Abstract
No validated biomarkers exist for acute graft-versus-host disease (GVHD). We screened plasma with antibody microarrays for 120 proteins in a discovery set of 42 patients who underwent transplantation that revealed 8 potential biomarkers for diagnostic of GVHD. We then measured by enzyme-linked immunosorbent assay (ELISA) the levels of these biomarkers in samples from 424 patients who underwent transplantation randomly divided into training (n = 282) and validation (n = 142) sets. Logistic regression analysis of these 8 proteins determined a composite biomarker panel of 4 proteins (interleukin-2-receptor-alpha, tumor-necrosis-factor-receptor-1, interleukin-8, and hepatocyte growth factor) that optimally discriminated patients with and without GVHD. The area under the receiver operating characteristic curve distinguishing these 2 groups in the training set was 0.91 (95% confidence interval, 0.87–0.94) and 0.86 (95% confidence interval, 0.79–0.92) in the validation set. In patients with GVHD, Cox regression analysis revealed that the biomarker panel predicted survival independently of GVHD severity. A panel of 4 biomarkers can confirm the diagnosis of GVHD in patients at onset of clinical symptoms of GVHD and provide prognostic information independent of GVHD severity.
Introduction
Acute graft-versus-host disease (GVHD) occurs in approximately half of patients following allogeneic hematopoietic cell transplantation (HCT), a curative therapy for several malignant and nonmalignant disorders. The diagnosis of acute GVHD is based on clinical criteria that may be confirmed by biopsy of one of the 3 target organs (skin, gastrointestinal tract, or liver). The severity of acute GVHD is graded clinically from I to IV using a standardized system that evaluates 3 principal target organs,1 with increased mortality rates with significant GVHD (grades II–IV).2
There is no validated diagnostic blood test for acute GVHD, although multiple blood proteins have been described as potential biomarkers in small studies.3-23 Differences in any single protein have lacked sufficient specificity and sensitivity to be of clinical use. Although recent mass spectrometric (MS) profiling of urine24,25 and serum26 demonstrate the presence of spectral patterns associated with GVHD, these approaches do not identify specific proteins. We have previously reported a quantitative analysis of several potential biomarkers for GVHD in the plasma of a small number of patients.27 The aim of the current study was to expand the search for GVHD biomarkers, to validate candidate proteins using high-throughput assays in a large number of patient samples, and to determine their significance with respect to clinical outcomes.
Methods
Study population
The entire study population consisted of 466 subjects who underwent allogeneic hematopoietic stem cell transplantation performed between 2001 and 2006 at the University of Michigan. Twenty-one patients who developed severe acute GVHD (maximum grade III-IV with 76% GVHD-related mortality at day 200, GVHD+severe) and 21 patients who never developed GVHD (GVHD−) comprised a discovery set, 282 patients (GVHD− = 166, GVHD+ = 116) comprised a training set, and 142 patients (GVHD− = 76, GVHD+ = 66) comprised a validation set. All samples were collected prospectively under protocols approved by the University of Michigan institutional review board. Blood was drawn on days 0, 7, 14, 21, 28, 56, and 100 after transplantation and within 24 hours of diagnosis of GVHD. GVHD was staged per modified Glucksberg criteria1 and was confirmed by biopsy in more than 95% of patients, including those with grade I GVHD. Patients with clinical signs of acute GVHD as their only major complication were selected, and patients with veno-occlusive disease (VOD), idiopathic pneumonia syndrome (IPS), or septic shock, who represent 15% of allogeneic recipients at our center, were excluded. We used diagnosis samples from GVHD+ patients that were taken at the time of diagnosis and we selected samples from GVHD− patients so that both groups of samples were balanced for time of acquisition. The 2 groups were also balanced for age, intensity of the conditioning regimen (reduced versus full), and donor source (related versus unrelated). GVHD prophylaxis was methotrexate and tacrolimus of standard duration for full-intensity conditioning or methotrexate and mycophenolate mofetil of standard duration for reduced-intensity conditioning.
Sample preparation
Plasma was obtained after Ficoll (Amersham, Piscataway, NJ) gradient centrifugation of 20 mL whole blood that had been collected in heparin-containing Vacutainers tubes (Becton Dickinson, Franklin Lakes, NJ) to prevent clotting. Samples were aliquoted without additives into cryovials and stored at −80°C.
Antibody array
We used commercial arrays (Schleicher & Schuell ‘S&S’, Whatman, Keane, NH) of dotted antibodies to 120 proteins (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and an indirect labeling procedure to competitively hybridize each individual plasma sample. A reference pool was formed by pooling equal amounts of all of the plasma samples to obtain the ratio of the intensities of the resulting fluorescence signals (test sample/reference), as detailed in Document S1.
Sequential ELISA
A sequential enzyme-linked immunosorbent assay (ELISA) protocol was used to maximize the number of measured analytes per sample. This sequential protocol measures multiple analytes per plasma sample by reusing the same aliquot consecutively in individual ELISA plates. Antibody pairs were purchased as follows: CRP from Bender Medsystems (Vienna, Austria), IL-8 from Becton Dickinson (BD OptEIA), CA19.9 from Bio-Quant (San Diego, CA), and all others from R&D systems (Duoset; Minneapolis, MN). Samples and standards were analyzed in duplicate according to a previously described protocol.28 Briefly, 3 capture antibodies for 3 analytes were coated in 3 different 96-well Immulon 4HBX plates (Thermo Fisher Scientific, Waltham, MA) overnight at 4°C. After plate I was prepared (washed and blocked for 1 hour), standards and samples were diluted as necessary (IL-2Rα undiluted; HGF ½; IL-8 ¼, and TNFR1  ) and added for 2 hours at room temperature (RT) while mixing on an orbital shaker. Plate II was prepared so that samples were transferred to plate II from plate I at the end of its incubation period. Plate I was then washed, and biotinylated antibody was added and incubated for 2 hours. Streptavidin-conjugated horseradish peroxidase (SA-HRP) was diluted at 1:10 000 and then added for 30 minutes. The TMB color substrate (Becton Dickinson) was prepared and incubated for 10 to 30 minutes, the reaction terminated by addition of 1.5 M sulfuric acid, and the absorbance immediately determined using a SpectraMax 190 plate reader (Molecular Devices, Sunnyvale, CA). Consecutive cycles proceeded similarly for each plate. Interassay variability coefficients (CVs) among the 11 plates used for analyzing the 428 samples were 1.48% for IL-2Rα, 3.02% for TNFR1, 1.49% for IL-8, and 1.23% for HGF.
) and added for 2 hours at room temperature (RT) while mixing on an orbital shaker. Plate II was prepared so that samples were transferred to plate II from plate I at the end of its incubation period. Plate I was then washed, and biotinylated antibody was added and incubated for 2 hours. Streptavidin-conjugated horseradish peroxidase (SA-HRP) was diluted at 1:10 000 and then added for 30 minutes. The TMB color substrate (Becton Dickinson) was prepared and incubated for 10 to 30 minutes, the reaction terminated by addition of 1.5 M sulfuric acid, and the absorbance immediately determined using a SpectraMax 190 plate reader (Molecular Devices, Sunnyvale, CA). Consecutive cycles proceeded similarly for each plate. Interassay variability coefficients (CVs) among the 11 plates used for analyzing the 428 samples were 1.48% for IL-2Rα, 3.02% for TNFR1, 1.49% for IL-8, and 1.23% for HGF.
Statistical methods
ELISA data in the discovery set were monotonically transformed using the logarithm of the marker value plus one tenth of its median value over all samples. GVHD− and GVHD+ groups were compared in the discovery set using 2-sample t tests. Raw values from ELISA assays were used in the training and validation sets. Differences in patient characteristics between the training and validation sets were assessed with Kruskal-Wallis tests for continuous values and chi-square tests of association for categoric values. Protein level differences between groups were assessed with Wilcoxon rank sum tests. Area under the curve (AUC) and corresponding 95% confidence intervals were computed nonparametrically. Logistic regression was used to develop a composite panel of biomarkers that discriminated between GVHD+ and GVHD− patients. Overall survival was modeled using Cox regression methods and nonrelapse mortality was modeled using cumulative incidence regression methods as described in Fine and Gray.29
Results
Identification of potential biomarkers of acute GVHD in a discovery set
We hypothesized that samples from patients whose GVHD was severe would be most likely to yield informative biomarkers. We therefore performed a discovery study that compared samples from 21 patients with severe acute GVHD (GVHD+severe) with samples from 21 patients without GVHD who were similar in age, intensity of the conditioning regimen (reduced versus full), donor source (related versus unrelated), and time of sample acquisition (Table S2). Samples from patients with VOD, IPS, or septic shock were excluded. We analyzed these samples using an antibody microarray that targeted diverse classes of proteins including acute-phase reactants, cytokines, angiogenic factors, tumor markers, leukocyte adhesion molecules, and metalloproteinases or their inhibitors (Table S1). The 35 biomarkers that demonstrated the most significant differences between groups are shown in Figure S1. For full information on these microarrays, please see the online supplement.
