Abstract
The polycomb repressive complex (PRC) 2 contains 3 core proteins, EZH2, SUZ12, and EED, in which the SET (suppressor of variegation–enhancer of zeste-trithorax) domain of EZH2 mediates the histone methyltransferase activity. This induces trimethylation of lysine 27 on histone H3, regulates the expression of HOX genes, and promotes proliferation and aggressiveness of neoplastic cells. In this study, we demonstrate that treatment with the S-adenosylhomocysteine hydrolase inhibitor 3-deazaneplanocin A (DZNep) depletes EZH2 levels, and inhibits trimethylation of lysine 27 on histone H3 in the cultured human acute myeloid leukemia (AML) HL-60 and OCI-AML3 cells and in primary AML cells. DZNep treatment induced p16, p21, p27, and FBXO32 while depleting cyclin E and HOXA9 levels. Similar findings were observed after treatment with small interfering RNA to EZH2. In addition, DZNep treatment induced apoptosis in cultured and primary AML cells. Furthermore, compared with treatment with each agent alone, cotreatment with DZNep and the pan-histone deacetylase inhibitor panobinostat caused more depletion of EZH2, induced more apoptosis of AML, but not normal CD34+ bone marrow progenitor cells, and significantly improved survival of nonobese diabetic/severe combined immunodeficiency mice with HL-60 leukemia. These findings indicate that the combination of DZNep and panobinostat is effective and relatively selective epigenetic therapy against AML cells.
Introduction
Deregulated epigenome, especially alterations in methylation of DNA and histone proteins, coupled to genetic mutations and silencing of tumor suppressor genes, are critical to the development and sustaining the biology of transformed cells, including acute leukemia cells.1,2 This has motivated the use of novel agents that target deregulated epigenetic mechanisms in acute myeloid leukemia (AML).3 Lysine-specific histone deacetylation, H3 lysine (K) 27 trimethylation (3Me), and DNA methylation are the important mechanisms involved in the epigenetic silencing of genes, including tumor suppressor genes (TSGs) such as p16.4,5 Polycomb group proteins are multiprotein complexes that epigenetically silence gene expression, including TSGs.5-7 EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2) that also includes SUZ12, EED, and YY1. EZH2 acts as a histone lysine methyltransferase (KMTase), which mediates 3Me of K27 on H3 to silence expression of PRC2 target genes involved in lineage differentiation.8,9 EZH2 has been shown to be abundantly expressed in purified hematopoietic stem cells (HSCs), where it preserves HSC potential and prevents HSC exhaustion.10 EZH2 regulates cell proliferation by promoting S-phase entry and G2-M transition, and it is highly expressed in tumor versus normal tissues.11-13 EZH2-mediated cell-cycle progression promoted by gene repression also involves histone deacetylation by histone deacetylase-1 (HDAC-1), with which EZH2 interacts through its PRC2-binding partner EED.14-17 EZH2 is overexpressed in a variety of malignancies, including prostate, breast, and bladder cancers, and hematologic malignancies with poor prognosis.11-13,18-20 Knockdown of EZH2 by small interfering RNA (siRNA) has been demonstrated to inhibit breast cancer cell proliferation, whereas pharmacologic inhibition of EZH2 resulted in apoptosis of breast cancer, but not normal cells.21 EZH2 was shown to directly interact with and regulate the activity of the DNA methyltransferases (DNMTs) DNMT1, DNMT3a, and DNMT3b.22,23 DNMTs function to transfer a methyl group from S-adenosyl-methionine to the 5′ position of cytosine in the CpG dinucleotides in the promoters of genes, thereby maintaining a consistent pattern of epigenetic gene silencing of TSGs in cancer cells.24,25 DNA methylation by DNMTs also recruits HDAC activity to the promoters of silenced genes. Similar to the PRC2 complex, DNMT1 has a direct interaction with histone deacetylases HDAC1 and HDAC2.26,27 Although genes methylated in cancer cells are packaged with nucleosomes containing the 3Me H3K27 mark, genes silenced in cancer by 3Me H3K27 have been shown to be independent of promoter DNA methylation, thus highlighting that 3Me H3K27 could potentially be an independent mechanism for silencing TSGs.28-30 Consistent with this, DNA methylation and transcriptional silencing of cancer genes have been shown to persist despite the depletion of EZH2, suggesting that simultaneously inhibiting DNMT1 and EZH2 would be more effective in reversing 3Me H3K27 and DNA methylation.29,31 We had previously reported that treatment with the pan-HDAC inhibitor (HDI) panobinostat (PS, also known as LBH589; Novartis Pharmaceutical) depletes the levels of EZH2, SUZ12, and EED with concomitant depletion of 3Me H3K27 in cultured and primary AML cells.19 Within the PRC2 complex, EZH2 bound and recruited the DNMT1. PS treatment disrupted the binding between DNMT1 and EZH2, and attenuated DNMT1 levels and its binding to the EZH2-targeted gene promoters, p16 and JunB.31 In addition, treatment with PS has also been shown to deplete the leukemia-associated oncoproteins accompanied by growth arrest and apoptosis of leukemia cells.32
3-Deazaneplanocin A (DZNep) is the cyclopentanyl analog of 3-deazaadenosine that inhibits the activity of S-adenosyl-l-homocysteine (AdoHcy) hydrolase, the enzyme responsible for the reversible hydrolysis of AdoHcy to adenosine and homocysteine.33,34 This results in the intracellular accumulation of AdoHcy, which leads to inhibition of the S-adenosyl-l-methionine–dependent KMTase activity. DZNep has also been shown to inhibit the activity of the methyltransferases, resulting in undermethylation of mRNAs.35 DZNep has been reported to inhibit the 7-methyl-guanosine–capping structure of mature mRNA in vitro and in vivo.34 Recently, DZNep was reported to deplete the expression levels of the PRC2 complex in breast cancer cells with concomitant loss of 3Me H3K27 mark and derepression of epigenetically silenced targets.21,35 Combined treatment with DZNep and the HDAC inhibitor, trichostatin A, was shown to derepress PRC2-targeted genes, for example, the F-box protein FBXO32, a component of the stem cell factor ubiquitin protein E3 ligase complex. This was shown to induce apoptosis of breast and colorectal, but not normal cells, suggesting that EZH2 antagonists and HDAC inhibitors would be an effective combination epigenetic therapy.21,35 However, the antileukemia apoptotic activity of DZNep alone or in combination with PS had not been previously determined. In this study, we show that treatment with DZNep depletes PRC2 complex proteins and induces apoptosis of cultured and primary AML cells. In addition, our findings show that the combined treatment of DZNep and PS causes more depletion of PRC2 complex proteins and induces synergistic apoptosis of cultured and primary AML cells.
Methods
Reagents
PS was kindly provided by Novartis Pharmaceuticals. DZNep was acquired from the National Cancer Institute. Entinostat (SNDX-275) was kindly provided by Syndax Pharmaceuticals. Anti-EZH2, anti-SUZ12, anti-EED, anti-p21, anti-acetylated K27 histone H3, monoclonal anti-trimethylated K27 histone H3 antibody, anti-HOXA9, and anti–β-actin antibodies were obtained, as previously described.19 Monoclonal anti-p27 was purchased from BD Transduction Laboratories. Polyclonal antitrimethylated K9 histone H3, polyclonal antitrimethylated K79 histone H3, polyclonal trimethylated K4 histone H3, and polyclonal were purchased from Millipore. Monoclonal anti-DNMT1 antibody was obtained from Abcam. Monoclonal anti-FBXO32 and polyclonal anti-CCAAT/enhancer–binding protein α (CEBPα) were acquired from Santa Cruz Biotechnology. Monoclonal anti-cyclin E, anti-poly(adenosine 5′-diphosphate [ADP]-ribose) polymerase (PARP), and anti-histone H3 antibody were purchased from Cell Signaling Technology. Monoclonal fluorescein isothiocyanate–conjugated anti-CD11b and fluorescein isothiocyanate–conjugated isotype control antibody were obtained from BD Biosciences.
Cell lines and cell culture
AML HL-60 cells were obtained and maintained, as previously described.19 OCI-AML-3 cells were cultured in α minimum essential medium with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% nonessential amino acids. Logarithmically growing cells were exposed to the designated concentrations of DZNep and/or PS. After these treatments, cells or cell pellets were washed free of the drug(s) before the performance of the studies.
Primary AML blasts
Primary AML samples and normal CD34+ mononuclear cells were obtained with informed consent in accordance with the Declaration of Helsinki as part of a clinical protocol approved by the Institutional Review Board of the Medical College of Georgia. Peripheral blood or bone marrow aspirate samples were collected and separated for mononuclear cells, as previously described.19,31,32 Banked, delinked, and deidentified donor peripheral blood CD34+ mononuclear cells that were procured, but not used for engraftment in the recipients, were purified by immunomagnetic beads conjugated with anti-CD34 antibody before utilization in the cell viability assay and immunoblot analysis (StemCell Technologies). Cells were determined to be greater than 80% CD34+ by immunostaining with a PE-conjugated CD34 antibody (StemCell Technologies), followed by flow cytometry. To isolate CD34+/CD38−/Lin− HSCs from primary AML bone marrow aspirate, a Human Primitive Hematopoietic Progenitor Cell Enrichment Kit (StemCell Technologies) was used following the manufacturer's protocol.
Cell-cycle analysis
After the designated treatments, cells were harvested and washed twice with 1× phosphate-buffered saline (PBS) and fixed in ethanol overnight. Fixed cells were washed twice with 1× PBS and stained with propidium iodide for 15 minutes at 37°C. Cell-cycle data were collected on a flow cytometer with a 488 nM laser and analyzed with ModFit 3.0 (Verity Software House), as previously described.36
Assessment of apoptosis of AML cells
Untreated or drug-treated cells were stained with annexin V (BD Pharmingen) and propidium iodide, and the percentage of apoptotic cells was determined by flow cytometry. To analyze synergism between DZNep and PS in inducing apoptosis, cells were treated with DZNep (100-750 nmol/L) and PS (5-20 nmol/L) at a constant ratio for 48 hours. The percentage of apoptotic cells was determined by flow cytometry, as previously described.36 The combination index (CI) for each drug combination was obtained by median dose effect of Chou and Talalay,37 using the CI equation within the commercially available software Calcusyn (Biosoft). CI values of less than 1.0 represent synergism of the 2 drugs in the combination.
Colony culture assay
After the designated treatments, cells were harvested and washed twice with 1× PBS, and approximately 500 cells were plated in complete Methocult (StemCell Technologies) and cultured for 7 to 10 days at 37°C in a 5% CO2 environment. Colony growth was measured as a percentage of the control cell colony growth, as previously described.38
Leukemia cell differentiation
Assessment of percentage of nonviable cells
After designated treatments, cells were stained with trypan blue (Sigma-Aldrich). The numbers of nonviable cells were determined by counting the cells that showed trypan blue uptake in a hemocytometer, and reported as percentage of untreated control cells.
RNA interference and nucleofection
For siRNA-mediated down-regulation of EZH2, an EZH2-specific Smart Pool (M-004218-01) was purchased from Dharmacon. Nonspecific control siRNA (no. 4611) was purchased from Ambion. All siRNA experiments were performed at a final concentration of 100 nM duplex siRNA. Nucleofection of siRNAs into OCI-AML3 and HL-60 cells was performed, as previously described.19
RNA isolation and reverse transcription–polymerase chain reaction
RNA was extracted from the cultured cells using an RNaqueous-4PCR (Applied Biosystems). Purified RNA was quantitated and reverse transcribed using Superscript II, according to the manufacturer's protocol (Invitrogen). Resulting cDNAs were used in subsequent polymerase chain reactions (PCR) for EZH2, SUZ12, EED, FBXO32, cyclin E, p16, p21, and p27. PCRs for β-actin were used as an internal loading control for the PCRs. Amplified products were resolved on a 2% agarose gel and recorded with an ultraviolet transilluminator. Horizontal scanning densitometry was performed with ImageQuant 5.2, and band intensity was compared with β-actin. Quantitative real-time PCR analysis for EZH2, SUZ12, and EED was performed on cDNA using TaqMan probes from Applied Biosystems. A TaqMan probe for glyceraldehyde-3-phosphate dehydrogenase was used to normalize relative expression of the mRNA.
Detection and analysis of hsa-miR-101 in AML cells
For detection of hsa-miR-101 in AML HL-60 and OCI-AML3 cells,39 microRNAs were isolated with a kit from Applied Biosystems. Total RNA was reverse transcribed with a stem loop primer included in the TaqMan hsa-miR-101 microRNA assay following the manufacturer's protocol (Applied Biosystems). Expression of hsa-miR-101 was detected by quantitative PCR (qPCR) with a TaqMan probe specific to hsa-miR-101. Relative expression of hsa-miR-101 was normalized against expression of 18S RNA.
Cell lysis and protein quantitation
Untreated or drug-treated cells were centrifuged, and the cell pellets were resuspended in 200 μL lysis buffer, as previously described.38
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting
One hundred micrograms of total cell lysate was used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Western blot analyses of DNMT1, EZH2, SUZ12, EED, BMI1, 3MeK27H3, 2MeK27H3, acetyl K27H3, PARP, FBXO32, cyclin E, p21, p27, and HOXA9 were performed on total cell lysates using specific antisera or monoclonal antibodies, as previously described.38 The expression level of either β-actin or histone H3 was used as the loading control for the Western blots. Blots were developed with a chemiluminescent substrate enhanced chemiluminescence (Amersham Biosciences). Immunoblot analyses were performed at least twice, and representative blots were subjected to densitometric analysis. Densitometry was performed using ImageQuant 5.2 (GE Healthcare).
Chromatin immunoprecipitation and polymerase chain reaction
OCI-AML3 and HL-60 cells were treated with DZNep for 24 hours. After drug exposure, the chromatin in the cells was cross-linked with formaldehyde for 10 minutes at 37°C. The cross-linking reaction was quenched with 1/20 volume of 2.5 M glycine for 5 minutes at room temperature, and then the cells were washed twice for 5 minutes in ice-cold 1× PBS. Cell lysis, sonication, and chromatin immunoprecipitation for EZH2 were performed, according to the manufacturer's protocol (Upstate Biotechnology). For quantitative assessment of HOXA9, CDH1, and WNT1 in the chromatin immunoprecipitates, a SYBR Green Mastermix from Applied Biosystems was used. Relative enrichment in the chromatin immunoprecipitates was normalized against HOXA9, CDH1, and WNT1 in the input samples.
In vivo model of AML
HL-60 cells (5 million) were injected into the tail vein of female nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice, and the mice were monitored for 7 days. The following treatments were administered in cohorts of 7 mice for each treatment: vehicle alone, 1 mg/kg DZNep, 10 mg/kg PS, and DZNep plus PS. Treatments were initiated on day 7. DZNep was administered twice per week (Tuesday-Thursday) intraperitoneally for 2 weeks, and then discontinued. PS was administered 3 days per week (Monday, Wednesday, and Friday) for 4 weeks. The survival of mice from the tail vein model is represented with a Kaplan-Meier survival plot. Because this combination had not previously been studied, we selected a dose of DZNep that had been determined to be safe in previously reported studies and combined it with a dose of PS that we had previously reported to be safe and biologically effective.34,40 NOD/SCID mice, human AML HL-60 cell xenograft studies were sanctioned by protocol approved by the Medical College of Georgia Institutional Animal Care and Use Committee.
Statistical analysis
Significant differences between values obtained in a population of leukemic cells treated with different experimental conditions were determined using the Student t test. P values of less than .05 were assigned significance.
Results
DZNep treatment induces cell-cycle arrest and apoptosis, and markedly reduces clonogenic survival of AML cells
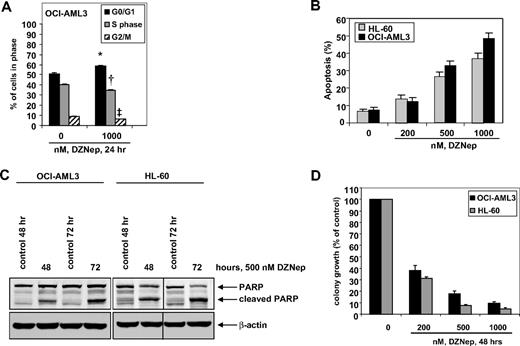
We first determined the effects of DZNep treatment on the cell cycle of OCI-AML3 and HL-60 cells. As demonstrated in Figure 1, treatment of OCI-AML3 cells with DZNep (1.0 μM) resulted in a significant increase in accumulation of cells in the G0/G1 phase (58.5%) with a concomitant decrease in the number of cells in S phase (35.2%) and G2/M phases (6.3%) of the cell cycle (P < .05). A lesser effect was seen with lower concentrations of DZNep (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). We next determined the effects of DZNep treatment on induction of apoptosis in OCI-AML3 and HL-60 cells. Exposure to DZNep dose dependently induced apoptosis to a greater extent in OCI-AML3 than HL-60 cells (Figure 1B). As shown in Figure 1C, DZNep treatment also induced caspase-dependent cleavage of PARP more in HL-60 than in OCI-AML3 cells (Figure 1C). In addition, treatment with DZNep (200 nM to 2.0 μM) for 48 hours, dose dependently, inhibited colony growth of OCI-AML3 and HL-60 cells (Figure 1D). We recently created and described the HDI-resistant human AML HL-60 (HL-60/LR) cells that are resistant to PS and HDIs.38 DZNep dose dependently induced apoptosis of HL-60/LR cells (supplemental Figure 1B).
Treatment with DZNep induces apoptosis in a dose- and time-dependent manner and markedly reduces clonogenic survival of AML cells. (A) OCI-AML3 cells were treated with the indicated concentrations of DZNep for 24 hours, then fixed and stained with propidium iodide, and cell-cycle status was determined by flow cytometry. *G0/G1 values significantly different from untreated cells; †S-phase values significantly different from untreated cells; ‡G2/M values significantly different from untreated cells. (B) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of DZNep for 72 hours. Then, the cells were stained with annexin V, and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent SEM. (C) OCI-AML3 and HL-60 cells were left untreated, or treated with 500 nmol/L DZNep, for 48 and 72 hours. After this, total cell lysates were prepared and immunoblot analysis was performed for PARP. The expression levels of β-actin in the lysates served as the loading control. A vertical line has been inserted to indicate a repositioned gel lane. (D) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 48 hours. After treatment, colony growth in semisolid media was assessed after 7 days. Bar graphs represent the mean percentage values ± SEM of untreated colony growth.
Treatment with DZNep induces apoptosis in a dose- and time-dependent manner and markedly reduces clonogenic survival of AML cells. (A) OCI-AML3 cells were treated with the indicated concentrations of DZNep for 24 hours, then fixed and stained with propidium iodide, and cell-cycle status was determined by flow cytometry. *G0/G1 values significantly different from untreated cells; †S-phase values significantly different from untreated cells; ‡G2/M values significantly different from untreated cells. (B) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of DZNep for 72 hours. Then, the cells were stained with annexin V, and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent SEM. (C) OCI-AML3 and HL-60 cells were left untreated, or treated with 500 nmol/L DZNep, for 48 and 72 hours. After this, total cell lysates were prepared and immunoblot analysis was performed for PARP. The expression levels of β-actin in the lysates served as the loading control. A vertical line has been inserted to indicate a repositioned gel lane. (D) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 48 hours. After treatment, colony growth in semisolid media was assessed after 7 days. Bar graphs represent the mean percentage values ± SEM of untreated colony growth.
Treatment with DZNep depletes EZH2 and SUZ12 protein levels in cultured and primary AML cells
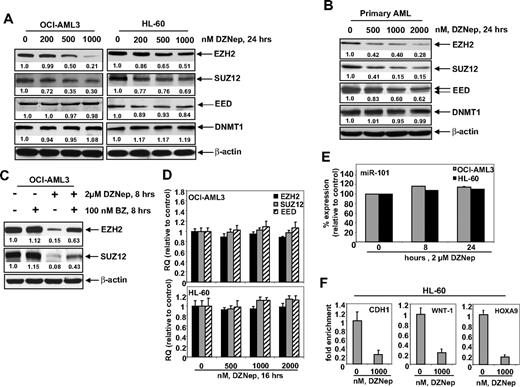
Treatment of DZNep has been demonstrated to deplete the expression levels of EZH2 and SUZ12 in breast and colon cancer cells. Consistent with these reports, treatment with DZNep dose dependently reduced the protein expression of EZH2 and SUZ12 in OCI-AML3 and HL-60 cells, as well as in a sample of primary AML blasts (Figure 2A-B). After exposure to DZNep, EED levels declined in the primary AML cells (Figure 2B), but only a slight decline in EED protein levels was observed in HL-60, and none was observed in OCI-AML3 cells (Figure 2A). DZNep treatment failed to produce any significant effect on the mRNA expression of EZH2, SUZ12, or EED either in HL-60 and OCI-AML3, as determined by qPCR (Figure 2D) or in primary AML cells (data not shown). However, cotreatment with the proteasome inhibitor bortezomib (BZ) significantly, but not completely, restored the DZNep mediated decline in the protein levels of EZH2 and SUZ12 (Figure 2C), but did not abrogate the antileukemic effect of DZNep (supplemental Figure 1C). Recently, the expression of EZH2 was shown to be inhibited by microRNA-101 in cancer cells.39 However, treatment with DZNep did not significantly alter the expression of miR-101 in HL-60 and OCI-AML3 cells (Figure 2E). DZNep treatment also diminished EZH2 binding to target gene promoters of HOXA9, CDH1, and WNT1 in HL-60 cells (Figure 2F) and CDH1 and WNT1 in OCI-AML3 (supplemental Figure 2).
Treatment with DZNep depletes expression of polycomb group proteins EZH2, SUZ12, and EED in cultured and primary AML cells. (A) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. After this, total cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, and DNMT1. The expression levels of β-actin in the lysates served as the loading control. (B) Primary AML cells were treated with the indicated concentrations of DZNep for 24 hours. At the end of treatment, cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, and DNMT1. The expression levels of β-actin in the lysates served as the loading control. (C) OCI-AML3 cells were treated with BZ and DZNep as indicated for 8 hours. After treatment, cell lysates were prepared and immunoblot analysis was performed for EZH2 and SUZ12. The expression levels of β-actin in the lysates served as the loading control. (D) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 16 hours. Then, total RNA was isolated and quantitative real-time PCR was performed with TaqMan probes for EZH2, SUZ12, and EED. The relative quantity (RQ) of each mRNA was normalized against glyceraldehyde-3-phosphate dehydrogenase expression. (E) HL-60 and OCI-AML3 cells were treated with 2 μM DZNep for 8 and 24 hours. Total RNA was isolated and reverse transcribed with a stem loop primer for hsa-miR-101. After reverse transcription, qPCR for hsa-miR-101 was performed, and expression of hsa-miR-101 was normalized against 18S RNA expression. (F) HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. Chromatin immunoprecipitation was performed with anti-EZH2 antibody. Immunoprecipitated DNA was used for qPCR of the WNT1, CHD1, and HOXA9 promoters. Fold enrichment data are relative to the control and are expressed as a ratio of the cycle threshold for the chromatin immunoprecipitation DNA versus the cycle threshold for the input samples.
Treatment with DZNep depletes expression of polycomb group proteins EZH2, SUZ12, and EED in cultured and primary AML cells. (A) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. After this, total cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, and DNMT1. The expression levels of β-actin in the lysates served as the loading control. (B) Primary AML cells were treated with the indicated concentrations of DZNep for 24 hours. At the end of treatment, cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, and DNMT1. The expression levels of β-actin in the lysates served as the loading control. (C) OCI-AML3 cells were treated with BZ and DZNep as indicated for 8 hours. After treatment, cell lysates were prepared and immunoblot analysis was performed for EZH2 and SUZ12. The expression levels of β-actin in the lysates served as the loading control. (D) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 16 hours. Then, total RNA was isolated and quantitative real-time PCR was performed with TaqMan probes for EZH2, SUZ12, and EED. The relative quantity (RQ) of each mRNA was normalized against glyceraldehyde-3-phosphate dehydrogenase expression. (E) HL-60 and OCI-AML3 cells were treated with 2 μM DZNep for 8 and 24 hours. Total RNA was isolated and reverse transcribed with a stem loop primer for hsa-miR-101. After reverse transcription, qPCR for hsa-miR-101 was performed, and expression of hsa-miR-101 was normalized against 18S RNA expression. (F) HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. Chromatin immunoprecipitation was performed with anti-EZH2 antibody. Immunoprecipitated DNA was used for qPCR of the WNT1, CHD1, and HOXA9 promoters. Fold enrichment data are relative to the control and are expressed as a ratio of the cycle threshold for the chromatin immunoprecipitation DNA versus the cycle threshold for the input samples.
DZNep treatment inhibits 3MeK27H3 levels associated with increase in pro-death FBOX32 and growth-inhibitory p16, p21, and p27 levels in AML cells
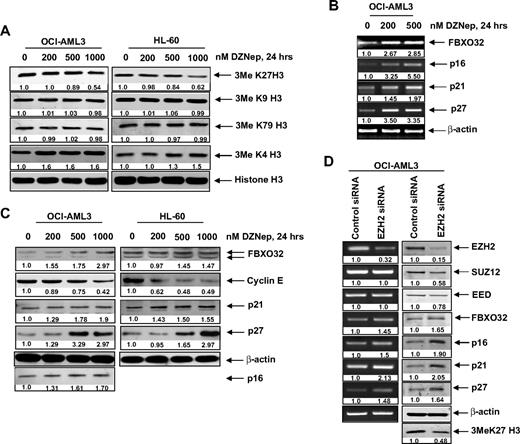
We next determined the effects of DZNep treatment on EZH2-mediated trimethylation of K27 on histone H3 (3MeK27H3), and on the other important histone H3 methylation marks. Treatment with DZNep was accompanied by an approximately 40% loss of levels of the repressive 3MeK27H3 mark in both OCI-AML3 as well as HL-60 cells (Figure 3A). In contrast, there was no significant effect on the 3MeK9H3 and 3MeK79H3 levels. Interestingly, DZNep treatment increased the levels of the permissive 3MeK4H3 mark (Figure 3A). DZNep-mediated reduction in the levels of EZH2 and 3MeK27H3 was associated with higher mRNA and protein levels of FBXO32 (Figure 3B), a gene previously shown to be repressed by EZH2-mediated 3MeK27H3.21 Concomitantly, treatment with DZNep reduced the levels of cyclin E, which is targeted by FBXO32. Furthermore, treatment with DZNep induced the mRNA and protein levels of the cell-cycle regulators p16, p21, and p27 (Figure 3B-C). However, in HL-60 cells, p16 levels were undetectable (Figure 3C). To determine whether the DZNep-mediated decline in EZH2 was mechanistically linked to up-regulation of the levels of p16, 21, p27, and FBXO32, we also determined the effect of siRNA to EZH2 in OCI-AML3 cells. Figure 3D (left panel) demonstrates that, compared with the control siRNA, treatment with siRNA to EZH2 caused decline in mRNA of EZH2, but not of SUZ12 and EED levels. This was associated with decline in protein levels of EZH2, SUZ12, and EED (Figure 3D right panel). Consequently, the disruption of the PRC2 complex caused by siRNA to EZH2 increased p16, p21, p27, and FBXO32 mRNA and protein levels in OCI-AML3 cells (Figure 3D). Reduced EZH2 levels also caused a decline in the levels of 3MeK27H3 in OCI-AML3 cells (Figure 3D).
