Abstract
The identification of novel factors regulating human T helper (Th)–cell differentiation into functionally distinct Th1 and Th2 subsets is important for understanding the mechanisms behind human autoimmune and allergic diseases. We have identified a protein of relevant evolutionary and lymphoid interest (PRELI), a novel protein that induces oxidative stress and a mitochondrial apoptosis pathway in human primary Th cells. We also demonstrated that PRELI inhibits Th2-cell development and down-regulates signal transducer and activator of transcription 6 (STAT6), a key transcription factor driving Th2 differentiation. Our data suggest that calpain, an oxidative stress–induced cysteine protease, is involved in the PRELI-induced down-regulation of STAT6. Moreover, we observed that a strong T-cell receptor (TCR) stimulus induces expression of PRELI and inhibits Th2 development. Our results suggest that PRELI is involved in a mechanism wherein the strength of the TCR stimulus influences the polarization of Th cells. This study identifies PRELI as a novel factor influencing the human primary Th-cell death and differentiation.
Introduction
Differentiation of human CD4+ T helper (Th) cells into functionally distinct Th1 and Th2 subsets plays a central role in the pathogenesis of human allergic and inflammatory diseases. The most potent stimuli influencing this process are the cytokines interleukin (IL)–12 and IL-4.1,2 Signal transducer and activator of transcription 6 (STAT6) is an important transducer of the IL-4–induced signaling pathway leading to Th2 differentiation.3
The strength of the T-cell receptor (TCR) signal also influences the direction of the Th1/Th2 polarization. A strong TCR stimulus tends to enhance Th1 differentiation, whereas weak TCR signals favor Th2 polarization.4,5 Th1 and Th2 cells also differ in terms of their susceptibility to apoptosis, with Th2 cells being more resistant to apoptotic cell death than Th1 cells.6 IL-4 prevents apoptosis in several cell types, but the molecular mechanisms involved are largely unknown. In T cells, STAT6 does not seem to be required for the antiapoptotic effect of IL-4,7 indicating that the IL-4– induced Th2 differentiation and protection from apoptosis represent distinct signaling cascades emanating from the same receptor. In an earlier study by our group, IL-4 was shown to inhibit caspase-3 activity during the early phases of human Th2 differentiation while up-regulating the expression of the antiapoptotic proteins BCL-2 and BCL-XL.8 Other studies have also demonstrated that the IL-4 signaling pathway regulates apoptosis through regulation of mitochondrial proteins,9 and by maintaining the mitochondrial membrane potential (Δψm).10
The activity of STAT6 is tightly regulated through several mechanisms (for a review, see Hebenstreit et al11 ), including tyrosine and serine phosphorylation and methylation. Inhibition of STAT6 signaling is controlled directly by specific phosphatases, as well as indirectly by suppressor of cytokine signaling (SOCS) proteins. In addition, several studies indicate that STAT6 is negatively regulated by proteolytic degradation. In mouse mast cells, STAT6 is proteolytically cleaved by both a protease belonging to the elastase family and by activated calpain. This cleavage generates truncated products of 65 and 70 kDa, which then function as dominant negative forms of STAT6.12 In mouse T-cell lines, activated STAT6 is completely degraded by calpain.13 The physiologic role of calpain-mediated cleavage of STAT6 is currently unclear. Calpains are calcium-dependent proteases implicated in cell proliferation, apoptosis, and differentiation.14
In this study we identify PRELI (protein of relevant evolutionary and lymphoid interest) as a novel activation-induced gene in human primary Th cells. PRELI has previously been reported to be highly expressed in adult lymph nodes and peripheral blood leukocytes. In addition, based on its high expression levels in fetal liver and germinal center B lymphocytes, PRELI has been proposed to be important for B-cell development.15 To date, the cellular role of PRELI has been obscure. It is an evolutionarily conserved protein that is, in mice, coexpressed with Rab24, a member of the Rab GTPase family,16 which is thought to be involved in autophagy-related processes.17 We report for the first time that PRELI is also highly induced in human primary CD4+ Th cells in response to TCR activation. Functionally, this study characterizes PRELI as a novel regulator of the mitochondria-mediated apoptosis pathway in human primary Th cells. In addition, we demonstrate that PRELI regulates STAT6 and Th2 differentiation. Based on our findings, we propose that PRELI is involved in a mechanism in which the strength of the TCR stimulus dictates the polarization of Th cells.
Methods
Plasmid constructs
The PRELI cDNA was amplified with polymerase chain reaction (PCR) from human Th2 cells with PRELI-forward and PRELI-reverse primers (Table 1) and cloned into the pFLAG-CMV-2 vector (Kodak, New Haven, CT), creating a pFlag-PRELI construct. The pIRES2-H2Kk-PRELI construct was made by amplifying PRELI with PRELI-forward and PRELI-reverse-2 primers (Table 1), using pFlag-PRELI as a template and subsequently ligating the PCR product to the pIRES2-H2Kk vector.18
Sequences of primers, probes, shRNA, and siRNA oligonucleotides used
| PCR | |
| PRELI-Fwd1 | 5′-CGGAATTCCATGGTGAAGTATTTCCTGGGCCA-3′ |
| PRELI-Rev1 | 5′-TGCTCTAGACTACACAAACTGTTGCTGCTGCTG-3′ |
| PRELI-Rev2 | 5′-CGCGGATCCCTACACAAACTGTTGCTGCTGCTG-3′ |
| Taqman RT-PCR | |
| EF1α-Probe | 5′-(FAM)-AGCGCCGGCTATGCCCCTG-(TAMRA)-3′ |
| EF1α-Fwd | 5′-CTGAACCATCCAGGCCAAAT-3′ |
| EF1α-Rev | 5′-GCCGTGTGGCAATCCAAT-3′ |
| PRELI-Probe | 5′-(FAM)-CCGGGAAGCCTGGGTCTCCTCTAGCT-(TAMRA)-3′ |
| PRELI-Fwd | 5′-TGGCTGGACTGAAATCC-3′ |
| PRELI-Rev | 5′-CAGCTCTGGAGACACCAAAT-3′ |
| STAT6-Probe | 5′-(FAM)-CAGGACACCATCAAACCACTGCCAAA-(TAMRA)-3′ |
| STAT6-Fwd | 5′-TGGGCCGTGGCTTCAC-3′ |
| STAT6-Rev | 5′-CCGGAGACAGCGTTTGGT-3′ |
| siRNA/shRNA sequence | |
| Scramble | 5′-GCGCGCUUUGUAGGAUUCGUU-3′ |
| PRELI-siRNA1 | 5′-CCAAGACUAUGAAGGGUUUU-3′ |
| PRELI-siRNA2 | 5′-CCAAGGACCUCGCCAGCAAUU-3′ |
| PRELI-shRNA | 5′-GGAGAAGGCAAAGGAGACGUU-3′ |
| PCR | |
| PRELI-Fwd1 | 5′-CGGAATTCCATGGTGAAGTATTTCCTGGGCCA-3′ |
| PRELI-Rev1 | 5′-TGCTCTAGACTACACAAACTGTTGCTGCTGCTG-3′ |
| PRELI-Rev2 | 5′-CGCGGATCCCTACACAAACTGTTGCTGCTGCTG-3′ |
| Taqman RT-PCR | |
| EF1α-Probe | 5′-(FAM)-AGCGCCGGCTATGCCCCTG-(TAMRA)-3′ |
| EF1α-Fwd | 5′-CTGAACCATCCAGGCCAAAT-3′ |
| EF1α-Rev | 5′-GCCGTGTGGCAATCCAAT-3′ |
| PRELI-Probe | 5′-(FAM)-CCGGGAAGCCTGGGTCTCCTCTAGCT-(TAMRA)-3′ |
| PRELI-Fwd | 5′-TGGCTGGACTGAAATCC-3′ |
| PRELI-Rev | 5′-CAGCTCTGGAGACACCAAAT-3′ |
| STAT6-Probe | 5′-(FAM)-CAGGACACCATCAAACCACTGCCAAA-(TAMRA)-3′ |
| STAT6-Fwd | 5′-TGGGCCGTGGCTTCAC-3′ |
| STAT6-Rev | 5′-CCGGAGACAGCGTTTGGT-3′ |
| siRNA/shRNA sequence | |
| Scramble | 5′-GCGCGCUUUGUAGGAUUCGUU-3′ |
| PRELI-siRNA1 | 5′-CCAAGACUAUGAAGGGUUUU-3′ |
| PRELI-siRNA2 | 5′-CCAAGGACCUCGCCAGCAAUU-3′ |
| PRELI-shRNA | 5′-GGAGAAGGCAAAGGAGACGUU-3′ |
Fwd indicates forward; Rev, reverse; FAM, 6-carboxyfluorescein; and TAMRA, 6-carboxytetramethylrhodamine.
Oligonucleotide microarrays
Pooled cord blood CD4+ cells from at least 6 healthy neonates were activated with plate-bound α-CD3 (0.5 μg/well) and α-CD28 (0.5 μg/mL) and cultured with 2.5 ng/mL of IL-12 (Th1), 10 ng/mL of α-IL-12 plus 10 ng/mL of IL-4 (Th2), or no cytokines (Th0). After 48 hours, 40 U/mL of IL-2 was added to the cultures. Samples were harvested at 0, 0.5, 1, 2, 4, 6, 12, 24, 48 and 72 hours. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA), and 100 ng of RNA was taken for microarray sample preparation, which was performed according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). The samples were hybridized to Affymetrix U133 Plus 2.0 arrays containing probes for approximately 47 000 transcripts (known genes and expressed sequence tags). The Affymetrix signals were derived using the robust multiarray average procedure19 in affy, a Bioconductor software package (http://www.bioconductor.org).
Cell isolation, transfection, and culture
Hek293 cells (ATCC, Manassas, VA) were cultured in Dulbecco modified eagle medium (DMEM; Sigma-Aldrich, St Louis, MO) supplemented with pen/strep, 2 mM l-glutamine, 1 mM sodium pyruvate, and 10% fetal calf serum (FCS). HeLa cells (ATCC) were cultured in modified eagle medium (MEM) with Earle salts (both Invitrogen, Carlsbad, CA), supplemented with pen/strep, 2 mM l-glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, and 10% FCS. For transfections, the cells were plated on cover slips (precoated with poly-l-lysine) in 6-well plates at a density of 0.7 × 106 cells/2 mL of DMEM (Hek293) or 0.4 × 106 cells/2 mL of MEM (HeLa) without antibiotics. After 24 hours, the cells were transfected with 1.5 μg (Hek293) or 4 μg (HeLa) of empty pFlag vector or pFlag-PRELI using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Human mononuclear cells were isolated from cord blood from healthy neonates or peripheral blood (buffy coats) from healthy blood donors (Finnish Red Cross) using Ficoll-Paque isolation (Amersham Pharmacia Biotech, Piscataway, NJ). CD4+ cells were further purified using DYNAL magnetic beads (Invitrogen). Cord blood CD4+ cells from several individuals were pooled after the isolation. CD4+ cells were activated, unless otherwise indicated, with plate-bound α-CD3 (0.5 μg/well) and soluble α-CD28 (0.5 μg/mL; both from Immunotech, Marseille, France)8 and cultured at a density of approximately 2 × 106 cells/mL in 24-well plates in Yssel medium (Iscove modified Dulbecco medium [IMDM; Invitrogen] supplemented with Yssel medium concentrate, pen/strep, and 1% AB-serum). When indicated, Th1 differentiation was induced at the time of activation with 2.5 ng/mL of IL-12, and Th2 polarization with 10 ng/mL of IL-4 (both from R&D Systems, Minneapolis, MN). In 7-day cultures, 40 U/mL of IL-2 (R&D Systems) was added to the cultures after 48 hours of priming. When indicated, the cells were cultured in the presence of 0.2 to 1 mM H2O2 (Merck, Darmstadt, Germany), 1.25 mM N-acetyl-cysteine or 5 μM calpastatin peptide (both from Calbiochem, San Diego, CA).
In PRELI overexpression experiments, buffy coat CD4+ cells were suspended in Optimem I (5 × 106 cells/100 μL; Invitrogen) and transfected with pIRES-H2Kk-PRELI or pIRES-H2Kk vectors using the nucleofection technique (Amaxa, Cologne, Germany).18 Nucleofected cells were incubated at 37°C for 16 hours (2.5 × 106 cells/mL), after which dead cells were removed, and the transfected cells were enriched as previously described.18 Briefly, dead cells, apoptotic cells, and debris were depleted from the cultures using magnetic-activated cell-sorting (MACS) Dead Cell Removal Microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Subsequently, the remaining cells were incubated with MACSelect Kk MicroBeads coated with H-2Kk–antibody (Miltenyi Biotec) for 15 minutes to magnetically label the transfected cells. The magnetic separation of H-2Kk–positive cells was done with a positive selection column placed in the magnetic field of a MACS separator. After the enrichment, cell viability and purity (H-2Kk–positivity) were determined by flow cytometry. The purified H-2Kk–positive cells were activated and cultured as described earlier in this section. In PRELI knockdown studies, naive or peripheral blood CD4+ cells, nucleofected with scrambled or PRELI-specific siRNA oligos (1.5 μg/4 × 106 cells/transfection; Sigma-Aldrich; Table 1), were incubated at 37°C for 24 hours (2 × 106 cells/mL) and subsequently activated and cultured in Yssel medium as described earlier in this section.
Research involving the use of human blood from unknown donors was permitted by the Finnish Ethics Committee.
Immunofluorescent staining
To study the localization of PRELI in Hek293 or HeLa cells, the cells were washed with the culture medium 24 hours after transfection and incubated with 20 to 200 nM MitotrackerRed CMXRos (Molecular Probes, Eugene, OR) for 30 minutes at 37°C. The cells were washed with cold phosphate-buffered saline (PBS), fixed with methanol for 5 minutes at −20°C, washed twice with PBS, and blocked with goat serum (Invitrogen) for 15 minutes. The cells were stained for Flag-PRELI with 25 μg/mL mouse α-Flag-M2 antibody (Sigma-Aldrich) for 1 hour. The cells were washed with PBS (3 × 5 minutes), incubated with 5 μg/mL goat α-mouse-fluorescein isothiocyanate (FITC) antibody (Caltag Laboratories, Burlingame, CA) for 45 minutes, and washed with PBS. To study the effect of PRELI on the production of reactive oxygen species (ROS) in HeLa cells, the cells were treated in a similar manner except that, instead of being incubated with Mitotracker, they were incubated with 20 μM dihydrorhodamine 123 (DHR123; Molecular Probes) for 30 minutes at 37°C. Alexa Fluor 568-conjugated goat α-mouse antibody (5 μg/mL; Molecular Probes) was used as a secondary antibody to detect overexpressed PRELI. Cells were analyzed at room temperature with a Zeiss LSM 510 META confocal microscope (Carl Zeiss, Jena, Germany) using Plan-Apochromat 63×/1.4 oil differential interference contrast (DIC) objective. FITC and DHR123 were excited with a 488-nm laser line and emissions were collected with a 500- to 530-nm bandpass filter. MitotrackerRed and Alexa568 were excited with a 543-nm laser line and emissions were collected with a 560-nm longpass filter. Images were analyzed with LSM5 software (Carl Zeiss).
To examine the effect of PRELI on Δψm and ROS production in primary Th cells, the nucleofected cells were harvested after 24 hours of activation and incubated with 50 nM Mitoprobe DilC15 (Molecular Probes) in PBS or 20 μM DHR123 in Yssel medium, respectively (∼0.3 × 106 cells/500 μL) for 30 minutes at 37°C, and subsequently washed with PBS. Staining with annexin V–FITC (BD Pharmingen, San Jose, CA) and α-active caspase-3-PE antibody (BD Pharmingen) was performed according to manufacturers' instructions. Propidium iodide (PI; Clontech, Mountain View, CA) was mixed with the cells (5 μL/0.3 × 106 cells in 500 μL PBS) 20 seconds before measurement. To measure the expression of chemoattractant receptor-homologous molecule expressed on Th2 cells (CRTH2) on polarized Th cells, 0.3 to 0.5 × 106 cells were first washed with PBS, then with MACS buffer (0.5% bovine serum albumin, 2 mM ethylenediaminetetraacetic acid in PBS, pH 7.2) and subsequently incubated with 5 μL of CRTH2-phosphatidylethanolamine (PE) antibody for 10 minutes at 4°C. Next, the cells were washed twice with MACS buffer and resuspended in 1% formalin/PBS. Samples were measured with a FACSCalibur fluorescence-activated cell sorter and analyzed with CellQuest Pro (both from BD Biosciences, San Jose, CA).
Intracellular cytokine staining
Cytokine staining and analysis was performed as previously described.18 The antibodies used were α-IFNγ-FITC (3 μL/100μL permeabilization buffer) and α-IL4-PE (3 μL; both from Caltag Laboratories) or α-IL4-PE (1 μL; BD Pharmingen).
Taqman reverse transcription–polymerase chain reaction
Samples were prepared for Taqman reverse transcription–polymerase chain reaction (RT-PCR) analysis as described previously.20 Gene expression levels were measured using the TaqMan ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA).21 The housekeeping gene elongation factor 1α (EF1α) was used as a reference transcript. The primers and probes used (MedProbe, Oslo, Norway; Table 1) were designed using Primer Express software (Applied Biosystems).
Western blotting
The cell lysates were sonicated, and equal amounts of protein were run on 10% sodium dodecyl sulfate–polyacrylamide gels and transferred to a nitrocellulose membrane (Hybond enhanced chemiluminescence [ECL]; GE Healthcare, Piscataway, NJ). Proteins were detected using the following antibody dilutions: 1:1000 sheep α-PRELI-antiserum (kind gift from Dr Elisabeth Fox, Leeds, United Kingdom), 1:500 mouse α-STAT6 (BD Biosciences), 1:1000 rabbit α-phospho-STAT6 (Tyr641; Cell Signaling Technology, Beverly, MA), 1:200 rabbit α-STAT1 p84/p91 (E-23; Santa Cruz Biotechnology, Santa Cruz, CA), 1:200 rabbit α-STAT4 (C-20; Santa Cruz Biotechnology), 1:500 rabbit α-calpain 1 large subunit (μ-type; Cell Signaling Technology) or 1:20 000 mouse α-β-actin (Sigma-Aldrich). Horseradish peroxidase–conjugated secondary antibodies were: 1:10 000 goat α-mouse IgG (Santa Cruz Biotechnology), 1:10 000 goat α-rabbit Ig (BD Biosciences) or 1:20 000 donkey α-sheep (Jackson ImmunoResearch Laboratories, West Grove, PA). The protein bands were visualized with ECL (GE Healthcare).
Results
PRELI is up-regulated in response to T-cell activation in human primary T helper cells
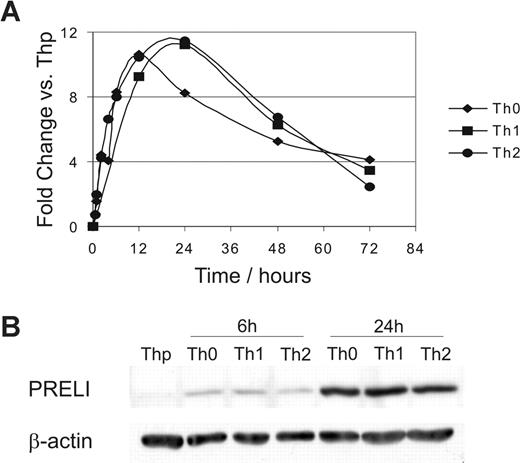
We identified PRELI as an activation-induced gene in primary human Th cells using Affymetrix oligonucleotide arrays. This study was conducted to identify novel genes involved in the regulation of early human primary Th-cell differentiation and activation (unpublished results). The expression of PRELI mRNA was highly increased when naive CD4+ cells (Thp) were subjected to TCR activation. However, it was not differentially regulated by the Th1/Th2 polarizing cytokines IL-12 or IL-4. PRELI expression increased for the first 24 hours after activation and decreased thereafter (Figure 1A). A similar induction profile was also obtained at the protein level (Figure 1B and data not shown).
PRELI is up-regulated in response to activation in human primary T helper cells at the mRNA and protein level. (A) Expression of PRELI mRNA. Naive CD4+ cells were activated and cultured in the absence (Th0) or presence of IL-12 (Th1) or IL-4 (Th2). The figure represents the fold change of PRELI expression in Th0, Th1, and Th2 cells versus nonactivated (Thp) cells (Affymetrix probe set (224232_s_at). Representative data from 3 independent experiments. (B) Expression of PRELI at protein level. Naive CD4+ cells were cultured as in panel A for 6 and 24 hours. Samples from 5 donors (neonates) were pooled.
PRELI is up-regulated in response to activation in human primary T helper cells at the mRNA and protein level. (A) Expression of PRELI mRNA. Naive CD4+ cells were activated and cultured in the absence (Th0) or presence of IL-12 (Th1) or IL-4 (Th2). The figure represents the fold change of PRELI expression in Th0, Th1, and Th2 cells versus nonactivated (Thp) cells (Affymetrix probe set (224232_s_at). Representative data from 3 independent experiments. (B) Expression of PRELI at protein level. Naive CD4+ cells were cultured as in panel A for 6 and 24 hours. Samples from 5 donors (neonates) were pooled.
PRELI is localized in mitochondria and reduces the mitochondrial membrane potential (Δψm)
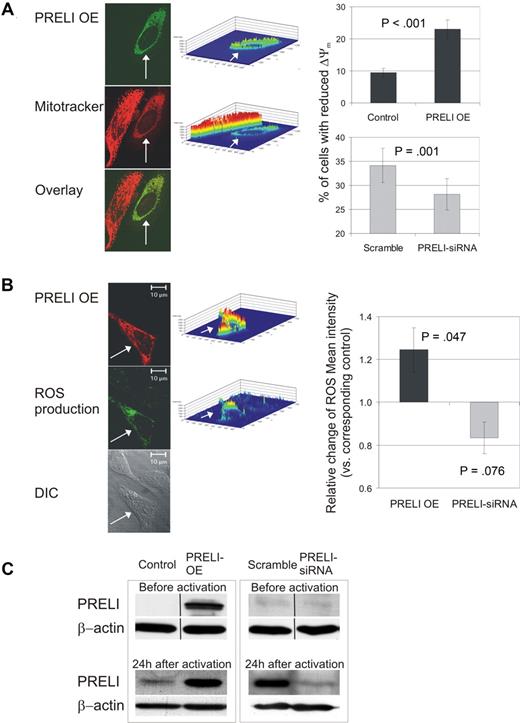
To clarify the cellular function of PRELI, we first studied its localization. PRELI contains an MSF1 domain whose function is unknown but may have a role in intramitochondrial protein sorting. MitotrackerRed CMXRos costaining was used to determine whether PRELI localizes to mitochondria. Exogenously expressed PRELI was localized in the mitochondria of HeLa, Hek293, and primary CD4+ cells (Figure 2A and data not shown). This is consistent with the description of mitochondrial localization of PRELI in HeLa cells and a mitochondrial targeting sequence in the N-terminal domain of the PRELI protein.16 Interestingly, the fluorescence intensity of the membrane potential sensitive Mitotracker was lower in HeLa cells overexpressing pFlag-PRELI than in cells not overexpressing PRELI, indicating that the Δψm was reduced by PRELI (Figure 2A). To study this in human primary CD4+ cells, we used 2 approaches: PRELI overexpression and knockdown by specific siRNAs. Consistent results were obtained with both of these approaches using flow cytometric analysis. Overexpression of PRELI increased the proportion of cells with reduced Δψm, whereas silencing PRELI significantly decreased the number of these cells, confirming that PRELI reduces the Δψm in human primary Th cells (Figure 2A). To verify the overexpression and knockdown of PRELI in our experiments, we analyzed the levels of PRELI protein by Western blotting before and 24 hours after the activation. In the overexpression experiments, the transfected cells were enriched before the culture. Figure 2C shows the representative levels of PRELI protein in the overexpression and knockdown experiments. In this study, we often observed that the effect of PRELI overexpression had a larger effect than knockdown (eg, Figure 2A,B). This may be due to the fact that, in the overexpression method, the transfected cells are purified prior to the activation and polarization, unlike in the siRNA approach. In addition, PRELI is already overexpressed at the time of the activation, whereas the effects of knockdown start to appear after the activation, that is, when the endogenous PRELI is induced (Figure 2C). This can also partly account for the larger effects of PRELI overexpression than of the siRNA approach.
PRELI localizes in mitochondria, alters the Δψm, and induces mitochondrial ROS production. (A) PRELI localizes in mitochondria and reduces Δψm. HeLa cells were stained for pFlag-PRELI (transfected cell indicated by white arrow). Mitochondria were visualized with MitotrackerRed CMXRos. Intensity profiles (middle panel) visualize FITC (PRELI overexpression) and Mitotracker fluorescence intensities in these cells. CD4+ cells (right panel) were nucleofected with pIRES-H2Kk (control) or PRELI overexpression vector (black bars) and subsequently sorted. Cells were also nucleofected with scramble or PRELI-siRNAs ( ). Subsequently, the cells were activated (with α-CD3 0.1 or 0.5 μg/well and α-CD28) for 24 hours, stained with Mitoprobe DilC15 and analyzed by flow cytometry. Two separate cell populations with high and low Mitoprobe DilC15 intensities seen in the flow cytometric analysis depict cells with active and reduced membrane potentials, respectively (histograms not shown). Thus, the percentage of cells with reduced membrane potential stands for the proportion of cells with low Mitoprobe DilC15 intensity in a sample. Bars represent the mean values plus or minus SEM of 7 independent experiments. (B) PRELI induces mitochondrial ROS production. Overexpressed pFlag-PRELI (Alexa 568, red) and mitochondrial ROS production stained with DHR123 (green). CD4+ cells (right panel), nucleofected and activated as in panel A, were stained with DHR123 and analyzed by flow cytometry. Bars represent the average fold changes plus or minus SEM of ROS intensity in PRELI overexpression or PRELI-siRNA samples compared with the corresponding control sample (pIRES-H2Kk or scramble). The values of the control samples were set as 1. Overexpression and siRNA data are obtained from 7 and 4 independent experiments, respectively. (A,B) Δψm and ROS were measured from living cell populations according to forward scatter/side scatter (FSC/SSC) plots (overexpression experiments) or total cell population (siRNA experiments). Statistical significances were calculated using the 2-tailed paired t test. (C) Representative levels of PRELI protein in overexpression and knockdown experiments. In overexpression experiments the peripheral blood CD4+ cells, transfected with pIRES-H2Kk-PRELI or pIRES-H2Kk control vector, were enriched 16 hours after the nucleofection and subsequently activated for 24 hours. In PRELI knockdown experiments, the cells nucleofected with scrambled- or PRELI-siRNA oligos were incubated for approximately 24 hours and subsequently activated for 24 hours. Samples were harvested for Western blotting before (in overexpression experiments after the enrichment) and after the activation. Vertical lines have been inserted to indicate a repositioned gel lane. OE indicates overexpression.
). Subsequently, the cells were activated (with α-CD3 0.1 or 0.5 μg/well and α-CD28) for 24 hours, stained with Mitoprobe DilC15 and analyzed by flow cytometry. Two separate cell populations with high and low Mitoprobe DilC15 intensities seen in the flow cytometric analysis depict cells with active and reduced membrane potentials, respectively (histograms not shown). Thus, the percentage of cells with reduced membrane potential stands for the proportion of cells with low Mitoprobe DilC15 intensity in a sample. Bars represent the mean values plus or minus SEM of 7 independent experiments. (B) PRELI induces mitochondrial ROS production. Overexpressed pFlag-PRELI (Alexa 568, red) and mitochondrial ROS production stained with DHR123 (green). CD4+ cells (right panel), nucleofected and activated as in panel A, were stained with DHR123 and analyzed by flow cytometry. Bars represent the average fold changes plus or minus SEM of ROS intensity in PRELI overexpression or PRELI-siRNA samples compared with the corresponding control sample (pIRES-H2Kk or scramble). The values of the control samples were set as 1. Overexpression and siRNA data are obtained from 7 and 4 independent experiments, respectively. (A,B) Δψm and ROS were measured from living cell populations according to forward scatter/side scatter (FSC/SSC) plots (overexpression experiments) or total cell population (siRNA experiments). Statistical significances were calculated using the 2-tailed paired t test. (C) Representative levels of PRELI protein in overexpression and knockdown experiments. In overexpression experiments the peripheral blood CD4+ cells, transfected with pIRES-H2Kk-PRELI or pIRES-H2Kk control vector, were enriched 16 hours after the nucleofection and subsequently activated for 24 hours. In PRELI knockdown experiments, the cells nucleofected with scrambled- or PRELI-siRNA oligos were incubated for approximately 24 hours and subsequently activated for 24 hours. Samples were harvested for Western blotting before (in overexpression experiments after the enrichment) and after the activation. Vertical lines have been inserted to indicate a repositioned gel lane. OE indicates overexpression.
PRELI localizes in mitochondria, alters the Δψm, and induces mitochondrial ROS production. (A) PRELI localizes in mitochondria and reduces Δψm. HeLa cells were stained for pFlag-PRELI (transfected cell indicated by white arrow). Mitochondria were visualized with MitotrackerRed CMXRos. Intensity profiles (middle panel) visualize FITC (PRELI overexpression) and Mitotracker fluorescence intensities in these cells. CD4+ cells (right panel) were nucleofected with pIRES-H2Kk (control) or PRELI overexpression vector (black bars) and subsequently sorted. Cells were also nucleofected with scramble or PRELI-siRNAs ( ). Subsequently, the cells were activated (with α-CD3 0.1 or 0.5 μg/well and α-CD28) for 24 hours, stained with Mitoprobe DilC15 and analyzed by flow cytometry. Two separate cell populations with high and low Mitoprobe DilC15 intensities seen in the flow cytometric analysis depict cells with active and reduced membrane potentials, respectively (histograms not shown). Thus, the percentage of cells with reduced membrane potential stands for the proportion of cells with low Mitoprobe DilC15 intensity in a sample. Bars represent the mean values plus or minus SEM of 7 independent experiments. (B) PRELI induces mitochondrial ROS production. Overexpressed pFlag-PRELI (Alexa 568, red) and mitochondrial ROS production stained with DHR123 (green). CD4+ cells (right panel), nucleofected and activated as in panel A, were stained with DHR123 and analyzed by flow cytometry. Bars represent the average fold changes plus or minus SEM of ROS intensity in PRELI overexpression or PRELI-siRNA samples compared with the corresponding control sample (pIRES-H2Kk or scramble). The values of the control samples were set as 1. Overexpression and siRNA data are obtained from 7 and 4 independent experiments, respectively. (A,B) Δψm and ROS were measured from living cell populations according to forward scatter/side scatter (FSC/SSC) plots (overexpression experiments) or total cell population (siRNA experiments). Statistical significances were calculated using the 2-tailed paired t test. (C) Representative levels of PRELI protein in overexpression and knockdown experiments. In overexpression experiments the peripheral blood CD4+ cells, transfected with pIRES-H2Kk-PRELI or pIRES-H2Kk control vector, were enriched 16 hours after the nucleofection and subsequently activated for 24 hours. In PRELI knockdown experiments, the cells nucleofected with scrambled- or PRELI-siRNA oligos were incubated for approximately 24 hours and subsequently activated for 24 hours. Samples were harvested for Western blotting before (in overexpression experiments after the enrichment) and after the activation. Vertical lines have been inserted to indicate a repositioned gel lane. OE indicates overexpression.
). Subsequently, the cells were activated (with α-CD3 0.1 or 0.5 μg/well and α-CD28) for 24 hours, stained with Mitoprobe DilC15 and analyzed by flow cytometry. Two separate cell populations with high and low Mitoprobe DilC15 intensities seen in the flow cytometric analysis depict cells with active and reduced membrane potentials, respectively (histograms not shown). Thus, the percentage of cells with reduced membrane potential stands for the proportion of cells with low Mitoprobe DilC15 intensity in a sample. Bars represent the mean values plus or minus SEM of 7 independent experiments. (B) PRELI induces mitochondrial ROS production. Overexpressed pFlag-PRELI (Alexa 568, red) and mitochondrial ROS production stained with DHR123 (green). CD4+ cells (right panel), nucleofected and activated as in panel A, were stained with DHR123 and analyzed by flow cytometry. Bars represent the average fold changes plus or minus SEM of ROS intensity in PRELI overexpression or PRELI-siRNA samples compared with the corresponding control sample (pIRES-H2Kk or scramble). The values of the control samples were set as 1. Overexpression and siRNA data are obtained from 7 and 4 independent experiments, respectively. (A,B) Δψm and ROS were measured from living cell populations according to forward scatter/side scatter (FSC/SSC) plots (overexpression experiments) or total cell population (siRNA experiments). Statistical significances were calculated using the 2-tailed paired t test. (C) Representative levels of PRELI protein in overexpression and knockdown experiments. In overexpression experiments the peripheral blood CD4+ cells, transfected with pIRES-H2Kk-PRELI or pIRES-H2Kk control vector, were enriched 16 hours after the nucleofection and subsequently activated for 24 hours. In PRELI knockdown experiments, the cells nucleofected with scrambled- or PRELI-siRNA oligos were incubated for approximately 24 hours and subsequently activated for 24 hours. Samples were harvested for Western blotting before (in overexpression experiments after the enrichment) and after the activation. Vertical lines have been inserted to indicate a repositioned gel lane. OE indicates overexpression.
PRELI induces production of ROS by mitochondria and apoptotic cell death in human primary T helper cells
Various death or stress signals induce the loss of Δψm, resulting in mitochondrial membrane permeabilization, generation of ROS and the subsequent release of cytochrome c and caspase-3 activation, thereby leading to the activation of apoptotic cell death. Mitochondria are the major source of ROS but they can also serve as their targets during the apoptotic process.22 Our finding that PRELI altered the Δψm suggested that overexpression of PRELI could influence ROS production by mitochondria. To study this, HeLa cells were transfected with pFlag-PRELI and the ROS produced by the mitochondria were stained with DHR123. The production of ROS was slightly induced in the cells where PRELI was overexpressed (Figure 2B). To confirm these results, CD4+ cells overexpressing PRELI or the control plasmid were activated, stained with DHR123, and analyzed by flow cytometry. PRELI overexpression slightly increased the ROS production also in primary Th cells (Figure 2B). This was further confirmed in experiments where PRELI was specifically knocked down using siRNA oligos (siRNA1 or 2) or a PRELI-specific short hairpin RNA (shRNA) plasmid (Figure 2B and data not shown).
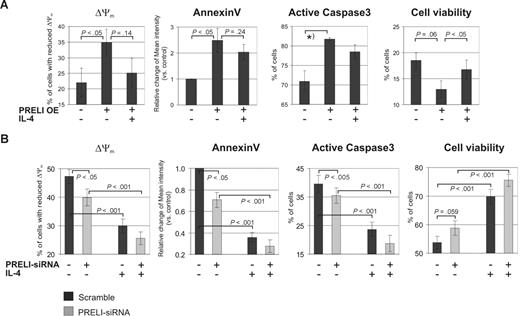
These findings led us to investigate whether PRELI plays a role in primary Th cell apoptosis. Furthermore, because IL-4 is known to protect cells from apoptosis by regulating proteins mediating the mitochondrial apoptosis pathway8,9 and by maintaining the mitochondrial membrane potential,10 we also investigated the combined effects of IL-4 and PRELI on Δψm and apoptosis. Human primary CD4+ cells nucleofected with the PRELI overexpression vector or PRELI-siRNA oligos (and their corresponding controls) were activated and cultured with or without IL-4 for 24 hours. The cells were stained with PI to determine cell viability (PI-negative cells) by flow cytometry. Early apoptosis was studied by detecting annexin V intensities in PI-negative cell populations. In parallel, the cells were also stained with active–caspase-3 antibody and Mitoprobe (Δψm). The annexin V intensity, as well as the number of cells with active caspase-3 and reduced Δψm, was higher in the samples overexpressing PRELI than in the control cells (Figure 3A), indicating that PRELI induces the mitochondrial apoptosis pathway in human Th cells. In addition, PRELI reduced cell viability. IL-4 partly inhibited the effects of PRELI, indicating that while PRELI and IL-4 have opposing effects on the mitochondrial apoptosis pathway, IL-4 cannot completely reverse the effect of PRELI. Consistent results were obtained in PRELI knockdown experiments (Figure 3B). The removal of PRELI significantly decreased annexin V intensity as well as the number of cells with reduced Δψm. Furthermore, the number of cells with active caspase-3 was slightly decreased. The presence of IL-4 further down-regulated these parameters, indicating that removal of PRELI and IL-4 have additive effects on the mitochondrial apoptosis pathway. Consistent with this, the cell viability was improved by PRELI-siRNAs and further up-regulated by IL-4. IL-4 treatment alone significantly increased cell viability and down-regulated the other parameters, as expected. The fact that the effect of PRELI on caspase-3 in the knockdown experiments was rather small may indicate that its influence is, at least partially, due to a caspase-independent apoptosis pathway, that has been previously described in human primary T lymphocytes. This pathway is characterized by ROS production, loss of Δψm, and phosphatidylserine exposure.23,24 Taken together, these results suggest that while PRELI and IL-4 counter-regulate the mitochondrial apoptosis pathway in human primary Th cells, the addition of IL-4 only partly reverses the effect of PRELI.
PRELI induces the mitochondrial apoptosis pathway in Th cells and is partly inhibited by IL-4. (A) Overexpression of PRELI induces mitochondria-mediated apoptosis and this is partly counteracted by IL-4. CD4+ cells nucleofected with pIRES-H2Kk or PRELI overexpression vector were sorted and activated as in Figure 2A, cultured with or without IL-4 for 24 hours, and stained and analyzed by flow cytometry. (B) Knockdown of PRELI and IL-4 treatment has additive effects on the mitochondrial apoptosis pathway. CD4+ cells nucleofected with scramble or PRELI-siRNAs were cultured, stained, and analyzed as in panel A. (A,B) Cell viability = PI-negative cells. Bars represent the mean values plus or minus SEM of 4 to 5 independent experiments, except that caspase-3 was measured in 2 independent overexpression experiments that included IL-4 treatment. In addition, caspase-3 was measured in 5 independent PRELI overexpression experiments without the IL-4 treatment (data not shown). In these experiments PRELI significantly increased the proportion of cells with active caspase-3 (*P < .05). In graphs of annexin V, bars represent the average fold changes plus or minus SEM of annexin V intensity between the untreated control sample and the other samples. The values of the untreated control samples were set as 1. Δψm was measured from living cell populations according to FSC/SSC plots (overexpression experiments) or total cell population (siRNA experiments). The percentage of cells with reduced Δψm was determined as described in Figure 2A. Statistical significances were calculated using the 2-tailed paired t test. OE indicates overexpression.
PRELI induces the mitochondrial apoptosis pathway in Th cells and is partly inhibited by IL-4. (A) Overexpression of PRELI induces mitochondria-mediated apoptosis and this is partly counteracted by IL-4. CD4+ cells nucleofected with pIRES-H2Kk or PRELI overexpression vector were sorted and activated as in Figure 2A, cultured with or without IL-4 for 24 hours, and stained and analyzed by flow cytometry. (B) Knockdown of PRELI and IL-4 treatment has additive effects on the mitochondrial apoptosis pathway. CD4+ cells nucleofected with scramble or PRELI-siRNAs were cultured, stained, and analyzed as in panel A. (A,B) Cell viability = PI-negative cells. Bars represent the mean values plus or minus SEM of 4 to 5 independent experiments, except that caspase-3 was measured in 2 independent overexpression experiments that included IL-4 treatment. In addition, caspase-3 was measured in 5 independent PRELI overexpression experiments without the IL-4 treatment (data not shown). In these experiments PRELI significantly increased the proportion of cells with active caspase-3 (*P < .05). In graphs of annexin V, bars represent the average fold changes plus or minus SEM of annexin V intensity between the untreated control sample and the other samples. The values of the untreated control samples were set as 1. Δψm was measured from living cell populations according to FSC/SSC plots (overexpression experiments) or total cell population (siRNA experiments). The percentage of cells with reduced Δψm was determined as described in Figure 2A. Statistical significances were calculated using the 2-tailed paired t test. OE indicates overexpression.
PRELI down-regulates Th2 cell differentiation
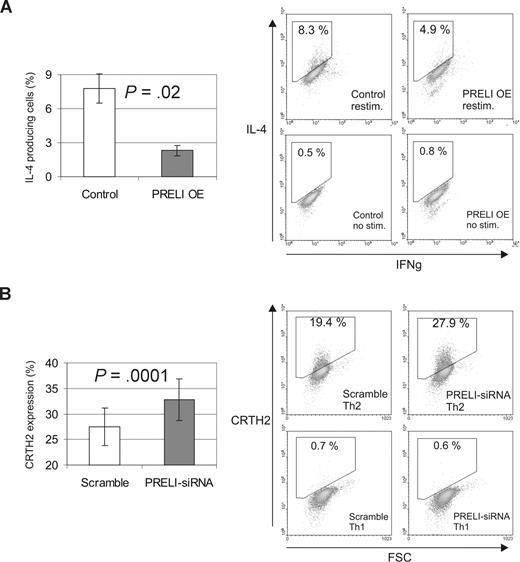
After establishing that PRELI and IL-4 counterregulate the mitochondrial apoptosis pathway in primary Th cells, we wanted to investigate the role of PRELI on Th-cell differentiation. For this, CD4+ cells nucleofected with a control or PRELI overexpression vector were polarized into Th1 and Th2 cells for 7 days. Subsequently, we measured the production of IFNγ and IL-4, the hallmark cytokines produced by Th1 and Th2 cells, respectively. Our results show that PRELI influences the differentiation of Th2 cells. The number of IL-4-producing Th2 cells was significantly reduced when PRELI was overexpressed in the cells (Figure 4A). Consistent data were obtained in PRELI knockdown experiments. Since the number of IL-4–producing cells after the first round of human Th2 cell polarization is typically less than 10%, as previously reported,25 we analyzed the effect of PRELI on the expression of another Th2 cell marker; the cell surface molecule CRTH2. CRTH2 is preferentially expressed on Th2 cells and its expression correlates strongly with the production of IL-4 and IL-13 in human Th2-skewed cultures.26,27 As expected, depletion of PRELI increased the number of CRTH2-positive Th2 cells compared with the control cells (Figure 4B). No reproducible effect on IFNγ production by Th1 cells could be detected in experiments involving PRELI overexpression (n = 3, P = .28) or knockdown (n = 9, P = .42; data not shown).
PRELI down-regulates Th2 cell polarization. (A) Overexpression of PRELI down-regulates the number of IL-4 producing Th2 cells. CD4+ cells nucleofected with a control (pIRES-H2Kk) or PRELI overexpression vector were polarized into Th2 cells for 7days. Subsequently, the cells were stimulated with phorbol myristate acetate + ionomycin to induce the cytokine production or not stimulated (negative controls). Subsequently, the production of IL-4 and IFNγ was determined by intracellular cytokine staining and flow cytometric analysis. (B) Knockdown of PRELI induces the expression of a Th2 cell surface marker CRTH2. CD4+ cells nucleofected with scramble or PRELI-siRNAs were cultured under Th1 and Th2 polarizing conditions for 6 to 7 days. After this, CRTH2 expression was analyzed by flow cytometry. Th1 cells were used as negative controls. (A,B) Bars represent the mean values plus or minus SEM, calculated from independent experiments (overexpression, n = 3; siRNA, n = 8). Statistical significances were determined using the 2-tailed paired t test. Dot plot figures show representative data from the experiments. OE indicates overexpression.
PRELI down-regulates Th2 cell polarization. (A) Overexpression of PRELI down-regulates the number of IL-4 producing Th2 cells. CD4+ cells nucleofected with a control (pIRES-H2Kk) or PRELI overexpression vector were polarized into Th2 cells for 7days. Subsequently, the cells were stimulated with phorbol myristate acetate + ionomycin to induce the cytokine production or not stimulated (negative controls). Subsequently, the production of IL-4 and IFNγ was determined by intracellular cytokine staining and flow cytometric analysis. (B) Knockdown of PRELI induces the expression of a Th2 cell surface marker CRTH2. CD4+ cells nucleofected with scramble or PRELI-siRNAs were cultured under Th1 and Th2 polarizing conditions for 6 to 7 days. After this, CRTH2 expression was analyzed by flow cytometry. Th1 cells were used as negative controls. (A,B) Bars represent the mean values plus or minus SEM, calculated from independent experiments (overexpression, n = 3; siRNA, n = 8). Statistical significances were determined using the 2-tailed paired t test. Dot plot figures show representative data from the experiments. OE indicates overexpression.
STAT6 is down-regulated by PRELI and oxidative stress in human primary T helper cells
To dissect the mechanism behind PRELI's effect on Th2 differentiation, we looked for any possible effect on STAT6, the key transcription factor involved in driving Th2 differentiation. Our results show that PRELI down-regulates the amount of STAT6 at the protein level. Removal of PRELI from cord blood or peripheral blood CD4+ cells led to up-regulation of STAT6 protein levels, whereas STAT6 mRNA levels were not notably affected (Figure 5A). This indicates that PRELI regulates STAT6 at the posttranscriptional level. We also investigated the effect of PRELI on the Th1-specific transcription factors STAT1 and STAT4. The depletion of PRELI had no reproducible effect on STAT1 or STAT4 proteins in 7 (P = .97) or 6 (P = .35) independent experiments, respectively (Figure 5A and data not shown).
PRELI and oxidative stress down-regulate STAT6 and activate calpain, a known STAT6 protease. (A) Knockdown of PRELI leads to increase of STAT6 at the protein level. Nucleofected CD4+ cells were activated for 24 hours and subsequently harvested for Taqman RT-PCR analysis and Western blotting. Protein bands were quantified with a microcomputer imaging device (MCID) and normalized against β-actin. Bars represent the average fold changes plus or minus SEM, calculated from independent experiments (STAT6 protein, n = 10; STAT6 mRNA, n = 4). (B) PRELI does not influence the tyrosine (Y641) phosphorylation of STAT6. CD4+ cells nucleofected with pIRES-H2Kk or the PRELI overexpression vector were activated and cultured in the presence of IL-4 for 1 hour and subsequently harvested for Western blotting. Protein bands were quantified and normalized, as in panel A. Representative data from 5 independent experiments are shown. (C) STAT6 is down-regulated at the protein level by oxidative stress in human primary Th cells. Cord blood CD4+ cells were activated for 20 hours and subsequently exposed to increasing concentrations of H2O2 for 4 hours. The protein and mRNA levels of STAT6 were analyzed by Western blotting and Taqman RT-PCR analysis. A portion of the cells was stained with DHR123, to assess ROS production, and analyzed by flow cytometry. Representative data from 4 independent experiments are shown. (D) Neutralization of intracellular ROS increases the level of STAT6. CD4+ cells were activated and cultured with or without 1.25 mM NAC for 24 hours, after which the cells were harvested for Western blotting. STAT6 protein bands were quantified and normalized as in panel A. Representative data from 6 independent experiments are shown. (E) Oxidative stress induces calpain activation in human Th cells. Exposure of the cells to H2O2 was performed as in panel C (black arrow, inactive procalpain [80 kDa]; gray arrow, active calpain [75 kDa]). The active and inactive calpain bands were quantified with an MCID and the active versus inactive calpain ratios were calculated in each sample. Subsequently, the active/inactive calpain ratio of the control (untreated) sample was set as 1 and the fold changes of the ratios of the other samples were calculated in relation to the control sample. Bars represent the mean values plus or minus SEM of the fold changes of different replicates. Representative data from 2 experiments are shown. (F) STAT6 is up-regulated in response to calpain inhibition. CD4+ cells were activated and cultured for 24 hours with or without calpastatin peptide (5 μM) and subsequently harvested for Western blotting. STAT6 protein bands were quantified with an MCID and normalized against β-actin. Vertical lines have been inserted to indicate a repositioned gel lane. The relative ratios of active versus inactive calpain were determined as described in panel E. Representative data from 5 independent experiments are shown. (G) Overexpression of PRELI in Hek293 cells induces activation of calpain. Hek293 cells transfected with a control vector (empty pFlag) or pFlag-PRELI were harvested for Western blotting 24 hours after transfection. The relative ratios of active versus inactive calpain were determined as described in panel E. Data are representative of 2 independent experiments. (A,B,D,F) Statistical significances were determined using the 2-tailed paired t test. OE indicates overexpression.
PRELI and oxidative stress down-regulate STAT6 and activate calpain, a known STAT6 protease. (A) Knockdown of PRELI leads to increase of STAT6 at the protein level. Nucleofected CD4+ cells were activated for 24 hours and subsequently harvested for Taqman RT-PCR analysis and Western blotting. Protein bands were quantified with a microcomputer imaging device (MCID) and normalized against β-actin. Bars represent the average fold changes plus or minus SEM, calculated from independent experiments (STAT6 protein, n = 10; STAT6 mRNA, n = 4). (B) PRELI does not influence the tyrosine (Y641) phosphorylation of STAT6. CD4+ cells nucleofected with pIRES-H2Kk or the PRELI overexpression vector were activated and cultured in the presence of IL-4 for 1 hour and subsequently harvested for Western blotting. Protein bands were quantified and normalized, as in panel A. Representative data from 5 independent experiments are shown. (C) STAT6 is down-regulated at the protein level by oxidative stress in human primary Th cells. Cord blood CD4+ cells were activated for 20 hours and subsequently exposed to increasing concentrations of H2O2 for 4 hours. The protein and mRNA levels of STAT6 were analyzed by Western blotting and Taqman RT-PCR analysis. A portion of the cells was stained with DHR123, to assess ROS production, and analyzed by flow cytometry. Representative data from 4 independent experiments are shown. (D) Neutralization of intracellular ROS increases the level of STAT6. CD4+ cells were activated and cultured with or without 1.25 mM NAC for 24 hours, after which the cells were harvested for Western blotting. STAT6 protein bands were quantified and normalized as in panel A. Representative data from 6 independent experiments are shown. (E) Oxidative stress induces calpain activation in human Th cells. Exposure of the cells to H2O2 was performed as in panel C (black arrow, inactive procalpain [80 kDa]; gray arrow, active calpain [75 kDa]). The active and inactive calpain bands were quantified with an MCID and the active versus inactive calpain ratios were calculated in each sample. Subsequently, the active/inactive calpain ratio of the control (untreated) sample was set as 1 and the fold changes of the ratios of the other samples were calculated in relation to the control sample. Bars represent the mean values plus or minus SEM of the fold changes of different replicates. Representative data from 2 experiments are shown. (F) STAT6 is up-regulated in response to calpain inhibition. CD4+ cells were activated and cultured for 24 hours with or without calpastatin peptide (5 μM) and subsequently harvested for Western blotting. STAT6 protein bands were quantified with an MCID and normalized against β-actin. Vertical lines have been inserted to indicate a repositioned gel lane. The relative ratios of active versus inactive calpain were determined as described in panel E. Representative data from 5 independent experiments are shown. (G) Overexpression of PRELI in Hek293 cells induces activation of calpain. Hek293 cells transfected with a control vector (empty pFlag) or pFlag-PRELI were harvested for Western blotting 24 hours after transfection. The relative ratios of active versus inactive calpain were determined as described in panel E. Data are representative of 2 independent experiments. (A,B,D,F) Statistical significances were determined using the 2-tailed paired t test. OE indicates overexpression.
In addition, we investigated whether the IL-4 induced tyrosine (Y641) phosphorylation of STAT6 was affected in response to PRELI in CD4+ cells. Peripheral blood CD4+ cells overexpressing PRELI or the empty control vector were activated and cultured in the presence of IL-4 for 30 minutes, 1 hour or 24 hours. Subsequently, the levels of total STAT6 and phosphorylated STAT6 were determined by Western blotting. The amount of phosphorylated STAT6 was also reduced to the same extent as that of the total protein (Figure 5B). This indicates that PRELI down-regulates levels of the STAT6 protein with no additional effect on its IL-4–dependent phosphorylation.
