7TlDQTRYpFAfFmJ32PbwAIETVnWjeSliSnAR9qmroQECD295gGuRG-C2TKXsTD3jFZnxPyQkZNQ7THcZD2ikZ8qRKbxThozWiWVvO57HEO1vr298FLC5Qy~iOb3XsNWo~LbD76eMbQKcnX8C13AWChJq~KP58uCmU0SG-tcD~I8P3q-7xxMN5QNuFYgqmvq~YjH7c2DbXMa2lZFJyQNUpQyOaqc7O9XzOVJ4m1CObUQ7w8yTow5Xb2KQ4EbDXv6qAA-459bO94zzQIs7fDv08f0cqcu~K0xqGhw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)