DZNep treatment depletes trimethylation of K27 on histone H3 and induces the expression of cell-cycle regulatory genes p16, p21, and p27, as well as the cell death regulator FBXO32 in AML cells. (A) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. After this, nuclear extracts were prepared and immunoblot analysis was performed for 3MeK27 histone H3, 3MeK9 histone H3, 3MeK79 histone H3, and 3MeK4 histone H3. The expression levels of histone H3 in the extracts served as the loading control. (B) OCI-AML3 cells were treated with the indicated concentrations of DZNep for 24 hours. After treatment, total RNA was isolated and RT-PCR was performed for FBXO32, p16, p21, and p27. A β-actin–specific reaction and expression levels served to ensure equal loading. (C) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. After this, total cell lysates were prepared and immunoblot analysis was performed for FBXO32, cyclin E, p16, p21, and p27. The expression levels of β-actin in the lysates served as the loading control. (D) OCI-AML3 cells were transfected with scrambled control or EZH2 siRNA for 48 hours. Then, total RNA was isolated and RT-PCR was performed for EZH2, SUZ12, EED, FBXO32, p16, p21, and p27. A β-actin–specific reaction and expression levels served to ensure equal loading. Alternatively, total cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, FBXO32, p16, p21, p27, and 3MeK27 histone H3. The expression level of β-actin in the lysates served as the loading control.
DZNep treatment depletes trimethylation of K27 on histone H3 and induces the expression of cell-cycle regulatory genes p16, p21, and p27, as well as the cell death regulator FBXO32 in AML cells. (A) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. After this, nuclear extracts were prepared and immunoblot analysis was performed for 3MeK27 histone H3, 3MeK9 histone H3, 3MeK79 histone H3, and 3MeK4 histone H3. The expression levels of histone H3 in the extracts served as the loading control. (B) OCI-AML3 cells were treated with the indicated concentrations of DZNep for 24 hours. After treatment, total RNA was isolated and RT-PCR was performed for FBXO32, p16, p21, and p27. A β-actin–specific reaction and expression levels served to ensure equal loading. (C) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep for 24 hours. After this, total cell lysates were prepared and immunoblot analysis was performed for FBXO32, cyclin E, p16, p21, and p27. The expression levels of β-actin in the lysates served as the loading control. (D) OCI-AML3 cells were transfected with scrambled control or EZH2 siRNA for 48 hours. Then, total RNA was isolated and RT-PCR was performed for EZH2, SUZ12, EED, FBXO32, p16, p21, and p27. A β-actin–specific reaction and expression levels served to ensure equal loading. Alternatively, total cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, FBXO32, p16, p21, p27, and 3MeK27 histone H3. The expression level of β-actin in the lysates served as the loading control.
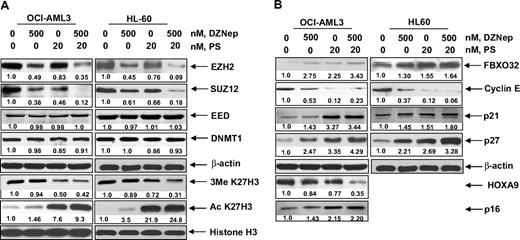
Cotreatment with DZNep and PS exerts superior anti-EZH2 effect, induces more p16, p21, and p27, as well as synergistically induces apoptosis of AML cells
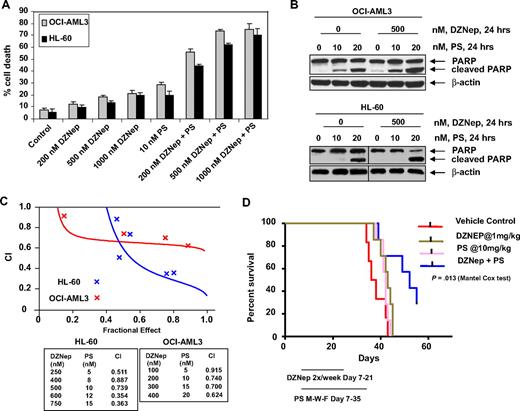
We next determined the combined effects of DZNep and PS in AML cells. Figure 4A demonstrates that, compared with each agent alone, combined treatment with DZNep (200 to 1000 nM) and PS induced more loss of cell viability, as shown by increased percentage of OCI-AML3 and HL-60 cells with uptake of trypan blue. In addition, cotreatment with DZNep and PS induced more PARP cleavage in OCI-AML3 and HL-60 cells, a hallmark of caspase-3 and -7 activities (Figure 4B). This was associated with synergistically increased apoptosis of OCI-AML3 and HL-60 cells, represented by the combination indices of less than 1.0 (Figure 4C). We also observed synergistic apoptotic effects, after cotreatment with DZNep and SNDX-275, a class I–specific HDAC inhibitor (supplemental Figure 3A-C). We next determined whether the combination of DZNep and PS would also exert increased in vivo antileukemia activity. Figure 4D demonstrates that the survival of NOD/SCID mice with AML due to HL-60 cells was significantly higher, if treated with DZNep and PS compared to treatment with PS, DZNep, or vehicle alone (P < .05). Cotreatment with DZNep and PS also did not increase the weight loss in the mice (data not shown). Median survival was as follows: control, 36 days; PS, 42 days; DZNep, 43 days; and DZNep plus PS, 52 days. The superior in vitro and in vivo antileukemia activity of the combination of DZNep and PS was also associated with greater decline in EZH2 and SUZ12, but not EED and DNMT1 levels (Figure 5A). Similar effects were observed in cells treated with DZNep and SNDX-275 (supplemental Figure 3D). This was associated with decline in 3MeK27H3 and increase in hyperacetylated K27H3 levels, after treatment of OCI-AML3 and HL-60 cells with DZNep plus PS (Figure 5A). Marked decline in the repressive 3MeK27H3 mark in OCI-AML3 cells due to cotreatment with DZNep and PS led to an increase in the levels of p16, p21, p27, and FBXO32, with concomitant decline in the levels of HOXA9 (Figure 5B). EZH2 is known to repress HOXA9, whereas cyclin E is one of the targets of FBXO32.21 These results underscore the link between DZNep plus PS-mediated inhibition of the repressive histone epigenetic marks and derepression of genes that are associated with growth-inhibitory and apoptotic antileukemia activity of the combination of DZNep and PS.
Cotreatment with PS and DZNep synergistically induces apoptosis of cultured AML cells and significantly prolongs survival of mice implanted with AML cells. (A) OCI-AML3 and HL-60 cells were treated with DZNep and/or PS as indicated for 48 hours. At the conclusion of treatment, cell death was assessed by trypan blue dye uptake in a hemocytometer. Columns represent the mean of 3 independent experiments; bars represent SEM. (B) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep and/or PS for 24 hours. Cell lysates were prepared and immunoblot analysis was performed for PARP cleavage. The levels of β-actin in the lysates served as the loading control. Vertical lines have been inserted to indicate a repositioned gel lane. (C) Analysis of dose-effect relationship for DZNep (100-750 nmol/L) and PS (5-20 nmol/L) for the apoptotic effects after 48 hours of exposure in OCI-AML3 and HL-60 cells was performed according to the median dose-effect method of Chou and Talalay. After this, the CI values were calculated. CI < 1, CI = 1, and CI > 1 represent synergism, additivity, and antagonism of the 2 agents, respectively. (D) Female NOD/SCID mice were injected in the lateral tail vein with HL-60 cells. The cells were allowed to engraft for 7 days before initiation of treatment. Mice were treated intraperitoneally with dimethylsulfoxide, 1 mg/kg DZNep 2 days per week for 2 weeks, and/or 10 mg/kg PS 3 days per week for 4 weeks. n = 7 per group. Survival of the mice in all groups (vehicle, DZNep alone, PS alone, and combination) is represented by Kaplan-Meier plot.
Cotreatment with PS and DZNep synergistically induces apoptosis of cultured AML cells and significantly prolongs survival of mice implanted with AML cells. (A) OCI-AML3 and HL-60 cells were treated with DZNep and/or PS as indicated for 48 hours. At the conclusion of treatment, cell death was assessed by trypan blue dye uptake in a hemocytometer. Columns represent the mean of 3 independent experiments; bars represent SEM. (B) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep and/or PS for 24 hours. Cell lysates were prepared and immunoblot analysis was performed for PARP cleavage. The levels of β-actin in the lysates served as the loading control. Vertical lines have been inserted to indicate a repositioned gel lane. (C) Analysis of dose-effect relationship for DZNep (100-750 nmol/L) and PS (5-20 nmol/L) for the apoptotic effects after 48 hours of exposure in OCI-AML3 and HL-60 cells was performed according to the median dose-effect method of Chou and Talalay. After this, the CI values were calculated. CI < 1, CI = 1, and CI > 1 represent synergism, additivity, and antagonism of the 2 agents, respectively. (D) Female NOD/SCID mice were injected in the lateral tail vein with HL-60 cells. The cells were allowed to engraft for 7 days before initiation of treatment. Mice were treated intraperitoneally with dimethylsulfoxide, 1 mg/kg DZNep 2 days per week for 2 weeks, and/or 10 mg/kg PS 3 days per week for 4 weeks. n = 7 per group. Survival of the mice in all groups (vehicle, DZNep alone, PS alone, and combination) is represented by Kaplan-Meier plot.
Cotreatment with PS enhances DZNep-mediated depletion of EZH2, SUZ12, and EED protein and induction of p16, p27, and FBXO32 protein in AML cells. (A-B) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep and PS for 24 hours. Then, total cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, DNMT1, 3MeK27H3, acetyl K27H3, FBXO32, cyclin E, p16, p21, p27, and HOXA9. The expression levels of β-actin in the lysates served as the loading control.
Cotreatment with PS enhances DZNep-mediated depletion of EZH2, SUZ12, and EED protein and induction of p16, p27, and FBXO32 protein in AML cells. (A-B) OCI-AML3 and HL-60 cells were treated with the indicated concentrations of DZNep and PS for 24 hours. Then, total cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, DNMT1, 3MeK27H3, acetyl K27H3, FBXO32, cyclin E, p16, p21, p27, and HOXA9. The expression levels of β-actin in the lysates served as the loading control.
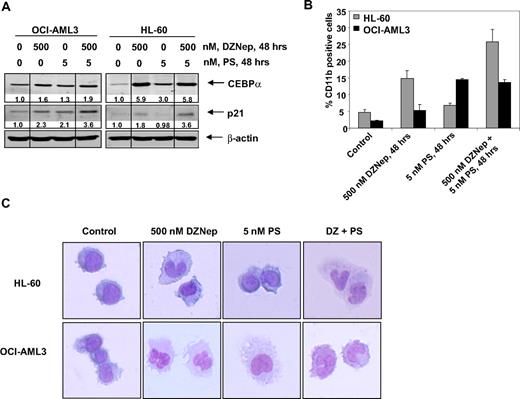
Differentiation effects of cotreatment with DZNep and PS in AML cells
We next determined the ability of DZNep to induce differentiation in AML cells. Exposure to DZNep (500 nM) or to PS (5 nM) for 48 hours induced CEBPα and p21 levels, which are known to be associated with growth arrest and differentiation in AML cells (Figure 6A and supplemental Figure 4). CD11b is a marker of myelomonocytic differentiation in AML cells, and its levels were also induced by DZNep or PS treatment in HL-60 and OCI-AML3 cells (Figure 6B). Cotreatment with DZNep and PS induced more p21 expression in HL-60 and OCI-AML3 cells, whereas more CEBPα expression was observed in OCI-AML3, but not HL-60 cells (Figure 6A). In contrast, compared with treatment with either agent alone, cotreatment with DZNep PS induced more CD11b expression in HL-60, but not OCI-AML3 cells (Figure 6B). Wright staining and light-microscopic evaluation demonstrated that treatment with DZNep also dose dependently induced nuclear and/or cytoplasmic morphologic features of differentiation (Figure 6C and supplemental Figure 4). In addition, combined treatment with DZNep and PS was associated with morphologic differentiation in both HL-60 and OCI-AML3 cells (Figure 6C).
Treatment with DZNep and/or PS induces differentiation of AML cells. (A) Immunoblot analysis of HL-60 and OCI-AML3 cells treated for 48 hours with the indicated concentrations of DZNep and/or PS. The expression levels of β-actin in the lysates served as the loading control. Vertical lines have been inserted to indicate repositioned gel lanes. (B) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of DZNep and/or PS for 48 hours. After this, cells were washed and stained with CD11b antibody, and the percentages of CD11b+ cells were determined by flow cytometry. (C) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of DZNep and/or PS for 72 hours. After treatment, the cells were cytospun onto glass slides, Wright stained, and observed with a microscope to assess cellular morphology.