To clarify the mechanism by which PRELI down-regulates STAT6, we examined whether STAT6 was regulated by the cellular redox state. Exposure of cells to H2O2 is often used as a model of oxidative stress. We observed that the amount of STAT6 protein decreased in a dose-dependent manner in response to increasing cellular ROS concentration, whereas the expression of STAT6 mRNA was not similarly affected (Figure 5C). This indicates that STAT6 is down-regulated in human primary Th cells by oxidative stress at the posttranscriptional level. Levels of STAT1 and STAT4 were not similarly affected by H2O2 as the level of STAT6 was (Figure 5C and data not shown), excluding the possibility of nonspecific, general protein degradation induced by the H2O2 treatment.
Consistent results were obtained when cellular ROS were neutralized by an antioxidant N-acetyl-cysteine (NAC). The amount of STAT6 was higher in the cells treated with NAC compared with the nontreated cells, confirming that ROS down-regulate STAT6 protein levels (Figure 5D). Down-regulation of ROS by NAC was verified by DHR123-staining and flow cytometric analysis (data not shown). Again, STAT1 or STAT4 were not affected in the same way as STAT6 (Figure 5D and data not shown).
Calpain, an oxidative stress–induced cysteine protease that down-regulates STAT6, is regulated by PRELI
STAT6 has been reported to be negatively regulated by the cysteine protease calpain in mouse T-cell lines and mast cells.12,13 Calpain, in turn, is activated by oxidative stress.28,29 Therefore, we wanted to investigate whether calpain was a mediator of PRELI-induced down-regulation of STAT6 in human primary Th cells.
Oxidative stress (H2O2) induces the cleavage/activation of inactive procalpain (80 kDa) to active calpain (75 kDa) in cord blood CD4+ cells (Figure 5E). In addition, as reported in other cell types, we observed that STAT6 is down-regulated by calpain in human primary Th cells. Calpastatin, an endogenous calpain inhibitor, prevents the autoproteolytic cleavage of the inactive procalpain to an active form, leading to the accumulation of inactive procalpain in the cell.30 In response to calpastatin treatment, the amount of inactive calpain in the cells was increased, indicating that this inhibitor reduced calpain activation. In addition, levels of STAT6 were also increased in response to calpain inhibition, supporting the conclusion that STAT6 is down-regulated by calpain in primary Th cells (Figure 5F). Furthermore, overexpression of PRELI in Hek293 cells clearly induced the cleavage/activation of calpain (Figure 5G). Taken together, these results suggest that down-regulation of STAT6 by PRELI could be mediated by oxidative stress and calpain activation.
The strength of the TCR stimulus influences the expression of PRELI and Th2 differentiation
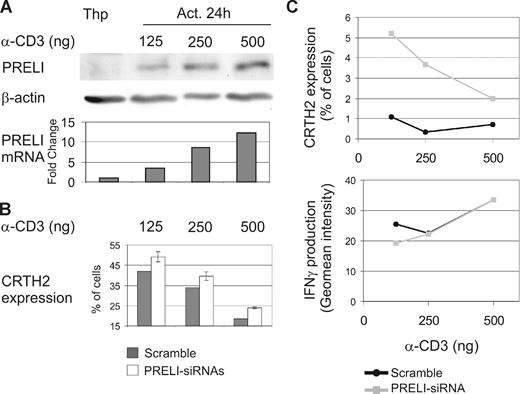
It is well established that the strength of the TCR signal influences Th1/Th2 polarization. A strong TCR stimulus promotes Th1 differentiation, while weaker TCR stimuli favor Th2 polarization.4,5 Interestingly, the intracellular levels of PRELI also correlated with the strength of the TCR activation. CD4+ cells were activated with different concentrations of α-CD3 (125, 250, or 500 ng/well) and α-CD28 for 24 hours and subsequently harvested for Western blotting and Taqman RT-PCR analysis. The extent of PRELI induced, at both the protein and mRNA levels, was clearly proportional to the concentration of α-CD3 used for stimulation (Figure 6A). In addition, CD4+ cells nucleofected with scrambled- or PRELI-siRNA oligos were activated as described earlier in this section and cultured under Th1- or Th2-polarizing conditions for7 days. The expression of CRTH2, a Th2 cell marker, was then analyzed by flow cytometry. Our results show that Th2 differentiation was inhibited when primary Th cells were activated with a strong TCR stimulus, whereas Th2 differentiation was enhanced when PRELI was knocked down (Figure 6B). These results indicate that the strength of the TCR activation, as well as the amount of PRELI in cells, influences the ability of a naive CD4+ cell to differentiate into Th2 cell (Figures 4 and 6B). Importantly, our results suggest that PRELI is a mediator, at least in part, in determining the effects of the strength of TCR stimulus on Th2 differentiation.
A strong TCR stimulus induces the expression of PRELI, and they both inhibit Th2 cell development. (A) PRELI is induced at the mRNA and protein levels by increasing strength of the TCR stimulus. Naive CD4+ cells were activated with different concentrations of α-CD3 (125, 250, or 500 ng/well) and α-CD28 for 24 hours and subsequently harvested for Western blotting and Taqman RT-PCR analysis. Representative data from 4 independent experiments are shown. (B) The strength of the TCR stimulus and the amount of PRELI in cells influence Th2 differentiation. CD4+ cells nucleofected with scrambled- or PRELI-siRNA oligos (1 or 2) were activated as in panel A and cultured under Th1- or Th2-polarizing conditions for 7 days. The cells were stained with CRTH2-PE and analyzed by flow cytometry. Th1 cells were used as negative controls. Representative data from 2 independent experiments are shown. White bars represent the mean value plus or minus SEM, calculated from PRELI-siRNA1 and PRELI-siRNA2 cultures from the representative experiments. (C) PRELI influences the relative proportion of Th cells under conditions that enable both Th1 and Th2 polarization. CD4+ cells nucleofected and activated (as described earlier in this paragraph) were cultured in the presence of both IL-12 (2.5 ng/mL) and IL-4 (10 ng/mL). After 7 days of culture, expression of CRTH2 was analyzed (as described earlier in this paragraph). Production of IFNγ was measured by intracellular cytokine staining. Representative data from 2 biologic replicates with PRELI-siRNA2 are shown.
A strong TCR stimulus induces the expression of PRELI, and they both inhibit Th2 cell development. (A) PRELI is induced at the mRNA and protein levels by increasing strength of the TCR stimulus. Naive CD4+ cells were activated with different concentrations of α-CD3 (125, 250, or 500 ng/well) and α-CD28 for 24 hours and subsequently harvested for Western blotting and Taqman RT-PCR analysis. Representative data from 4 independent experiments are shown. (B) The strength of the TCR stimulus and the amount of PRELI in cells influence Th2 differentiation. CD4+ cells nucleofected with scrambled- or PRELI-siRNA oligos (1 or 2) were activated as in panel A and cultured under Th1- or Th2-polarizing conditions for 7 days. The cells were stained with CRTH2-PE and analyzed by flow cytometry. Th1 cells were used as negative controls. Representative data from 2 independent experiments are shown. White bars represent the mean value plus or minus SEM, calculated from PRELI-siRNA1 and PRELI-siRNA2 cultures from the representative experiments. (C) PRELI influences the relative proportion of Th cells under conditions that enable both Th1 and Th2 polarization. CD4+ cells nucleofected and activated (as described earlier in this paragraph) were cultured in the presence of both IL-12 (2.5 ng/mL) and IL-4 (10 ng/mL). After 7 days of culture, expression of CRTH2 was analyzed (as described earlier in this paragraph). Production of IFNγ was measured by intracellular cytokine staining. Representative data from 2 biologic replicates with PRELI-siRNA2 are shown.
The relative proportion of Th1 versus Th2 cells generated plays a critical role in determining the outcome of the host immune response to an infection. Identification of the various factors involved in regulating this balance remains an active area of investigation. It was therefore of interest to examine the consequence of PRELI depletion under conditions where differentiation of both Th1 and Th2 was simultaneously promoted. For this, we cultured CD4+ T cells nucleofected and activated as described earlier in this section in the presence of both IL-12 and IL-4 for 7 days and subsequently measured the CRTH2 expression and IFNγ production by these cells. As shown in Figure 6C (scramble), Th2 cell generation was very low at all concentrations of α-CD3. We interpret these findings to indicate that the effects of IL-12 override those of IL-4, at least at the level of Th differentiation. Interestingly, however, knockdown of PRELI resulted in a significant increase in Th2 cell generation compared with the scramble control. In contrast, PRELI silencing had no detectable effect on IFNγ-producing cell populations (Figure 6C). On one level, these results provide additional support for the specific effects of PRELI in regulating Th2 cell differentiation. In addition, however, they also suggest a role for this effect in defining the relative proportion of Th1 versus Th2 cells that can be obtained from naive precursor cells.
Discussion
In this study, we have identified PRELI as a novel, activation-induced regulator of T helper cell apoptosis and differentiation. Our results suggest that these new functions for PRELI are mediated by an overlapping mechanism, by the regulation of the cellular redox state.