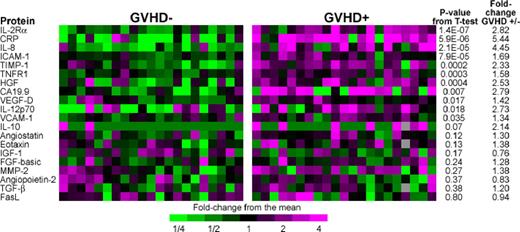
We selected 23 of these biomarkers to measure by sequential ELISA, based on the magnitude of differences between groups, their potential immunologic relevance, and availability of reliable reagents (Figure 1). Eight proteins (IL-2Rα, CRP, IL-8, ICAM-1, TIMP-1, TNFR1, HGF, CA19.9) gave P value less than .01 for 2-sample t tests comparing patients with and without GVHD, and all 8 showed larger fold differences between groups than the corresponding array assays. Leave one out cross validation (LOOCV) analysis of classifiers demonstrated that inclusion of between 7 and 11 biomarkers provided the best discrimination between groups (Figure S2). The AUC using the 8 most significant biomarkers in LOOCV was 0.94 (Figure S3).
ELISA heat map of discovery set samples. We performed sequential ELISA as described in “Antibody array methods” on samples from 21 GVHD− patients (left panel) and 21 GVHD+ patients (right panel) of the discovery set that were analyzed by microarray in Figure S1. Gray indicates that the sample was not assayed for that protein. Levels of PSA-ACT, IL-17, and IL-1β were not detectable and therefore do not appear in the figure. We measured tumor-necrosis-factor-receptor-1 (TNFR1) rather than TNF-α because of its close correlation to TNF-α and documented reproducibility of measurement in previously frozen samples.1 P values compare GVHD+ and GVHD− samples.
ELISA heat map of discovery set samples. We performed sequential ELISA as described in “Antibody array methods” on samples from 21 GVHD− patients (left panel) and 21 GVHD+ patients (right panel) of the discovery set that were analyzed by microarray in Figure S1. Gray indicates that the sample was not assayed for that protein. Levels of PSA-ACT, IL-17, and IL-1β were not detectable and therefore do not appear in the figure. We measured tumor-necrosis-factor-receptor-1 (TNFR1) rather than TNF-α because of its close correlation to TNF-α and documented reproducibility of measurement in previously frozen samples.1 P values compare GVHD+ and GVHD− samples.
Development of a diagnostic panel of GVHD by logistic regression
We next analyzed samples that had been collected prospectively from a separate group of 424 patients who underwent allogeneic HCT. These samples were randomly divided into a training set (GVHD− = 166, GVHD+ = 116) and a validation set (GVHD− = 76, GVHD+ = 66). GVHD+ groups included all GVHD grades (I-IV). The clinical characteristics of these patients are shown in Table 1. Patients in the training and validation sets were randomly chosen and there were no significant differences between sets for median age, nonmalignant disease, conditioning intensity, donor source, HLA match, or GVHD grade. The median day of sample acquisition was day 30 in the training set and day 29 in the validation set. Older recipients and recipients of grafts from donors who were not family members or who were less than 8/8 HLA identical matches were overrepresented in the GVHD groups.
Study population characteristics in the training set and validation set
| Characteristic . | Training cohort, N = 282 . | P value for difference between GVHD− and GVHD+ . | Validation cohort, N = 142 . | P value for difference between GVHD− and GVHD+ . | P value for difference between training and validation sets . | ||
|---|---|---|---|---|---|---|---|
| GVHD−, N = 166 . | GVHD+, N = 116 . | GVHD−, N = 76 . | GVHD+, N = 66 . | ||||
| Age, y | |||||||
| Median | 43 | 49 | .02 | 47 | 52 | .17 | .15 |
| Range | 1–68 | 1–71 | 4–69 | 1–66 | |||
| Diagnostic, % (no.) | |||||||
| Malignant* | 93 (154) | 97 (113) | .15 | 97 (74) | 97 (64) | .71 | .35 |
| Other† | 7 (12) | 3 (3) | 3 (2) | 3 (2) | |||
| Disease status at BMT,‡% (no.) | |||||||
| Low risk | 58 (96) | 49 (57) | .19 | 64 (49) | 56 (37) | .39 | .14 |
| High risk | 42 (70) | 51 (59) | 36 (27) | 44 (29) | |||
| Donor type, % (no.) | |||||||
| MRD | 68 (113) | 58 (68) | .13 | 75 (57) | 58 (38) | .04 | .66 |
| URD | 32 (53) | 42 (48) | 25 (19) | 42 (28) | |||
| Donor match, % (no.) | |||||||
| Matched | 95 (157) | 87 (101) | .04 | 93 (71) | 83 (55) | .10 | .46 |
| Mismatched | 5 (9) | 13 (15) | 7 (5) | 17 (11) | |||
| Conditioning regimen intensity, % (no.) | |||||||
| Full | 73 (122) | 63 (73) | .08 | 79 (60) | 61 (41) | .04 | .76 |
| Reduced | 27 (44) | 37 (43) | 21 (16) | 37 (25) | |||
| Grade at GVHD onset, % (no.) | |||||||
| 0 | 59 (166) | 0 | 54 (76) | 0 | |||
| 1 | 0 | 12 (34) | 0 | 11 (15) | .31 | ||
| 2 | 0 | 23 (64) | 0 | 24 (35) | |||
| 3–4 | 0 | 6 (18) | 0 | 11 (16) | |||
| Organ target at GVHD onset, % (no.) | |||||||
| Skin | 0 | 66 (71) | 0 | 58 (38) | |||
| Gut | 0 | 18 (20) | 0 | 24 (16) | .64 | ||
| Liver | 0 | 4 (5) | 0 | 4 (3) | |||
| Combined | 0 | 12 (14) | 0 | 14 (9) | |||
| Day after BMT of samples | |||||||
| Median | 30 | 30 | .36 | 30 | 29 | .85 | .85 |
| Range | 7–104 | 5–119 | 7–99 | 7–112 | |||
| Characteristic . | Training cohort, N = 282 . | P value for difference between GVHD− and GVHD+ . | Validation cohort, N = 142 . | P value for difference between GVHD− and GVHD+ . | P value for difference between training and validation sets . | ||
|---|---|---|---|---|---|---|---|
| GVHD−, N = 166 . | GVHD+, N = 116 . | GVHD−, N = 76 . | GVHD+, N = 66 . | ||||
| Age, y | |||||||
| Median | 43 | 49 | .02 | 47 | 52 | .17 | .15 |
| Range | 1–68 | 1–71 | 4–69 | 1–66 | |||
| Diagnostic, % (no.) | |||||||
| Malignant* | 93 (154) | 97 (113) | .15 | 97 (74) | 97 (64) | .71 | .35 |
| Other† | 7 (12) | 3 (3) | 3 (2) | 3 (2) | |||
| Disease status at BMT,‡% (no.) | |||||||
| Low risk | 58 (96) | 49 (57) | .19 | 64 (49) | 56 (37) | .39 | .14 |
| High risk | 42 (70) | 51 (59) | 36 (27) | 44 (29) | |||
| Donor type, % (no.) | |||||||
| MRD | 68 (113) | 58 (68) | .13 | 75 (57) | 58 (38) | .04 | .66 |
| URD | 32 (53) | 42 (48) | 25 (19) | 42 (28) | |||
| Donor match, % (no.) | |||||||
| Matched | 95 (157) | 87 (101) | .04 | 93 (71) | 83 (55) | .10 | .46 |
| Mismatched | 5 (9) | 13 (15) | 7 (5) | 17 (11) | |||
| Conditioning regimen intensity, % (no.) | |||||||
| Full | 73 (122) | 63 (73) | .08 | 79 (60) | 61 (41) | .04 | .76 |
| Reduced | 27 (44) | 37 (43) | 21 (16) | 37 (25) | |||
| Grade at GVHD onset, % (no.) | |||||||
| 0 | 59 (166) | 0 | 54 (76) | 0 | |||
| 1 | 0 | 12 (34) | 0 | 11 (15) | .31 | ||
| 2 | 0 | 23 (64) | 0 | 24 (35) | |||
| 3–4 | 0 | 6 (18) | 0 | 11 (16) | |||
| Organ target at GVHD onset, % (no.) | |||||||
| Skin | 0 | 66 (71) | 0 | 58 (38) | |||
| Gut | 0 | 18 (20) | 0 | 24 (16) | .64 | ||
| Liver | 0 | 4 (5) | 0 | 4 (3) | |||
| Combined | 0 | 12 (14) | 0 | 14 (9) | |||
| Day after BMT of samples | |||||||
| Median | 30 | 30 | .36 | 30 | 29 | .85 | .85 |
| Range | 7–104 | 5–119 | 7–99 | 7–112 | |||
GVHD indicates graft-versus-host disease.
Malignant disease included acute myelogenous leukemia (AML), myelodysplastic syndrome (MDS), acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML), non-Hodgkin lymphoma (NHL), Hodgkin disease (HD), multiple myeloma (MM), juvenile myelomonocytic leukemia (JMML), and chronic lymphocytic leukemia (CLL).
Nonmalignant disease included severe congenital neutropenia (SCN), severe aplastic anemia, Wiskott-Aldrich syndrome, β-thalassemia, essential thrombocythemia, myelofibrosis, Glanzmann thrombasthenia, congenital amegakaryocytic thrombocytopenia, and sickle cell anemia.
Low and high risk of disease status at BMT according to IBMTR guidelines.
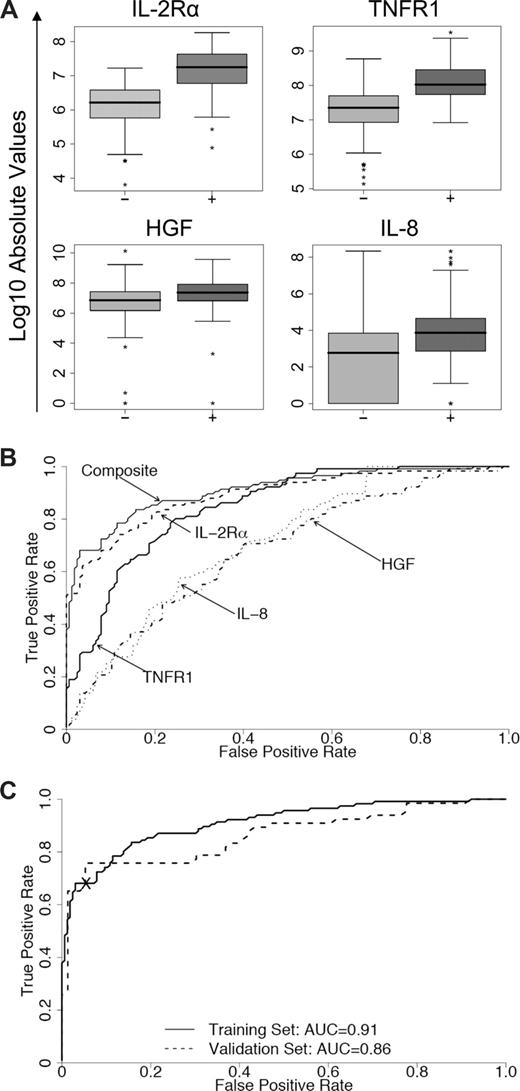
The median values and individual AUCs of the 8 biomarkers measured by sequential ELISA in the training set are presented in Table S3. Logistic regression determined that a linear combination of values for IL-2Rα, TNFR1, HGF, and IL-8 levels produced the best model to predict the occurrence of acute GVHD (Table 2). The distributions and ROC curves of these 4 biomarkers are shown in Figure 2A and 2B, respectively, with an AUC for the composite biomarker panel of 0.91 (95% CI, 0.87–0.94). When this model was applied to samples of the validation set, the corresponding AUC was 0.86 (95% CI, 0.79–0.92) (Figure 2C). Levels of IL-2Rα and TNFR1 contributed primarily to the accuracy of the model (P < .001 and P = .003, respectively). This 4-biomarker panel therefore effectively discriminated between patients with and without GVHD. We also used proportional odds logistic regression models to determine whether the 4-biomarker panel provided prognostic information regarding (1) the eventual maximum grade of GVHD and (2) involvement of specific target organs. HGF was the only marker that predicted maximum GVHD grade (P = .003; Table S4). The regression model determined that both HGF and IL-2Rα are associated with specific target organs (P = .03 and P = .04, respectively; Table S5). Figure S4 illustrates these relationships. After adjustment for the other biomarkers levels, analysis of variance across 3 groups (skin only, visceral only, and skin/visceral) demonstrates a significantly higher mean HGF level in patients with visceral-only and skin/visceral GVHD compared with skin-only GVHD. Likewise, the analysis of variance across groups demonstrates a significantly higher mean IL-2Rα level in patients with skin-only and skin/visceral GVHD compared with visceral-only GVHD.
Concentrations of 4 individual discriminator proteins, their receiver operating characteristic (ROC) curves, and the composite panel ROC curves. (A) Absolute values on a logarithmic scale of soluble IL-2Rα, TNFR1, HGF, and IL-8 in samples from the training with their medians and ranges. Patients without GVHD (−) versus with GVHD (+): P < .001. (B) Individual ROC curves for IL-2Rα (----), TNFR1 (—), HGF (.-.-.), and IL-8 (.....) and the composite panel (—). (C) ROC curves of the composite panel for the training set (—) and the validation set (- - - -). The cross separates patients into 2 groups based on their predicted probability of GVHD (high and low) with a specificity of 95% in the training set and 94% in the validation set.
Concentrations of 4 individual discriminator proteins, their receiver operating characteristic (ROC) curves, and the composite panel ROC curves. (A) Absolute values on a logarithmic scale of soluble IL-2Rα, TNFR1, HGF, and IL-8 in samples from the training with their medians and ranges. Patients without GVHD (−) versus with GVHD (+): P < .001. (B) Individual ROC curves for IL-2Rα (----), TNFR1 (—), HGF (.-.-.), and IL-8 (.....) and the composite panel (—). (C) ROC curves of the composite panel for the training set (—) and the validation set (- - - -). The cross separates patients into 2 groups based on their predicted probability of GVHD (high and low) with a specificity of 95% in the training set and 94% in the validation set.
Details for composite regression model
| Protein . | Coefficient estimate, ×10−3 . | Standard error, ×10−3 . | t value . | P . |
|---|---|---|---|---|
| Composite panel* | −4656 | 542 | 8.59 | <.001 |
| IL-2Rα | 3.43 | 0.52 | 6.60 | <.001 |
| TNFR1 | 0.48 | 0.16 | 3.00 | .003 |
| HGF | 0.11 | 0.06 | 1.69 | .09 |
| IL-8 | −0.45 | 0.38 | −1.18 | .24 |
| Protein . | Coefficient estimate, ×10−3 . | Standard error, ×10−3 . | t value . | P . |
|---|---|---|---|---|
| Composite panel* | −4656 | 542 | 8.59 | <.001 |
| IL-2Rα | 3.43 | 0.52 | 6.60 | <.001 |
| TNFR1 | 0.48 | 0.16 | 3.00 | .003 |
| HGF | 0.11 | 0.06 | 1.69 | .09 |
| IL-8 | −0.45 | 0.38 | −1.18 | .24 |
Predicted probability = ePanel / (1 + ePanel).
IL-2Rα indicates interleukin-2-receptor-alpha; TNFR1, tumor-necrosis-factor-1; HGF, hepatocyte growth factor; and IL-8, interleukin-8.
Composite panel = 1000 × (−4656 + 3.43 × IL2-Rα + 0.48 × TNFR1 + 0.11 × HGF − 0.45 × IL-8).
Biomarker panel and survival
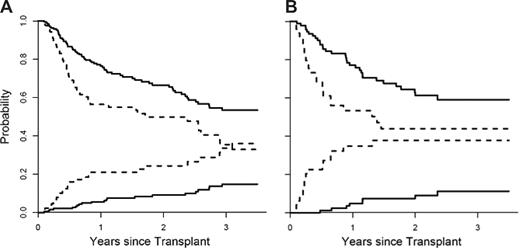
We next divided patients in the training set into 2 groups (high and low risk) based on their predicted probabilities for developing GVHD. We determined the threshold for high so that the false-positive rate did not exceed 5%. The nonrelapse mortality (NRM) and overall survival (OS) for both groups are shown in Figure 3A. When adjusted for age, donor type, HLA match, and intensity of conditioning, the differences in NRM between the 2 groups were highly significant (P = .001). When we applied the same definition to the validation set, the false-positive rate in the high-risk group was 6%. The NRM between groups was again significantly different when adjusted for all 4 variables (Figure 3B; P < .001). These 2 groups also experienced significantly different OS in both the training set (Figure 3A; P = .006, adjusted) and validation set (Figure 3B; P = .02, adjusted). Interestingly, the mortality from relapse at one year for all patients was similar in both the low- and high-risk groups (18% versus 17%; P = .54). Of note IL-2Rα predicted response to treatment at 4 weeks (P = .03) but the 4 biomarker panel did not (P = .17).
Nonrelapse mortality and overall survival stratified by the biomarker panel. (A) The cumulative incidence of nonrelapse mortality (NRM) and overall survival (OS), determined by Kaplan-Meier curve, is plotted according to the predicted probability of acute GVHD: low (—, n = 193) and high (----, n = 89). P = .001 and P = .006 (adjusted for age, donor type, HLA match, and intensity of conditioning) for differences in NRM and OS, respectively. The NRM at 3.5 years is 15% (95% CI, 9%-21%) for the low-risk group and 36% (95% CI, 24%-48%) for the high-risk group. OS at 3.5 years is 53% (95% CI, 45%-63%) for the low-risk group and 33% (95% CI, 22%-48%) in the high-risk group. (B) The cumulative incidence of NRM and OS of the 2 groups is plotted for the validation set: low (—, n = 93) and high (----, n = 49). P values less than .001 and .02 (adjusted as before) for differences in NRM and OS, respectively. The NRM at 3.5 years is 11% (95% CI, 4%-19%) for the low-risk group and 38% (95% CI, 23%-53%) for the high-risk group. OS at 3.5 years is 59% (95% CI, 49%-72%) for the low-risk group and 44% (95% CI, 31%-63%) for the high-risk group.
Nonrelapse mortality and overall survival stratified by the biomarker panel. (A) The cumulative incidence of nonrelapse mortality (NRM) and overall survival (OS), determined by Kaplan-Meier curve, is plotted according to the predicted probability of acute GVHD: low (—, n = 193) and high (----, n = 89). P = .001 and P = .006 (adjusted for age, donor type, HLA match, and intensity of conditioning) for differences in NRM and OS, respectively. The NRM at 3.5 years is 15% (95% CI, 9%-21%) for the low-risk group and 36% (95% CI, 24%-48%) for the high-risk group. OS at 3.5 years is 53% (95% CI, 45%-63%) for the low-risk group and 33% (95% CI, 22%-48%) in the high-risk group. (B) The cumulative incidence of NRM and OS of the 2 groups is plotted for the validation set: low (—, n = 93) and high (----, n = 49). P values less than .001 and .02 (adjusted as before) for differences in NRM and OS, respectively. The NRM at 3.5 years is 11% (95% CI, 4%-19%) for the low-risk group and 38% (95% CI, 23%-53%) for the high-risk group. OS at 3.5 years is 59% (95% CI, 49%-72%) for the low-risk group and 44% (95% CI, 31%-63%) for the high-risk group.
We next evaluated whether the biomarker panel provided additional prognostic information among those patients presenting with clinical signs of GVHD. Analysis of the 182 GVHD+ patients for individual population characteristics showed that both the maximum grade of GVHD and the levels of the biomarker panel were significantly associated with risk of death (P < .001; Table 3). When adjusted for age, donor relation, HLA match, intensity of conditioning regimen, and disease status at bone marrow transplantation (BMT), the hazard ratio for maximum grade of GVHD and the biomarker panel remained significant (P = .002 and P = .001, respectively). Patients with a maximum grade of GVHD III to IV had a hazard ratio of 2.35 compared with patients with a maximum grade I to II. Patients whose levels of the 4 biomarkers were 50% above the median had a hazard ratio of 2.46 compared with patients with median levels. In simultaneous Cox regression analyses using both maximum grade and biomarker panel levels, the hazard ratio for the biomarker panel remained relatively unchanged and continued to predict risk for death (P = .003, adjusted as before). The biomarker panel therefore predicted long-term survival independently of maximum GVHD grade. Levels of TNFR1 and HGF contributed primarily to the significance of the panel (P < .001 and P = .05, respectively).
Association of maximum GVHD grade and biomarker panel with overall survival
| . | Individual . | Simultaneous . | ||
|---|---|---|---|---|
| . | Hazard ratio . | P . | Hazard ratio . | P . |
| Unadjusted | ||||
| Maximum GVHD grade (1/2 vs 3/4) | 2.35 | <.001 | 2.11 | .001 |
| Biomarker panel | 2.46 | <.001 | 2.43 | <.001 |
| Adjusted* | ||||
| Maximum GVHD grade (1/2 vs 3/4) | 2.31 | .002 | 2.06 | .003 |
| Biomarker panel | 2.31 | .001 | 2.28 | .003 |
| . | Individual . | Simultaneous . | ||
|---|---|---|---|---|
| . | Hazard ratio . | P . | Hazard ratio . | P . |
| Unadjusted | ||||
| Maximum GVHD grade (1/2 vs 3/4) | 2.35 | <.001 | 2.11 | .001 |
| Biomarker panel | 2.46 | <.001 | 2.43 | <.001 |
| Adjusted* | ||||
| Maximum GVHD grade (1/2 vs 3/4) | 2.31 | .002 | 2.06 | .003 |
| Biomarker panel | 2.31 | .001 | 2.28 | .003 |
Adjusted for age, donor relation, donor matching, conditioning regimen, and status at transplantation.
Discussion
We used antibody microarrays that targeted 120 proteins to screen for biomarkers of GVHD in an unbiased fashion so that each protein had an equal chance of being chosen as a biomarker. Measuring levels by sequential ELISA, we tested a panel of 8 potential biomarkers first in a training set of 282 patients, then in a validation set of 142 additional patients. Using logistic regression models, we determined that a panel composed of 4 biomarkers (IL-2Rα, TNFR1, IL-8, and HGF) effectively discriminated between patients with and without GVHD. This biomarker panel was highly specific and predicted long-term survival independently of maximum GVHD grade.
A variety of MS-based proteomic approaches have recently been used in an attempt to diagnose GVHD, with promising results.24-26 An advantage of these approaches is that identification of proteins does not depend on the availability of antibodies as in the case of microarrays or ELISAs. These MS-based techniques do have inherent disadvantages such as the lack of identification of specific proteins, their labor intensity, and lack of speed. Newer technologies (eg, tandem MS) may make such techniques more attractive in the future. Protein arrays also hold great promise for noninvasive detection of new plasma biomarkers,30,31 although this technique has some disadvantages, such as the potential for high background and nonspecific interactions. We tried to circumvent such difficulties in this study by using array technology during the discovery phase and then using ELISA to quantitate the levels of individual proteins. Nonspecific interactions in the arrays may account for some of the discrepancies between array data and ELISA measurements (eg, PSA-ACT, MMP-2, IL1-β, IL-17). We then used a sequential ELISA approach rather than multiplex technique because of its feasibility and accuracy regarding low abundance proteins such as IL-2Rα.
The complex pathophysiology of GVHD32 suggests that plasma proteins involved in multiple processes such as T-cell alloreactivity, inflammation, and tissue damage and repair might be altered in the patient with the disease. All 4 biomarkers (IL-2Rα, TNFR1, IL-8, and HGF) that were identified in our analysis have been associated with GVHD in previous small studies.3-14 In this study of more than 450 patients, the 2 most discriminating proteins were cytokine receptors, presumably shed into the circulation by activated donor cells that mediate acute GVHD.33 Soluble IL-2Rα, the single best discriminator of GVHD, is a well-established marker of T-cell activation and suppression of donor T-cell responses is the mainstay of GVHD prophylaxis.34 TNFR1, the second best discriminator, is released from the surface of activated cells, particularly monocytes that secrete TNFα. TNFα is a principal mediator of GVHD in animal models,35 and its receptors have been identified as a biomarker in some clinical studies.9,10 The role of TNFα in clinical GVHD has recently been highlighted in a phase 2 study that observed a high percentage of complete responses when a soluble inhibitor of TNFα was added to steroids as primary treatment for GVHD.36 IL-8 is a potent chemoattractant that is likely to help direct migration of cellular effectors to GVHD target organs.37 We initially identified HGF in a discovery analysis of GVHD biomarkers27 ; this cytokine may be released by target organs as a physiologic response to GVHD damage, similar to cytokeratin-18 fragments that indicate epithelial apoptosis associated with intestinal and hepatic GVHD.20
This study did not include patients with a prior history of VOD, IPS, or septic shock, who represent 15% of allogeneic recipients at our center. We did not attempt to identify biomarkers that would differentiate GVHD from these other major transplantation-related complications because of the very small number of patients with these complications alone. The search for such biomarkers should however continue. Previous studies suggest that elevations of IL-2Rα and TNFR1 are associated not only with acute GVHD but also with VOD and IPS,5,9 and a recent report demonstrates that TNFR1 and IL-8 are significantly elevated in patients with IPS.38 However, patients with inflammatory bowel disease have biomarkers distinct from GVHD (IL-7, IL-12p40, TGF-β1, and PLGF),30 suggesting that biomarker panels can distinguish inflammatory processes with different etiologies. Additional discovery studies using unbiased newer technologies (eg, tandem mass spectrometry) may prove fruitful in this regard.
The biomarker panel provided a specific test for GVHD in the 85% of patients who do not have other major complications. A biomarker panel suggesting high risk at the onset of clinical symptoms was able to confirm GVHD with 95% specificity in this patient population. In current practice, tissue biopsies are performed to confirm the diagnosis of GVHD. If validated in prospective studies, a high-risk biomarker panel may obviate the need for an invasive procedure to confirm the diagnosis of GVHD. Furthermore, HGF was associated with visceral GVHD, thereby adding value to a 4-biomarker panel compared with a single biomarker, IL-2Rα alone, or 2 biomarkers, IL-2Rα and TNFR1.
The ability of the biomarker panel to predict NRM and OS independently of eventual maximum GVHD grade is particularly intriguing. This finding suggests that measurement of these 4 biomarkers at onset of GVHD may be useful to further identify high-risk groups and their outcomes. Levels of IL-2Rα alone at onset of GVHD were also associated with complete responses to treatment at 4 weeks (P = .03), although the composite panel was not. The reasons for this lack of correlation for the entire panel are unclear and may be due to the variety of treatments used to treat GVHD or change of supportive care over the 6 years of the study. Future prospective studies could focus on the value of this biomarker panel to refine grades of GVHD at the time of diagnosis by incorporating both biomarker panel and clinical grade (eg, grade II, high risk biomarker panel). If validated, a risk stratification of GVHD could ultimately guide the intensity and duration of GVHD treatment to minimize the toxicities of chronic steroid administration.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health (Bethesda, MD; Cancer Center P50 P01 CA039542-20), the Food and Drug Administration (Rockville, MD; 5R01FD002397-03-2), and the Doris Duke Charitable Foundation (New York, NY; 20020347). S.P. is a recipient of Fondation de France fellowship (Paris, France; Leukemia research, 2006-004309).
National Institutes of Health
Authorship
Contribution: S.P. and O.I.K. designed and planned the study and experiments, performed research, analyzed data, and wrote the paper; T.M.B. was the study statistician; R.K. designed experiments, analyzed data, and performed discovery statistics; S.G.C. and D.E.M. designed experiments, performed research, analyzed data, and edited the paper; S.W.C., C.L.K., K.R.C., P.R., and J.E.L. contributed to patient accrual, clinical data collection and quality assurance, research discussion, and paper editing; A.W. performed ELISA; D.B. performed database search; D.J. and J.W. managed human samples database; and S.M.H. and J.L.M.F. conceived and planned the study design and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sophie Paczesny, University of Michigan Comprehensive Cancer Center 6420, 1500 E Medical Center Drive, Ann Arbor, MI 48109-5942; e-mail: sophiep@umich.edu.
References
Author notes
*S.P. and O.I.K. contributed equally to this work.