Treatment with DZNep and/or PS induces differentiation of AML cells. (A) Immunoblot analysis of HL-60 and OCI-AML3 cells treated for 48 hours with the indicated concentrations of DZNep and/or PS. The expression levels of β-actin in the lysates served as the loading control. Vertical lines have been inserted to indicate repositioned gel lanes. (B) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of DZNep and/or PS for 48 hours. After this, cells were washed and stained with CD11b antibody, and the percentages of CD11b+ cells were determined by flow cytometry. (C) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of DZNep and/or PS for 72 hours. After treatment, the cells were cytospun onto glass slides, Wright stained, and observed with a microscope to assess cellular morphology.
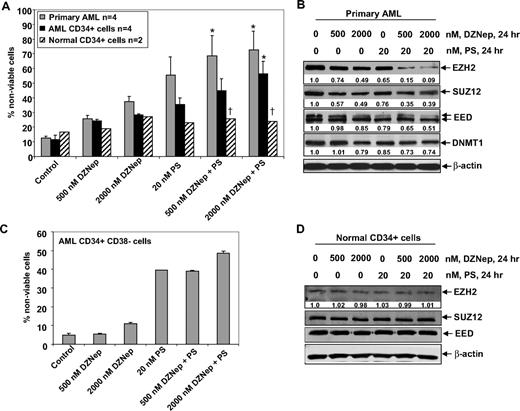
Cotreatment with DZNep and PS is selectively more lethal against primary AML versus normal bone marrow progenitor cells
Next, we compared the effects of DZNep and/or PS on the viability of primary AML versus normal CD34+ progenitor cells. Figure 7A demonstrates that exposure to DZNep or PS for 48 hours induced more loss of viability in 4 samples of AML blasts, as well as in the CD34+ enriched population of the AML progenitor cells, compared with the CD34+ normal progenitor cells (Figure 7A). Compared with treatment with either agent alone, cotreatment with DZNep (500 nM) and PS (20 nM) induced significantly more lethality of AML versus normal progenitor cells. Therefore, the antileukemia activity of DZNep was significantly enhanced when it was combined with PS (Figure 7A). This was associated with greater decline in EZH2, SUZ12, and EED levels in a representative sample of primary AML cells, after coexposure to DZNep and PS versus either agent alone (Figure 7B). Compared with treatment with 20 nM PS or 500 or 2000 nM DZNep alone, cotreatment with 2000 nM (but not 500 nM) DZNep and PS induced more loss of cell viability of CD34+/CD38−/Lin− leukemia stem cells (Figure 7C). As demonstrated in Figure 7D, neither DZNep nor PS treatment resulted in depletion of EZH2, SUZ12, or EED in normal CD34+ bone marrow progenitor cells. Collectively with the in vivo findings against AML in NOD/SCID mice due to HL-60 cells, these observations indicate that cotreatment with PS significantly enhances the anti-AML selectivity of DZNep.
Cotreatment with DZNep and PS exerts a greater antileukemia effect than either agent alone in primary AML cells. (A) Peripheral blood or bone marrow from 4 patients with AML, CD34+ cells enriched from 4 AML patients, and CD34+ cells from 2 normal donors were treated with the indicated concentrations of DZNep and/or PS for 48 hours. Then, the percentages of nonviable cells for each drug alone or drug combination were determined by trypan blue dye uptake in a hemocytometer. Columns represent the mean of the samples; bars represent SEM. *Values significantly greater (P < .05) than those after treatment with either agent alone at the indicated concentrations in the AML samples. †Values significantly less (P < .05) in normal CD34+ versus leukemia samples for the drug combinations. (B) Primary AML cells were treated with the indicated concentrations of DZNep and/or PS for 24 hours. After treatment, cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, and DNMT1. The expression levels of β-actin in the lysates served as the loading control. (C) CD34+/CD38−/Lin− cells enriched from the bone marrow of AML patients were treated with the indicated concentrations of DZNep and/or PS for 48 hours. The percentages of nonviable cells for each drug alone or drug combination were then determined by trypan blue dye uptake in a hemocytometer. Columns represent the mean of the samples; bars represent SEM. (D) Immunoblot analyses of CD34+ cells from a normal donor treated with the indicated concentrations of DZNep and/or PS for 24 hours. The expression levels of β-actin in the lysates served as the loading control.
Cotreatment with DZNep and PS exerts a greater antileukemia effect than either agent alone in primary AML cells. (A) Peripheral blood or bone marrow from 4 patients with AML, CD34+ cells enriched from 4 AML patients, and CD34+ cells from 2 normal donors were treated with the indicated concentrations of DZNep and/or PS for 48 hours. Then, the percentages of nonviable cells for each drug alone or drug combination were determined by trypan blue dye uptake in a hemocytometer. Columns represent the mean of the samples; bars represent SEM. *Values significantly greater (P < .05) than those after treatment with either agent alone at the indicated concentrations in the AML samples. †Values significantly less (P < .05) in normal CD34+ versus leukemia samples for the drug combinations. (B) Primary AML cells were treated with the indicated concentrations of DZNep and/or PS for 24 hours. After treatment, cell lysates were prepared and immunoblot analysis was performed for EZH2, SUZ12, EED, and DNMT1. The expression levels of β-actin in the lysates served as the loading control. (C) CD34+/CD38−/Lin− cells enriched from the bone marrow of AML patients were treated with the indicated concentrations of DZNep and/or PS for 48 hours. The percentages of nonviable cells for each drug alone or drug combination were then determined by trypan blue dye uptake in a hemocytometer. Columns represent the mean of the samples; bars represent SEM. (D) Immunoblot analyses of CD34+ cells from a normal donor treated with the indicated concentrations of DZNep and/or PS for 24 hours. The expression levels of β-actin in the lysates served as the loading control.
Discussion
Although CpG dinucleotide DNA methylation and the EZH2-mediated H3K27 trimethylation cooperate, individually the 3Me H3K27 and histone deacetylation have been shown to inhibit TSG expression independent of promoter DNA methylation.28-30 EZH2 overexpression was shown to promote cell proliferation by promoting S-phase entry and G2-M transition, whereas knockdown of EZH2 by siRNA or pharmacologically by DZNep inhibited cell proliferation and induced apoptosis of breast cancer, but not normal cells.21 In this study, we demonstrate for the first time that DZNep-mediated depletion of EZH2 and the 3Me H3K27 mark inhibits cell-cycle progression, as well as induces differentiation and apoptosis of AML cells. Importantly, we also observed that DZNep exerted relatively little effect on EZH2 and induced significantly less apoptosis in normal CD34+ progenitor cells, mitigating the concerns that EZH2 depletion may promote normal stem cell exhaustion.10 By inhibiting S-adenosyl-methionine–dependent histone KMTase activity of EZH2, treatment with DZNep somehow leads to depletion of EZH2.33,34 Although the precise mechanism by which DZNep promotes depletion of EZH2 is unclear, our findings demonstrate that the proteasome inhibitor BZ partially restores EZH2 levels in AML cells. DZNep treatment neither lowered the mRNA levels of EZH2 nor induced the levels of microRNA 101, which has been shown to abrogate the expression of EZH2 in prostate cancer cells,39 but diminished binding of EZH2 to its target promoters in AML cells. Furthermore, depletion of EZH2 also led to disruption of PRC2 complex and down-regulation of the levels of SUZ12 and EED in primary AML cells, as has also been previously reported for breast cancer cells.21 These findings are consistent with our previous observation in which treatment with PS was also discovered to promote proteasomal degradation of EZH2 and disrupt PRC2, associated with depletion of the levels of SUZ12 and EED.19,31 It is also noteworthy that in AML cells DZNep treatment inhibited 3Me H3K27, but not the 3Me H3K9 or 3Me H3K79 marks, while simultaneously increasing the levels of the permissive 3Me H3K4 mark (Figure 3A). Structure-activity basis of why treatment with DZNep exerts the differential effect on the activity of EZH2 versus the other H3KMTases responsible for the 3Me H3K9, 3Me H3K79, and 3Me H3K4 marks remains to be elucidated. It is also not clear whether the DZNep-mediated greater EZH2 depletion observed in the primary versus cultured AML cells is due to higher sensitivity of the primary AML blast progenitor cells to DZNep.
Among the gene expressions altered by DZNep in AML cells were those of p16, p21, and p27. Both mRNA and protein levels of p16, p21, and p27 were induced, although the precise mechanism(s) by which DZNep induces the levels of the 3 cell-cycle inhibitory proteins has not been elucidated.4,41 Consistent with previous reports, after treatment with DZNep, the levels of the E3 ubiquitin ligase FBXO32 were also induced in AML cells.42,43 Levels of FBXO32 have been previously shown to be repressed by EZH2-mediated 3MeK27H3.21 DZNep-induced FBXO32 levels were associated with decline in the levels of cyclin E, which is known to be targeted by FBXO32.44 Our findings demonstrating that treatment with the siRNA to EZH2 has similar effects on p16, p21, p27, cyclin E, and FBXO32 indicate that DZNep-mediated depletion of EZH2 is responsible for modulation of the cell-cycle regulatory genes. In addition, consistent with this, both present studies and previous reports have demonstrated that depletion of EZH2 inhibits cell-cycle progression through S and G2/M phases and inhibits clonogenic survival, as well as induces differentiation and apoptosis of transformed cells.18,45
Notably, compared with either agent alone, cotreatment with DZNep and the pan-HDAC inhibitor PS (LBH589) resulted in greater depletion of EZH2 and 3Me H3K27 than treatment with either agent alone. We have previously reported that depletion of EZH2 combined with pan-HDAC inhibition leads to depletion of HOXA9 expression, which is known to be leukemogenic and confers an aggressive phenotype in AML.19,43,44 In present studies, cotreatment with DZNep and PS cooperated in depleting HOXA9, which might have potentially contributed to the superior antileukemia activity of the combination. Consistent with this, cotreatment with DZNep and PS or SNDX-275 synergistically induced apoptosis of cultured AML cells. This was also associated with greater induction of p16, p21, p27, and FBXO32, as well as greater decline in cyclin E levels. These molecular perturbations due to treatment with DZNep and PS may serve as the predictive biomarkers of activity of this combination. Recently, F-box family of proteins have been shown to negatively regulate cell growth and survival, and the prosurvival phosphatidylinositol 3-kinase/AKT signaling inhibits FBXO32 levels.45,46 A related F-box family member FBXW7 was also shown to target mTOR for degradation.47 However, it is unclear how DZNep-mediated induction of FBXO32 or of the cell-cycle regulators p16 and p27 sensitizes AML cells to apoptosis induced by pan-HDAC inhibitor PS. The latter is known to trigger multiple mechanisms of cell death in transformed versus normal hemopoietic progenitor cells. It is possible that DZNep might either be interacting with and modulating several of these mechanisms, or circumventing potential mechanisms of resistance to apoptosis induced by PS.3,38,48 This was confirmed by our findings demonstrating that treatment with DZNep induced apoptosis of HL-60/LR cells that exhibit high levels of resistance to PS and other HDIs.38 This raises the possibility that cotreatment with DZNep and PS might prevent the emergence of resistance to HDIs in AML cells. Cotreatment with DZNep and PS also induced morphologic features of differentiation in HL-60 and OCI-AML3 cells, which was associated with increased expression of CEBPα. It is well recognized that CEBPα is required for the proper control of granulocytic differentiation, and in AML progenitor cells its expression is often abrogated.49 Restoration of CEBPα function may overcome the block in differentiation and may contribute to the anti-AML activity of treatment regimens, such as the combination of DZNep and PS.
Results of the in vivo studies presented in this work show that, compared with treatment with each agent alone, combined treatment with DZNep and PS improved survival of NOD/SCID mice with leukemia caused by the AML HL-60 cells. In a previous in vivo study in mice, the efficacy of DZNep as a nucleoside analog was tested against Ebola virus infection. In this study, the dose of DZNep used was similar to the dose used in present studies, and it was determined to be safe and to exhibit biologic activity.34,50 Notably, to date, neither DZNep nor one of its active analogues has been administered to humans. In contrast, the phase I/II studies of PS in patients with hematologic malignancies are currently active, and the findings to date have demonstrated that PS is safe to administer and is active against AML, non-Hodgkin lymphoma, and Hodgkin disease.51 Taken together with the preclinical and clinical experience with DZNep and PS alone, findings presented in this study clearly support the rationale to further test the in vivo anti-AML efficacy of the combinations of DZNep and PS. Our findings also highlight the potential biomarkers of activity of this combination, as well as create the rationale to develop and test epigenetic-targeted therapies against AML that incorporate pan-HDAC inhibitors with agents that abrogate EZH2 and PRC2.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health/National Cancer Institute R01 CA116629 (K.N.B.) and R01 CA123207 (K.N.B.). This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
National Institutes of Health
Authorship
Contribution: W.F., Y.W., A.S., R.R., J.C., R.B., and S.K. performed the in vitro studies with the cultured AML cells; P.F. and K.M.B. performed the in vivo HL-60 cell xenograft studies in NOD/SCID mice; C.U. and A.J. procured and assisted in performing the studies on primary AML and CD34+ normal progenitor cells; P.A., H.S., and V.E.M. provided reagents for the study; and K.N.B. planned and supervised the in vitro and in vivo studies and prepared the report.
Conflict-of-interest disclosure: P.A. is an employee of Novartis Institute for Biomedical Research, and K.N.B. has received clinical and laboratory research support from Novartis Institute for Biomedical Research. A.J. received research support from Novartis. The remaining authors declare no competing financial interests.
Correspondence: Kapil N. Bhalla, Medical College of Georgia Cancer Center, Medical College of Georgia, 1120 15th St, CN-2101 Augusta, GA 30912; e-mail: kbhalla@mcg.edu.