The mechanism by which PRELI affects the Δψm and ROS production is presently unknown. In Saccahromyces cerevisiae, Ups1p, the homologue of human PRELI (31% identity), regulates mitochondrial shape and the processing of the GTPase Mgm1p via an unknown mechanism.31 Because human PRELI was able to replace Ups1p in Mgm1p processing in yeast cells, it was considered to be likely that in human cells PRELI acts on optic atrophy 1 (OPA1), the human homologue of Mgm1p. OPA1 is an important regulator of mitochondrial membrane dynamics and apoptosis in human cells.32,33 Different isoforms and cleavage products of OPA1 have different effects on mitochondrial function and apoptosis, and their abundance varies greatly between organs. The differential processing of these isoforms plays an important role in the regulation of mitochondrial function and susceptibility to apoptosis. Interestingly, the variant that induces cytochrome c and caspase-mediated apoptosis is found predominantly in liver, kidney, and thymus.34 This resembles the expression of PRELI, which has also been suggested to be important for the development of vital and immunocompetent organs.15
Our preliminary results (unpublished data) suggest that PRELI does not affect the expression levels of OPA1 isoforms in human CD4+ cells. However, whether PRELI influences the function of OPA1, remains to be determined. It also needs to be emphasized that distinctions in OPA1 processing exist between yeast and humans.35 Therefore, the mechanisms involved in yeast do not necessarily apply to humans. Furthermore, the yeast homologue of PRELI, Ups1p, was suggested to have additional targets in mitochondria,31 suggesting that PRELI may also engage substrates other than OPA1.
Given that IL-4 partially neutralized the proapoptotic effects of PRELI, we expected that PRELI could possibly have greater impact on Th1 than Th2 differentiation. However, we found that PRELI negatively influences Th2 cell polarization, with no reproducible effect on IFNγ production by Th1 cells. Consistent with these findings, PRELI was found to down-regulate STAT6, the key molecule driving IL-4–induced Th2 differentiation. The effect of PRELI on STAT6 was restricted to the protein level. Importantly, our findings suggest that this function of PRELI was mediated through ROS-dependent activation of calpain. Taken together, our results suggest that PRELI, by influencing the intracellular redox state, influences Th2-cell development on 2 levels. It induces the mitochondrial apoptosis pathway in these cells, thus counteracting the protective effect of IL-4. In addition, it down-regulates STAT6 and Th2 differentiation.
It is known that Th1/Th2 cell differentiation is influenced by the strength of the activation stimulus. A strong TCR stimulus and enhanced calcium signaling increase the activity of calpains in T lymphocytes.36,37 These processes have all been shown to favor Th1 over Th2 differentiation.4,38,39 It has previously been suggested that strong TCR signals may influence T cell differentiation by promoting strong calcium signaling, calpain activation, and a subsequent degradation of STAT6.12,13,39 We showed that PRELI is highly up-regulated in response to T cell activation and that its expression correlates with the strength of the TCR stimulus. In addition, both a strong TCR stimulus and PRELI were shown to inhibit Th2 differentiation. Our data suggest that PRELI is involved in the mechanism through which the strength of the TCR stimulus influences the polarization of Th cells. Thus, it is possible that, in vivo, a strong TCR stimulus, besides enhancing calcium signaling and subsequent calpain activation, strongly induces the expression of PRELI, leading to increased degradation of STAT6 and decreased Th2 development. Similarly, a weaker activation would induce less PRELI and allow more potent Th2 differentiation. Importantly, our results also suggest a role for PRELI in influencing the relative proportions of Th cell types under conditions that enable both Th1 and Th2 polarization. Overall, this study identifies PRELI as a novel factor influencing human primary Th cell death and Th2 differentiation, thus providing new insights into the signaling mechanisms that control the selective activation of human Th cell subsets.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Antti Ellonen, Bhawna Gupta, Marjo Hakkarainen, Sarita Heinonen, and Mika Venojärvi for their expertise, contribution, and technical assistance in this study. This work was supported by the Academy of Finland (Helsinki, Finland); the Sigrid Jusélius Foundation (Helsinki, Finland); the Department of Biotechnology, Government of India (New Delhi, India); the National Technology Agency (TEKES; Helsinki, Finland); the Turku University Hospital Fund (Turku, Finland); the Väinö and Laina Kivi Foundation (Huittinen, Finland); the Pulmonary Association Heli (Helsinki, Finland); the Ida Montin Foundation (Helsinki, Finland); and the Finnish Cultural Foundation (Helsinki, Finland).
Authorship
Contribution: J.T., K.V.S.R., and R.L. designed the research; J.T., T.K., and H.L. performed experiments; J.T. analyzed results and made the figures; J.T., K.M.H., K.V.S.R., and R.L. interpreted data; J.T., K.V.S.R., and R.L. wrote the paper; K.M.H. and J.W. provided expertise and guidance; and K.M.H., K.V.S.R., and R.L. contributed reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Riitta Lahesmaa, Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, PO Box 123, FIN-20521, Turku, Finland; e-mail: riitta.lahesmaa@btk.fi.

![Figure 5. PRELI and oxidative stress down-regulate STAT6 and activate calpain, a known STAT6 protease. (A) Knockdown of PRELI leads to increase of STAT6 at the protein level. Nucleofected CD4+ cells were activated for 24 hours and subsequently harvested for Taqman RT-PCR analysis and Western blotting. Protein bands were quantified with a microcomputer imaging device (MCID) and normalized against β-actin. Bars represent the average fold changes plus or minus SEM, calculated from independent experiments (STAT6 protein, n = 10; STAT6 mRNA, n = 4). (B) PRELI does not influence the tyrosine (Y641) phosphorylation of STAT6. CD4+ cells nucleofected with pIRES-H2Kk or the PRELI overexpression vector were activated and cultured in the presence of IL-4 for 1 hour and subsequently harvested for Western blotting. Protein bands were quantified and normalized, as in panel A. Representative data from 5 independent experiments are shown. (C) STAT6 is down-regulated at the protein level by oxidative stress in human primary Th cells. Cord blood CD4+ cells were activated for 20 hours and subsequently exposed to increasing concentrations of H2O2 for 4 hours. The protein and mRNA levels of STAT6 were analyzed by Western blotting and Taqman RT-PCR analysis. A portion of the cells was stained with DHR123, to assess ROS production, and analyzed by flow cytometry. Representative data from 4 independent experiments are shown. (D) Neutralization of intracellular ROS increases the level of STAT6. CD4+ cells were activated and cultured with or without 1.25 mM NAC for 24 hours, after which the cells were harvested for Western blotting. STAT6 protein bands were quantified and normalized as in panel A. Representative data from 6 independent experiments are shown. (E) Oxidative stress induces calpain activation in human Th cells. Exposure of the cells to H2O2 was performed as in panel C (black arrow, inactive procalpain [80 kDa]; gray arrow, active calpain [75 kDa]). The active and inactive calpain bands were quantified with an MCID and the active versus inactive calpain ratios were calculated in each sample. Subsequently, the active/inactive calpain ratio of the control (untreated) sample was set as 1 and the fold changes of the ratios of the other samples were calculated in relation to the control sample. Bars represent the mean values plus or minus SEM of the fold changes of different replicates. Representative data from 2 experiments are shown. (F) STAT6 is up-regulated in response to calpain inhibition. CD4+ cells were activated and cultured for 24 hours with or without calpastatin peptide (5 μM) and subsequently harvested for Western blotting. STAT6 protein bands were quantified with an MCID and normalized against β-actin. Vertical lines have been inserted to indicate a repositioned gel lane. The relative ratios of active versus inactive calpain were determined as described in panel E. Representative data from 5 independent experiments are shown. (G) Overexpression of PRELI in Hek293 cells induces activation of calpain. Hek293 cells transfected with a control vector (empty pFlag) or pFlag-PRELI were harvested for Western blotting 24 hours after transfection. The relative ratios of active versus inactive calpain were determined as described in panel E. Data are representative of 2 independent experiments. (A,B,D,F) Statistical significances were determined using the 2-tailed paired t test. OE indicates overexpression.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/6/10.1182_blood-2008-07-166553/4/m_zh80020929350005.jpeg?Expires=1769105019&Signature=iyqvjjNUpd4mX-y5la2MsGwl9YPvyhXs39Jlq2MDg-VBE9xSttVcNYH0dAvf513nm2Or8Psz~anH~obs-6Spt9ldqs~tBpRhIY8n41f0ysMy4XLjKDntsrrBvwvjvKy7zNtXHU5wCxX3KeDh3-A2VxJ~eaxvRNd1A8KC~Mvi~TTYdzFvp~aF1~NSDft-NXVsyijZ8jhUIP5MT8KtFXWDhvVdRYqFKZXEuH-jFeWvCwXGS226AkemcPuRHcfQKQg9zGmZ7t4~d1Wy4IvNjQdUEb89EofLnAmQFwg4n39tljzNg0FmvhmcojqRNMqZFjzlNw6LgP0t2L6AZeraOSFjcQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal