In this issue of Blood, Paczesny and colleagues describe a set of 4 serum proteins that may help to confirm the diagnosis of acute GVHD and seem to be associated with worse survival independently of GVHD grade.
Acute graft-versus-host disease (GVHD), the main complication of allogeneic hematopoietic cell transplantation, is mainly diagnosed clinically and can be confirmed by biopsy of 1 of the 3 target organs (skin, gastrointestinal tract, or liver). The severity of acute GVHD is graded clinically from I to IV, with increased mortality rates in severe GVHD (grades II-IV).
Although multiple blood proteins have been described as potential biomarkers in previous smaller studies, no single protein or panel has emerged with sufficient specificity and sensitivity to enter clinical use. More recently, mass spectrometric profiling1-3 of urine and serum were reported to demonstrate the presence of spectral patterns associated with GVHD, but these approaches do not identify specific proteins.
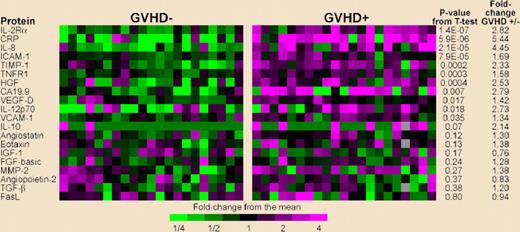
In this issue of Blood, Paczesny et al aimed to isolate candidate proteins using high-throughput assays on a large number of patient samples, and to determine their significance with respect to patient outcome. They screened plasma with antibody microarrays for 120 proteins in a discovery set of 42 transplantation patients that revealed 8 potential biomarkers for diagnostic of GVHD. Using enzyme-linked immunosorbent assays (ELISA), they then measured the levels of these biomarkers in samples from more than 400 transplantation patients divided into training and validation sets. Statistical analysis of these 8 proteins determined a panel of 4 proteins (IL-2-receptor–alpha, TNF-receptor-1, IL-8, and hepatocyte growth factor) that discriminated patients with and without GVHD. In patients with GVHD, Cox regression analysis revealed that the biomarker panel predicted survival independently of GVHD severity. This study is a major achievement in the field and brings proteomics nearly from the bench to the bedside. The authors have to be congratulated for the huge amount of work they have done and the refined analysis of their results.
However, as always in good science, this study raises more questions than it answers: As mentioned, other authors have used mass spectrometric profiling (these approaches do not identify specific proteins) while Paczesny et al used a predefined set of 120 proteins. In the first case, we are faced with lovely diagrams1-3 without knowing if they constitute proteins actually involved in inflammatory processes or allogeneic reactions. In the second case, we must guess regarding potentially important proteins, and thus end up with at least 3 candidates already reported to be involved in GVHD (IL-2-receptor-alpha, TNF-receptor-1, and IL-8) and 1 (hepatocyte growth factor) of yet uncertain significance. There is no doubt that future studies will refine our current knowledge.
In the study by Paczesny et al, patients with veno-occlusive disease (VOD), septic shock and idiopathic pneumonia syndrome were excluded. Thus, several real-life diagnostic challenge situations were excluded, that is, distinguishing VOD from liver GVHD and idiopathic pneumonia syndrome from acute lung GVHD. Finally, biomarkers were analyzed at a single time point and not sequentially, and the 4 proteins panel did not predict patient's response to treatment.
Without a doubt, future studies will refine our knowledge of the interrelationship between proteins and GVHD pathogenesis, severity of isolated target organ, and response to treatment. It would be of especially great clinical value to isolate a panel of biomarkers that could predict upfront patients with the worst prognostic (ie, those who will develop steroid-resistant GVHD).
ELISA heat map of discovery set samples. Gray indicates that the sample was not assayed for that protein. Levels of PSA-ACT, IL-17 and IL-1|gb were not detectable and therefore do not appear in the figure. Please see the complete figure in the article beginning on page 273.
ELISA heat map of discovery set samples. Gray indicates that the sample was not assayed for that protein. Levels of PSA-ACT, IL-17 and IL-1|gb were not detectable and therefore do not appear in the figure. Please see the complete figure in the article beginning on page 273.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal