Abstract
CD8+ T cells recognizing minor histocompatibility antigens (MiHAs) on leukemic stem and progenitor cells play a pivotal role in effective graft-versus-leukemia reactivity after allogeneic stem cell transplantation (SCT). Previously, we identified a hematopoiesis-restricted MiHA, designated LRH-1, which is presented by HLA-B7 and encoded by the P2X5 purinergic receptor gene. We found that P2X5 is significantly expressed in CD34+ leukemic subpopulations from chronic myeloid leukemia (CML) and acute myeloid leukemia (AML) patients. Here, we demonstrate that LRH-1–specific CD8+ T-cell responses are frequently induced in myeloid leukemia patients following donor lymphocyte infusions. Patients with high percentages of circulating LRH-1–specific CD8+ T cells had no or only mild graft-versus-host disease. Functional analysis showed that LRH-1–specific cytotoxic T lymphocytes (CTLs) isolated from 2 different patients efficiently target LRH-1–positive leukemic CD34+ progenitor cells from both CML and AML patients, whereas mature CML cells are only marginally lysed due to down-regulation of P2X5. Furthermore, we observed that relative resistance to LRH-1 CTL-mediated cell death due to elevated levels of antiapoptotic XIAP could be overcome by IFN-γ prestimulation and increased CTL-target ratios. These findings provide a rationale for use of LRH-1 as immunotherapeutic target antigen to treat residual or persisting myeloid malignancies after allogeneic SCT.
Introduction
Allogeneic stem cell transplantation (SCT) followed by donor lymphocyte infusions (DLIs) is a curative treatment option for patients with chronic myeloid leukemia (CML) or acute myeloid leukemia (AML).1,2 The therapeutic efficacy is attributed to the graft-versus-leukemia (GVL) response mediated by donor-derived T cells recognizing minor histocompatibility antigens (MiHAs) on malignant cells of the recipient.3,4 However, a significant number of patients with advanced CML and AML relapse due to persisting leukemic stem and progenitor cells.5 Adjuvant immunotherapy targeting hematopoietic cell–restricted MiHAs on these primitive leukemic cells may permanently eradicate the malignancy without inducing graft-versus-host disease (GVHD).6,7
Previously, we identified a hematopoiesis-restricted MiHA, designated LRH-1, which elicited an HLA-B7–restricted cytotoxic T lymphocyte (CTL) response in an advanced CML patient treated with DLI.8 Using isolated LRH-1–specific CTLs from this patient, we demonstrated that immunogenicity of LRH-1 resulted from differential expression of the P2X5 protein in recipient and donor cells as a consequence of a single nucleotide frameshift polymorphism. Interestingly, tetramer analysis showed a direct association between in vivo expansion of LRH-1–specific CD8+ T cells and the disappearance of Bcr-Abl–positive CML cells following DLI. Moreover, gene expression studies revealed that P2X5 is not expressed in nonhematopoietic tissues, including prominent GVHD target tissues such as skin, liver, colon, and small intestine.8 These findings illustrate that P2X5-encoded MiHA LRH-1 may play a significant role in inducing GVL-specific immunity against myeloid leukemia. Although we have shown that P2X5 mRNA is significantly expressed in leukemic CD34+CD38− and CD34+CD38+ subpopulations from most CML and AML patients,8 it remained to be investigated whether myeloid leukemic progenitor cells are susceptible to lysis by LRH-1–specific CTLs.
Here, we studied whether LRH-1–reactive CTLs specifically and efficiently target CD34+ leukemic progenitor cells from CML and AML patients. We demonstrate that LRH-1–specific CD8+ T-cell responses are frequently induced in myeloid leukemia patients following DLI. In addition, LRH-1–specific CTLs isolated from 2 different patients efficiently lyse LRH-1–expressing malignant CD34+ progenitor cells from both CML and AML patients. These findings illustrate that LRH-1 is an attractive target for eradication of residual or persisting myeloid leukemic progenitor cells after allogeneic SCT.
Methods
Transplantation patients
Seven patients, 5 females and 2 males, aged 20 to 54 years (median, 35 years) underwent transplantation for acute lymphoblastic leukemia (ALL, n = 2), acute myeloid leukemia (n = 2), chronic myeloid leukemia (n = 2), and multiple myeloma (n = 1). Donors were HLA-identical siblings in 5 cases and non–HLA-identical family members in 2 cases. Conditioning for transplantation was with cyclophosphamide (120 mg/kg) and total body irradiation (9 Gy) with the addition of idarubicin 42 mg/m2 in 3 patients and antithymocyte globulin in 2 cases. All grafts were partially T-cell depleted to 0.7 × 106 CD3+ cells/kg body weight of the recipient. DLI was given preemptively in 5 patients and therapeutically in 2 patients.
Cell isolation and cell culture
Peripheral blood (PB) and bone marrow (BM) samples have been collected after written informed consent obtained in accordance with the Declaration of Helsinki in ongoing clinical stem cell transplantation protocols approved by the Radboud University Nijmegen Medical Centre (RUNMC) and Leiden University Medical Center (LUMC) Institutional Review Boards. CD34+ CML and AML cells were isolated from PB and BM, respectively, obtained at diagnosis, by fluorescence activated cell sorting (FACS) on a Coulter Epics Elite Flow Cytometer (Beckman Coulter, Fullerton, CA). Primary T-cell cultures were generated from peripheral blood mononuclear cells (PBMCs) obtained 2 to 6 months after DLI from 2 CML patients (Pt; Pt1 and Pt2) and 1 AML patient (Pt3) who received a transplant of stem cells from an LRH-1–mismatched donor. After thawing, PBMCs were used partly for LRH-1/HLA-B7 tetramer staining. Remaining PBMCs were cultured in Iscove modified Dulbecco medium (IMDM; Invitrogen, Carlsbad, CA) supplemented with 10% human serum (HS; PAA Laboratories, Linz, Austria) and stimulated with irradiated (80 Gy) donor Epstein-Barr virus (EBV)–transformed lymphoblastoid cell line (LCL) pulsed with 10 μM LRH-1 peptide TPNQRQNVC at a ratio of 1:1. At days 1 and 4, 25 IU/mL IL-2 (Chiron, Emeryville, CA) was added to the cell cultures. After 2 to 3 weekly stimulations, these primary T-cell cultures were analyzed for the presence of LRH-1/HLA-B7-tetramer+ CD8+ T cells. LRH-1–specific CD8+ CTL culture TB1 was enriched to a more than 95% LRH-1–tetramer+ population from PBMCs of AML Pt3 by repeated LRH-1–specific stimulation. LRH-1–specific CD8+ CTL culture RP1 was isolated from CML Pt1 who was successfully treated with DLI after allogeneic SCT.8 Both CTL TB1 and RP1 recognize the 9-mer epitope TPNQRQNVC in the context of HLA-B*0702. The HLA-B7–alloreactive CD8+ CTL culture KOR18 was kindly provided by Prof Dr E. Goulmy (Department of Immunohematology, LUMC, Leiden, The Netherlands). CTL RP1, TB1, and KOR18 (0.5 × 106) were cultured in IMDM/10% HS containing irradiated (80 Gy) HLA-B7+ LRH-1+ EBV-LCLs (0.5 × 106), irradiated (60 Gy) allogeneic PBMCs (0.5 × 106) from 2 donors, 100 IU/mL IL-2, and 1 μg/mL PHA-M (Boehringer, Mannheim, Germany) in a total volume of 2 mL. Cell lines were cultured in IMDM/10% fetal calf serum (FCS; Integro, Zaandam, The Netherlands). KG1, KG1a, and K562 AML cell lines were retrovirally transduced with LZRS-HLA-B*0702-IRES-EGFP as described.8,9
Flow cytometry
PE-labeled LRH-1/HLA-B7 tetramers containing peptide TPNQRQNVC were produced as described.10 PBMCs or CTLs were incubated with 20 μg/mL HLA-B7/LRH-1 tetramer for 20 minutes at room temperature. After washing with PBS/0.5% BSA, cells were labeled with the appropriate concentration anti-CD8 (ProImmune, Oxford, United Kingdom), anti-CD3, and anti-CD45 (Beckman Coulter) for 15 minutes at 4°C. After washing, cells were resuspended in PBS/0.5% BSA and 7-amino-actinomycin D (7AAD; Sigma-Aldrich, St Louis, MO) was added. Cells were analyzed using the Coulter FC500 flow cytometer (Beckman Coulter). LRH-1–tetramer+ cells were further phenotyped using IgG2A, anti-CD45RA, anti-CD27, anti-Fas (Beckman Coulter), IgG1, anti-CD45RO (DAKO, Glostrup, Denmark), anti-CD127 (eBiosciences, San Diego, CA), anti-CD28 (Becton Dickinson, Franklin Lakes, NJ), and anti-CCR7 (R&D Systems, Minneapolis, MN). Relapse samples from CML Pt1 were phenotyped using anti-CD38, anti-CD117 (Becton Dickinson), anti-CD13, anti-CD33 (Beckman Coulter), and anti-CD133 (Miltenyi Biotec, Bergisch Gladbach, Germany).
Real-time quantitative reverse-transcription–PCR
Total RNA was isolated from cell samples using Trizol (Invitrogen) or the Zymo RNA isolation kit II (Zymo Research, Orange, CA). cDNA synthesis and polymerase chain reaction (PCR) amplification were performed as described.9,11,12 The hydroxymethylbilane synthase (HMBS) housekeeping gene was used to normalize gene expression. cDNA (1 μL) was amplified in a 50-μL reaction mixture containing 1.25 U AmpliTaq Gold (Applied Biosystems, Foster City, CA), 300 nM gene-specific forward and reverse primer, 150 to 300 nM gene-specific Taqman probe (150 nM for P2X5, and 300 nM for HMBS), 250 μM of each dNTP, 5 mM MgCl2, and 1× Taqman PCR buffer (Applied Biosystems). Alternatively, 1× SYBR green buffer was used for the apoptosis-regulating genes caspase-3, Smac, Bax, Bcl-xL, Bcl-2, Bak, and Bid. The following gene-specific primers and Taqman probes were used: P2X5: P2X5-F 5′-TCCTGGCGTACCTGGTCGT-3′, P2X5-R 5′-CTTCATTCTCAGCACAGACGTTC-3′, and P2X5-probe 5′-(TET)-TGGGTGTTCCTGATAAAGAAGGGTTACCA-(TAMRA)-3′; HMBS: HMBS-F 5′-GGCAATGCGGCTGCAA-3′, HMBS-R 5′-GGGTACCCACGCGAATCAC-3′, and HMBS-probe 5′-(VIC)-CTCATCTTTGGGCTGTTTTCTTCCGCC-(TAMRA)-3′; caspase-3: caspase-3-F 5′-GACAGACAGTGGTGTTGATGATGA-3′ and caspase-3-R 5′-TTTGAATTTCGCCAAGAATAATAAC-5′; Smac: Smac-F 5′-CACAATGGCGGCTCTGAAG-3′ and Smac-R 5′-CCACAACAGGAACACACAAACA-3′; Bax: Bax-F 5′-GAGCGGCGGTGATGGA-3′ and Bax-R 5′-TGGATGAAACCCTGAAGCAAA-3′; Bcl-xL: Bcl-xL-F 5′-CCACTTACCTGAATGACCACCTAGA-3′ and Bcl-xL-R 5′-CAGCGGTTGAAGCGTTCC-3′; Bcl-2: Bcl-2-F 5′-TCGCCCTGTGGATGACTGA-3′ and Bcl-2-R 5′-CAGAGACAGCCAGGAGAAATCA-3′; Bak: Bak-F 5′-TGAGTACTTCACCAAGATTGCCA-3′ and Bak-R 5′-AGTCAGGCCATGCTGGTAGAC-3′; and Bid: Bid-F 5′-CCTTGCTCCGTGATGTCTTTC-3 and Bid-R 5′-TCCGTTCAGTCCATCCCATTT-3′. Expression of apoptotic gene XIAP was determined using TaqMan gene expression assay Hs01597783_m1 (Applied Biosystems) with XIAP-probe 5′-(FAM)-GCCGGCTGTCCTGGCGCGAAAAGGT-(TAMRA)-3′ according to the manufacturer's instructions. PCR amplification was performed using an ABI Prism 7700 (Applied Biosystems) with the following PCR conditions: enzyme activation for 10 minutes at 95°C, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. P2X5 mRNA expression was quantified by determining calibration functions using JVM-2 as reference cell line. The level of P2X5 expression was calculated relative to the P2X5 expression in the JVM-2 cell line, which is susceptible to lysis by the LRH-1–specific CTLs. Previous experiments demonstrated that only target cells with a relative P2X5 mRNA expression level higher than 0.4 could be recognized by LRH-1–specific CTLs.8 Therefore, we arbitrarily set the cutoff level at 0.4 to discriminate between LRH-1–positive and –negative cell types. mRNA expression of apoptosis genes XIAP, caspase-3, Smac, Bax, Bcl-xL, Bcl-2, Bak, and Bid is shown in ΔΔCt values and was quantified relative to AML cell line KG1, which was set at 1 ΔΔCt value. ΔΔCt was calculated as follows: 2(−[ΔCtsample]−[ΔCtKG1]), in which ΔCt was normalized for HMBS by calculating ΔCt = Ctgene − CtHMBS per sample.
P2X5 genotyping
Genomic DNA was isolated from cell lines using the QIAamp DNA Blood mini kit (QIAGEN, Hilden, Germany). DNA was amplified as described,9 using the following gene-specific primers and Taqman probes: P2X5: P2X5-EXON3-F 5′-CCAAATCAAACCTCAGCACAGAC-3′, P2X5-EXON3-R 5′-CTCAGTGCCTCTCTGGTTCCTTA-3′, P2X5 5C allele-specific probe 5′-(FAM)-ATTGTGACCCCCAACCA-(MGB)-3′, and P2X5 4C allele-specific probe 5′-(VIC)-TGTGACCCCAACCAG-(MGB)-3′. After PCR amplification an end-plate read was performed using ABI Prism 7700 allelic discrimination software.
Cytotoxicity studies
Cytotoxicity assays were performed as described9,11,13 with some adaptations. Briefly, CD34+ progenitor cells were sorted from thawed PB samples obtained from chronic-phase CML patients (Pt8-Pt11). These samples contained 2% to 10% CD34+ leukemic progenitor cells, of which 8% to 15% were negative for CD38. Purified chronic-phase CD34+ CML cells were either directly labeled with 0.75 μM carboxyfluorescein diacetate succimidyl ester (CFSE; Molecular Probes Europe, Leiden, The Netherlands) and used in cytotoxicity assays, or first cultured for 1 week in the presence of a cytokine mixture (GM-CSF (100 ng/mL), G-CSF (100 ng/mL), IL-3 (50 ng/mL), SCF (25 ng/mL), and EPO (2 IU/mL). CD34+ accelerated phase CML relapse cells from Pt1 were cultured overnight in medium containing GM-CSF (100 ng/mL), G-CSF (100 ng/mL), IL-3 (50 ng/mL), SCF (25 ng/mL), and EPO (2 IU/mL) in the absence or presence of 100 U/mL IFN-γ. Thereafter CD34+ cells were sorted, labeled with 0.625 μM CFSE, and used in cytotoxicity assays in medium containing GM-CSF, G-CSF, IL-3, SCF, and EPO. PB or BM samples obtained from AML M0/1/2 patients (Pt12-Pt16) were cultured overnight in medium containing GM-CSF (100 ng/mL), G-CSF (100 ng/mL), IL-3 (50 ng/mL), SCF (25 ng/mL), and Flt3L (20 ng/mL) in the absence or presence of 100 U/mL IFN-γ. Thereafter CD34+ cells were sorted, labeled with 0.625 μM CFSE, and used in cytotoxicity assays in medium containing GM-CSF, G-CSF, IL-3, SCF, and Flt3L. EBV-LCLs were labeled with 1.5 μM CFSE. Subsequently, CFSE-labeled target cells were cocultured for 3 consecutive days with unlabeled effector cells in IMDM/10% FCS containing 25 U/mL IL-2 and the cytokines GM-CSF, G-CSF, IL-3, SCT, and EPO (for CD34+ CML cells) or Flt3L (for CD34+ AML cells). HLA-B*0702-IRES-EGFP–transduced AML cell lines were used as targets without CFSE labeling. After coculture, cells were harvested and 7AAD was added to identify the number of viable target cells. Target cell survival was calculated as follows: % survival = {[absolute no. viable target cells cocultured with CTLs]/[absolute no. viable target cells cultured in medium]} × 100%. Specific lysis was calculated as follows: % lysis = {100 − [% survival]}. To visualize target cell lysis, cytospins were made from CD34+ CML progenitor cells and CD34− cytokine-maturated CML cells on day 2 of coculture with LRH-1–specific CTL. Cells were morphologically examined after May-Grünwald-Giemsa staining.
Results
CD34+ myeloid leukemia cell lines are efficiently lysed by LRH-1–specific CTLs
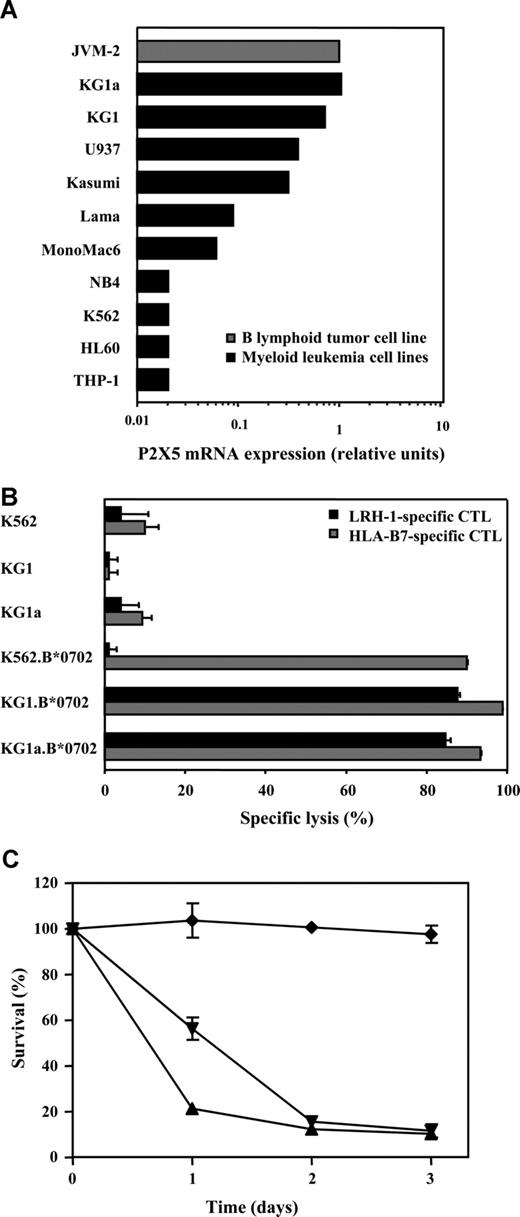
To investigate potential CTL recognition of myeloid malignancies, P2X5 gene expression levels were measured by quantitative reverse-transcription (RT)–PCR in a panel of malignant myeloid cell lines. Interestingly, relatively high P2X5 mRNA levels were detected in the immature CD34+CD38− myeloid cell line KG1 (0.71) and in the more undifferentiated subline KG1a (1.08) (Figure 1A). Lower P2X5 expression levels were found in U937 (0.39) and Kasumi (0.31). P2X5 expression was lowest in the more differentiated myeloid leukemia cell lines, such as Lama, MonoMac6, NB4, K562, HL-60, and THP-1. Next, we determined whether P2X5 mRNA expression in the KG1 cell lines results in susceptibility to LRH-1 CTL-mediated lysis. Because target cell recognition by LRH-1–specific CTLs is controlled by a single cytosine deletion polymorphism (rs5818907) in exon 3 of the P2X5 gene, we first performed P2X5 genotyping analysis. This analysis revealed that KG1 and KG1a cells are homozygous positive for the LRH-1–encoding allele (ie, C/C genotype). Since these cell lines do not express HLA-B7, they were retrovirally transduced with HLA-B*0702. Functional analysis revealed that LRH-1–specific CTLs efficiently kill HLA-B7–transduced KG1 and KG1a cells at a very low E/T ratio of 0.5:1 (Figure 1B,C). No cytotoxicity was observed against HLA-B7–transduced LRH-1–negative K562 cells (−/− genotype). By contrast, HLA-B7–transfected KG1 and K562 cell lines were effectively lysed by allo-HLA-B7–specific CTLs, indicating that HLA-B7 molecules were properly expressed (Figure 1B). These data demonstrate that P2X5 mRNA expression in human CD34+ myeloid leukemia cell lines results in LRH-1–specific CTL lysis.
P2X5 expression in KG1 AML cells results in recognition and lysis by LRH-1–specific CTLs. (A) P2X5 mRNA expression was determined by real-time quantitative RT-PCR in a panel of AML cell lines. (B) Specific cytotoxicity of LRH-1–specific CTL RP1 against AML cell lines KG1, KG1a, and K562 with and without transduction of HLA-B*0702. Specific lysis was determined after 2 days of coculture in a flow cytometry–based cytotoxicity assay at an E/T ratio of 0.5:1. (C) Survival up to 3 days of HLA-B*0702–transduced AML cell lines KG1 (▴), KG1a (▾), and K562 (♦) cocultured with LRH-1–specific CTL RP1 at an E/T ratio of 0.5:1. Data are displayed as mean plus or minus SD of triplicate wells.
P2X5 expression in KG1 AML cells results in recognition and lysis by LRH-1–specific CTLs. (A) P2X5 mRNA expression was determined by real-time quantitative RT-PCR in a panel of AML cell lines. (B) Specific cytotoxicity of LRH-1–specific CTL RP1 against AML cell lines KG1, KG1a, and K562 with and without transduction of HLA-B*0702. Specific lysis was determined after 2 days of coculture in a flow cytometry–based cytotoxicity assay at an E/T ratio of 0.5:1. (C) Survival up to 3 days of HLA-B*0702–transduced AML cell lines KG1 (▴), KG1a (▾), and K562 (♦) cocultured with LRH-1–specific CTL RP1 at an E/T ratio of 0.5:1. Data are displayed as mean plus or minus SD of triplicate wells.
LRH-1–specific CTL specifically target CD34+ CML and AML progenitor cells
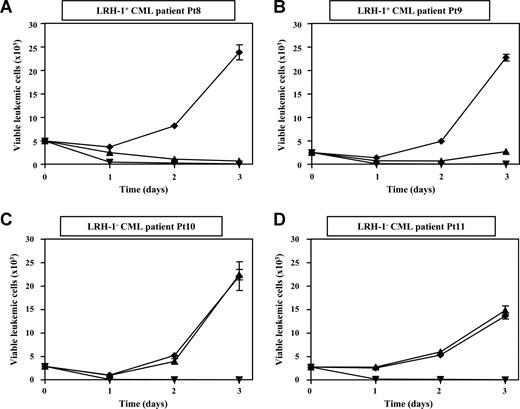
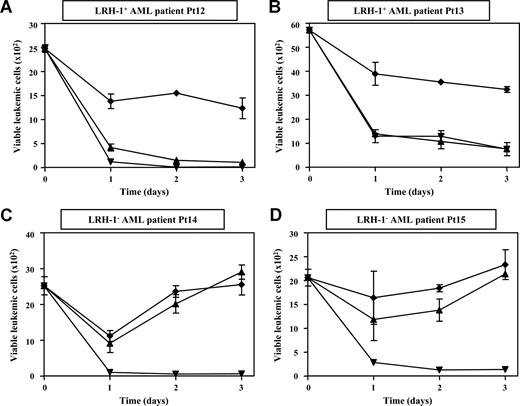
To investigate whether primary myeloid leukemic progenitor cells are susceptible to LRH-1 CTL-mediated killing, we performed cytotoxicity assays with CD34+ cells isolated from PB or BM of HLA-B7+ chronic-phase CML Pt8 to Pt11 and of HLA-B7+ AML Pt12 to Pt15. We observed that CD34+ progenitor cells from 2 LRH-1+ CML patients (Pt8 and Pt9), which had a P2X5 mRNA expression level of 4.68 and 1.86, respectively, were efficiently lysed by LRH-1–specific CTLs. Remarkably, targeting of CML progenitor cells by LRH-specific CTLs at a very low E/T ratio of 0.5:1 resulted in robust growth inhibition within 3 days (Figure 2A,B). No cytotoxicity and growth inhibition of CD34+ cells from 2 LRH-1− CML patients (Pt10 and Pt11) was observed, demonstrating specific targeting by LRH-1–reactive CTLs (Figure 2C,D). In addition, we observed that CD34+ cells from 2 LRH-1+ AML patients (Pt12 and Pt13) were efficiently lysed by LRH-1–specific CTLs at a very low E/T ratio of 0.5:1 (Figure 3A,B). No cytotoxicity and growth inhibition of CD34+ cells from 2 LRH-1− AML patients (Pt14 and Pt15) was observed, again demonstrating specific targeting by LRH-1–reactive CTLs (Figure 3C,D). By contrast, both LRH-1+ and LRH-1− CD34+ target cells of CML and AML patients were efficiently killed by allo-HLA-B7–specific CTLs.
Myeloid leukemic CD34+ progenitor cells from chronic-phase CML patients are efficiently lysed by LRH-1–specific CTLs. Survival of sorted CD34+ leukemic cells in flow cytometry–based cytotoxicity assays was determined from 4 HLA-B7+ patients after incubation with LRH-1–specific CTL RP1 (▴), HLA-B7–specific CTL KOR18 (▾; ie, positive control), or medium (♦) in the presence of growth factors. Proliferation of viable CFSE-labeled leukemic cells is shown from 2 LRH-1+ CML patients (A,B) and from 2 LRH-1− CML patients (C,D) in the absence or presence of CTLs at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
Myeloid leukemic CD34+ progenitor cells from chronic-phase CML patients are efficiently lysed by LRH-1–specific CTLs. Survival of sorted CD34+ leukemic cells in flow cytometry–based cytotoxicity assays was determined from 4 HLA-B7+ patients after incubation with LRH-1–specific CTL RP1 (▴), HLA-B7–specific CTL KOR18 (▾; ie, positive control), or medium (♦) in the presence of growth factors. Proliferation of viable CFSE-labeled leukemic cells is shown from 2 LRH-1+ CML patients (A,B) and from 2 LRH-1− CML patients (C,D) in the absence or presence of CTLs at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
Myeloid leukemic CD34+ progenitor cells from AML patients are efficiently lysed by LRH-1–specific CTLs. Survival of sorted CD34+ leukemic cells in flow cytometry–based cytotoxicity assays was determined from 4 HLA-B7+ patients after incubation with LRH-1–specific CTL RP1 (▴), HLA-B7-specific CTL KOR18 (▾; ie, positive control), or medium (♦) in the presence of growth factors. Proliferation of viable CFSE-labeled leukemic cells is shown from 2 LRH-1+ AML patients (A,B) and from 2 LRH-1− AML patients (C,D) in the absence or presence of CTLs at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
Myeloid leukemic CD34+ progenitor cells from AML patients are efficiently lysed by LRH-1–specific CTLs. Survival of sorted CD34+ leukemic cells in flow cytometry–based cytotoxicity assays was determined from 4 HLA-B7+ patients after incubation with LRH-1–specific CTL RP1 (▴), HLA-B7-specific CTL KOR18 (▾; ie, positive control), or medium (♦) in the presence of growth factors. Proliferation of viable CFSE-labeled leukemic cells is shown from 2 LRH-1+ AML patients (A,B) and from 2 LRH-1− AML patients (C,D) in the absence or presence of CTLs at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
Previously, we reported that mature myeloid cells such as monocytes and monocyte-derived dendritic cells (DCs) have a relatively low P2X5 mRNA expression, which is insufficient for recognition by LRH-1–specific CTLs.8 Here, we studied the effect of differentiation of CD34+ CML cells on LRH-1 CTL-mediated lysis. Therefore, CD34+ cells from 2 LRH-1+ CML patients (Pt8 and Pt9) were cultured for 7 days in the presence of differentiation-inducing growth factors and used in cytotoxicity assays. These cytokine-maturated CD34− CML cells from both patients were only marginally lysed by LRH-1–specific CTLs (Figure 4A,B). This decrease in responsiveness was associated with more than 10-fold down-regulation of P2X5 mRNA expression from 4.68 to 0.42 and from 1.86 to 0.14 in LRH-1+ CML Pt8 and Pt9, respectively (Figure 4C). Cytospin analysis confirmed this difference in susceptibility for cytotoxicity by LRH-1–specific CTLs between primary CD34+ CML progenitor cells and cytokine-maturated CML cells (Figure 4D). Allo-HLA-B7–specific CTLs showed equally effective lysis of both primitive and maturated CML cells. Together, these data demonstrate that LRH-1–specific CTLs effectively target primary leukemic progenitor cells from chronic phase CML and AML patients, whereas mature myeloid leukemic cells are not at all or only scarcely lysed.
Cytokine-maturated LRH-1+ CD34− CML cells are not lysed by LRH-1–specific CTLs. (A,B) Purified CD34+ CML cells were cultured for 1 week with growth factors before incubation with LRH-1–specific CTL RP1 (▴), HLA-B7-specific CTL KOR18 (▾; ie, positive control), or medium (♦) only. Survival of CFSE-labeled cytokine-maturated LRH-1+ CD34− CML cells from 2 LRH-1+ CML patients was determined in the absence or presence of CTLs at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells. (C) P2X5 mRNA expression was determined by real-time quantitative RT-PCR in sorted CD34+ CML cells (d0; ■) from 2 LRH-1+ CML patients and in the cytokine-matured CD34− progeny of these cells (d7;  ). Cytokine-maturation of CD34+ CML cells results in a down-regulation of P2X5 expression, confirming the marginal susceptibility for LRH-1 CTL-mediated lysis. The arbitrary threshold of 0.4 is indicated with a dashed horizontal line. (D) Cytospins of CD34+ progenitor cells (top panel) and cytokine-maturated CD34− cells (bottom panel) from LRH-1+ CML Pt8. Target cells were cocultured with CTLs at an E/T ratio of 0.5:1 for 2 days, after which the cells were analyzed. Morphologically, the CD34+ progenitor cells consisted of myeloblasts, whereas the cytokine-matured CD34− progeny of these cells consisted of promyelocytes, myelocytes, and granulocytes. The progenitor cells were clearly lysed by LRH-1–specific CTL RP1, whereas the more differentiated cells were no longer attacked. In contrast, both cell populations were susceptible to the allo-HLA-B7 control CTL KOR18. Imaging was performed with a Zeiss Axioskop microscope (Zeiss, Sliedrecht, The Netherlands) equipped with a Sony 3CCD color video camera system. Pictures were taken using a 40×/0.75 objective.
). Cytokine-maturation of CD34+ CML cells results in a down-regulation of P2X5 expression, confirming the marginal susceptibility for LRH-1 CTL-mediated lysis. The arbitrary threshold of 0.4 is indicated with a dashed horizontal line. (D) Cytospins of CD34+ progenitor cells (top panel) and cytokine-maturated CD34− cells (bottom panel) from LRH-1+ CML Pt8. Target cells were cocultured with CTLs at an E/T ratio of 0.5:1 for 2 days, after which the cells were analyzed. Morphologically, the CD34+ progenitor cells consisted of myeloblasts, whereas the cytokine-matured CD34− progeny of these cells consisted of promyelocytes, myelocytes, and granulocytes. The progenitor cells were clearly lysed by LRH-1–specific CTL RP1, whereas the more differentiated cells were no longer attacked. In contrast, both cell populations were susceptible to the allo-HLA-B7 control CTL KOR18. Imaging was performed with a Zeiss Axioskop microscope (Zeiss, Sliedrecht, The Netherlands) equipped with a Sony 3CCD color video camera system. Pictures were taken using a 40×/0.75 objective.
Cytokine-maturated LRH-1+ CD34− CML cells are not lysed by LRH-1–specific CTLs. (A,B) Purified CD34+ CML cells were cultured for 1 week with growth factors before incubation with LRH-1–specific CTL RP1 (▴), HLA-B7-specific CTL KOR18 (▾; ie, positive control), or medium (♦) only. Survival of CFSE-labeled cytokine-maturated LRH-1+ CD34− CML cells from 2 LRH-1+ CML patients was determined in the absence or presence of CTLs at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells. (C) P2X5 mRNA expression was determined by real-time quantitative RT-PCR in sorted CD34+ CML cells (d0; ■) from 2 LRH-1+ CML patients and in the cytokine-matured CD34− progeny of these cells (d7;  ). Cytokine-maturation of CD34+ CML cells results in a down-regulation of P2X5 expression, confirming the marginal susceptibility for LRH-1 CTL-mediated lysis. The arbitrary threshold of 0.4 is indicated with a dashed horizontal line. (D) Cytospins of CD34+ progenitor cells (top panel) and cytokine-maturated CD34− cells (bottom panel) from LRH-1+ CML Pt8. Target cells were cocultured with CTLs at an E/T ratio of 0.5:1 for 2 days, after which the cells were analyzed. Morphologically, the CD34+ progenitor cells consisted of myeloblasts, whereas the cytokine-matured CD34− progeny of these cells consisted of promyelocytes, myelocytes, and granulocytes. The progenitor cells were clearly lysed by LRH-1–specific CTL RP1, whereas the more differentiated cells were no longer attacked. In contrast, both cell populations were susceptible to the allo-HLA-B7 control CTL KOR18. Imaging was performed with a Zeiss Axioskop microscope (Zeiss, Sliedrecht, The Netherlands) equipped with a Sony 3CCD color video camera system. Pictures were taken using a 40×/0.75 objective.
). Cytokine-maturation of CD34+ CML cells results in a down-regulation of P2X5 expression, confirming the marginal susceptibility for LRH-1 CTL-mediated lysis. The arbitrary threshold of 0.4 is indicated with a dashed horizontal line. (D) Cytospins of CD34+ progenitor cells (top panel) and cytokine-maturated CD34− cells (bottom panel) from LRH-1+ CML Pt8. Target cells were cocultured with CTLs at an E/T ratio of 0.5:1 for 2 days, after which the cells were analyzed. Morphologically, the CD34+ progenitor cells consisted of myeloblasts, whereas the cytokine-matured CD34− progeny of these cells consisted of promyelocytes, myelocytes, and granulocytes. The progenitor cells were clearly lysed by LRH-1–specific CTL RP1, whereas the more differentiated cells were no longer attacked. In contrast, both cell populations were susceptible to the allo-HLA-B7 control CTL KOR18. Imaging was performed with a Zeiss Axioskop microscope (Zeiss, Sliedrecht, The Netherlands) equipped with a Sony 3CCD color video camera system. Pictures were taken using a 40×/0.75 objective.
LRH-1 CTL-mediated killing of CML blasts is enhanced by IFN-γ pretreatment.
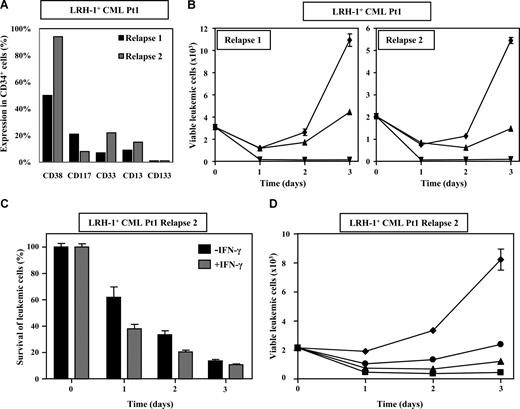
LRH-1–specific CTLs were initially isolated from CML Pt1 after treatment with DLI for relapse of the disease in accelerated phase.8 To determine whether CML progenitor cells from this patient were responsive to LRH-1 CTL-mediated killing, we performed cytotoxicity assays with CD34+ cells obtained at early relapse 11 months after SCT and at late relapse 4 years after DLI.8 Phenotypic analysis of CD34+ cells showed that these relapse samples were composed of different subpopulations based on expression of CD38, CD117, CD33, CD13, and CD133 (Figure 5A). However, both relapse samples similarly expressed P2X5 mRNA (ie, 2.39 for relapse 1 and 2.88 for relapse 2), and the P2X5 mRNA levels were comparable with those observed in CD34+ cells of chronic phase CML patients (Pt8 and Pt9). Unexpectedly, we observed that CD34+ cells from accelerated phase CML Pt1 (Figure 5B) were less efficiently lysed by LRH-1–specific CTLs than CD34+ cells from chronic phase CML and AML patients. Nevertheless, overnight stimulation of CD34+ cells of CML Pt1 with 100 U/mL IFN-γ enhanced susceptibility to lysis by LRH-1–specific CTLs (Figure 5C). This improved killing was not associated with an increase of P2X5 mRNA levels, but correlated with up-regulation of HLA-B7 and adhesion molecules such as CD54 at the cell surface of the leukemic blasts (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Although CD34+ CML cells from this patient were less susceptible to lysis by LRH-specific CTLs, targeting of IFN-γ–treated CML cells at a relatively high E/T ratio of 10:1 resulted in robust growth inhibition within 3 days (Figure 5D). Together, these data indicate that lysis of myeloid leukemic progenitor cells that are relatively refractory to alloreactive CTLs can be further augmented by higher CTL-target ratios and in the presence of inflammatory cytokines.
Myeloid leukemic CD34+ progenitor cells from accelerated-phase CML Pt1 are efficiently lysed by LRH-1–specific CTLs under inflammatory conditions and with relatively high E/T ratios. (A) Subpopulations of CD34+ cells in relapse samples 1 and 2 from CML Pt1 were phenotyped with antibodies directed against CD38, CD117, CD33, CD13, and CD133. (B) Survival of sorted CD34+ leukemic cells from relapses 1 and 2 from Pt1 was determined in flow cytometry–based cytotoxicity assays after incubation with LRH-1–specific CTL RP1 (▴), HLA-B7–specific CTL KOR18 (▾; ie, positive control), or medium (♦) in the presence of growth factors. Proliferation of viable CFSE-labeled leukemic cells is shown in the absence or presence of CTLs at an E/T ratio of 3:1. Data are depicted as mean plus or minus SD of triplicate wells. (C)usb Pretreatment with IFN-γ increases susceptibility to LRH-1 CTL-mediated lysis. Percentage survival of CD34+ cells from relapse 2 with or without preincubation with inflammatory cytokine IFN-γ in the presence of LRH-1–specific CTL RP1 was determined in flow cytometry–based cytotoxicity assays. Survival is calculated compared with medium after coculture at an E/T ratio of 10:1. Data are depicted as mean plus or minus SD of triplicate wells. (D) Higher E/T ratios result in improved lysis of IFN-γ–pretreated CD34+ cells from relapse 2. Survival of sorted CD34+ leukemic cells from relapse 2 from Pt1 was determined in flow cytometry–based cytotoxicity assays after incubation with LRH-1–specific CTL RP1 at E/T ratios 10:1 (■), 3:1 (▴), and 0.5:1 (●) or medium (♦) in the presence of growth factors. Data are depicted as mean plus or minus SD of triplicate wells.
Myeloid leukemic CD34+ progenitor cells from accelerated-phase CML Pt1 are efficiently lysed by LRH-1–specific CTLs under inflammatory conditions and with relatively high E/T ratios. (A) Subpopulations of CD34+ cells in relapse samples 1 and 2 from CML Pt1 were phenotyped with antibodies directed against CD38, CD117, CD33, CD13, and CD133. (B) Survival of sorted CD34+ leukemic cells from relapses 1 and 2 from Pt1 was determined in flow cytometry–based cytotoxicity assays after incubation with LRH-1–specific CTL RP1 (▴), HLA-B7–specific CTL KOR18 (▾; ie, positive control), or medium (♦) in the presence of growth factors. Proliferation of viable CFSE-labeled leukemic cells is shown in the absence or presence of CTLs at an E/T ratio of 3:1. Data are depicted as mean plus or minus SD of triplicate wells. (C)usb Pretreatment with IFN-γ increases susceptibility to LRH-1 CTL-mediated lysis. Percentage survival of CD34+ cells from relapse 2 with or without preincubation with inflammatory cytokine IFN-γ in the presence of LRH-1–specific CTL RP1 was determined in flow cytometry–based cytotoxicity assays. Survival is calculated compared with medium after coculture at an E/T ratio of 10:1. Data are depicted as mean plus or minus SD of triplicate wells. (D) Higher E/T ratios result in improved lysis of IFN-γ–pretreated CD34+ cells from relapse 2. Survival of sorted CD34+ leukemic cells from relapse 2 from Pt1 was determined in flow cytometry–based cytotoxicity assays after incubation with LRH-1–specific CTL RP1 at E/T ratios 10:1 (■), 3:1 (▴), and 0.5:1 (●) or medium (♦) in the presence of growth factors. Data are depicted as mean plus or minus SD of triplicate wells.
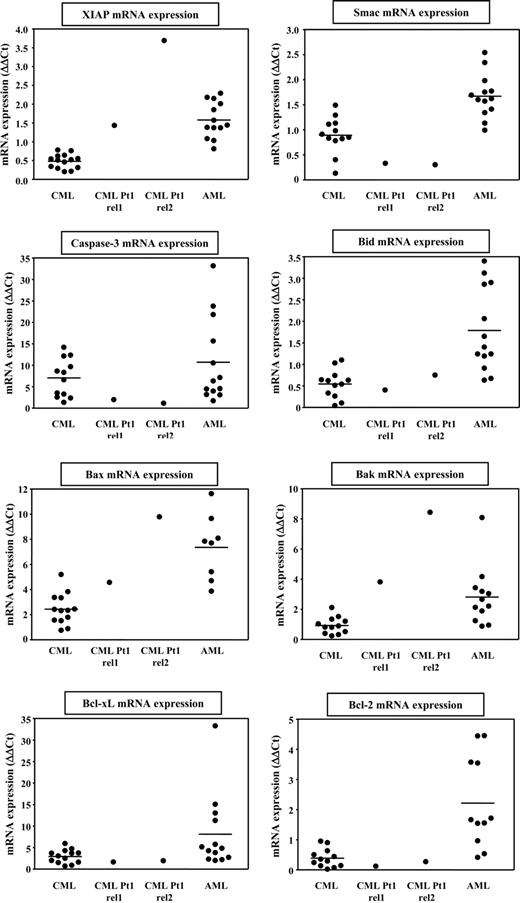
Elevated XIAP expression counteracts LRH-1 CTL-mediated cell death of leukemia progenitor cells
To gain insight into mechanisms that could explain the relative resistance of CML relapse cells from Pt1 to LRH-1 CTL-mediated lysis, we determined expression levels of several apoptosis-regulating genes in CD34+ leukemia cells from CML and AML patients by quantitative RT-PCR. Interestingly, expression of the antiapoptotic gene XIAP in CD34+ cells from Pt1 was much higher than in CD34+ cells from other CML patients (Figure 6). By contrast, we observed relatively low levels of caspase-3 and the XIAP-inhibitor Smac in the CD34+ cells from Pt1 compared with the other samples (Figure 6). For Bid, Bcl-xL, and Bcl-2, the expression levels measured in CD34+ cells from Pt1 were in the same range as found in CD34+ CML cells from other patients (Figure 6). The levels of proapoptotic genes Bax and Bak were relatively high in the CD34+ cells from Pt1, but this seems not to result in higher susceptibility to LRH-1 CTL-mediated lysis. These results suggest that elevated expression of the downstream antiapoptotic mediator XIAP could be involved in inhibiting apoptotic cell death of CD34+ CML cells from Pt1 targeted by LRH-1–specific CTLs. By contrast, elevated XIAP expression has no effect on HLA-B7 CTL-mediated killing. This may be explained by the difference in the avidity of the CTL-target interaction between LRH-1–specific CTLs and HLA-B7–specific CTLs in conjunction with differences in antigen density between specific LRH-1.B7 complexes and all HLA-B7 molecules on CD34+ cells. Our observation that hardly any IFN-γ is produced by LRH-1–specific CTLs upon recognition of CML relapse cells from Pt1, while significant IFN-γ is released by HLA-B7–specific CTLs, supports this hypothesis (Figure S2A). In addition to CML samples, we also assessed expression of apoptosis mediators in CD34+ cells from AML patients. In general, higher levels of proapoptotic as well as antiapoptotic genes were found in CD34+ AML cells compared with CD34+ CML cells. Furthermore, the expression was more heterogeneous, which might be due to a more diverse genetic background of the AML patients.
Antiapoptotic gene XIAP is highly expressed in CD34+ CML relapse cells from Pt1 compared with CML and AML cells from other patients. Expression of apoptosis-regulating genes was determined by real-time quantitative RT-PCR in CML cells from Pt1 obtained at relapse 1 and relapse 2, and a panel of CML and AML samples. Expression is depicted as ΔΔCt values and was quantified relative to AML cell line KG1, which was set at 1 ΔΔCt, and normalized for the expression of housekeeping gene HMBS.
Antiapoptotic gene XIAP is highly expressed in CD34+ CML relapse cells from Pt1 compared with CML and AML cells from other patients. Expression of apoptosis-regulating genes was determined by real-time quantitative RT-PCR in CML cells from Pt1 obtained at relapse 1 and relapse 2, and a panel of CML and AML samples. Expression is depicted as ΔΔCt values and was quantified relative to AML cell line KG1, which was set at 1 ΔΔCt, and normalized for the expression of housekeeping gene HMBS.
LRH-1–specific CD8+ T cells are frequently expanded in myeloid leukemia patients after DLI
Immunogenicity of LRH-1 is controlled by a single cytosine deletion polymorphism (rs5818907) located in exon 3 of the P2X5 gene.8 Hence, CD8+ T cells from a homozygous LRH-1− (−/− genotype) donor can elicit an allogeneic CTL response directed against P2X5-expressing cells from homozygous (C/C genotype) or heterozygous (C/− genotype) LRH-1+ patients. To determine the population frequency of the LRH-1–positive and –negative alleles, we genotyped 90 unrelated individuals by allele-specific PCR. The genotype frequencies were 4% C/C, 50% C/−, and 46% −/−, and the LRH-1+ and LRH-1− alleles were present at frequencies of 0.29 and 0.71, respectively (Table 1). Using the formula pxq3 + 0.75xp2xq2, where p and q represent allele frequencies,14 we calculated that between siblings the recipient disparity for the LRH-1+ allele is 13.6% (P = .29 and q = 0.71), whereas recipient disparity for the LRH-1− allele is 4.9% (P = .71 and q = 0.29). In addition, we experimentally determined the LRH-1 disparity in 51 HLA-B7–matched SCT donor-recipient pairs by genotyping analysis. In these 51 donor-recipient pairs, we identified 11 LRH-1+ patients (= 22%) who had received a transplant from a LRH-1− donor.
Genotype and allele frequencies of LRH-1
| Allele . | No. . | Percentage . |
|---|---|---|
| Genotype frequency | ||
| −/− | 41 | 46 |
| C/− | 45 | 50 |
| C/C | 4 | 4 |
| Allele frequency | ||
| − | 127 | 71 |
| C | 53 | 29 |
| Allele . | No. . | Percentage . |
|---|---|---|
| Genotype frequency | ||
| −/− | 41 | 46 |
| C/− | 45 | 50 |
| C/C | 4 | 4 |
| Allele frequency | ||
| − | 127 | 71 |
| C | 53 | 29 |
Next, we investigated the in vivo immunogenicity of LRH-1 in these mismatched patients following DLI. PBMCs from 7 of in total 11 LRH-1–mismatched recipients, including CML Pt1, could be analyzed for the presence of circulating LRH-1–specific CD8+ T cells using tetramer staining (Table 2). Remarkably, we detected in 3 of 7 patients a significant percentage of LRH-1–specific CD8+ T cells between 10 and 25 weeks after DLI (Table 2). These LRH-1–tetramer+ cells could not be detected (≤ 0.01%) before DLI (data not shown). Previously, we reported that in CML Pt1 emergence of LRH-1–specific CD8+ T cells coincided with the disappearance of Bcr-Abl–positive CML cells.8 Here, we report that in addition to this CML patient, LRH-1–tetramer+ CD8+ T cells could be detected in 2 other myeloid leukemia patients following treatment with DLI. One of these patients was also a CML patient (Pt2) who was treated with imatinib mesylate (Glivec) in combination with low-dose DLI (5 × 106 CD3+ cells/kg body weight) for relapsed disease 4.5 years after allogeneic SCT. A PBMC sample obtained at 11 weeks after DLI contained a relative low frequency of 0.1% LRH-1–tetramer+ CD8+ T cells (Table 2). Specific detection of LRH-1–specific CTLs in this patient was verified after in vitro stimulation of the PBMC sample with peptide-pulsed donor EBV-LCLs. After 2 rounds of stimulation, the percentage of LRH-1–tetramer+ CD8+ T cells increased up to 0.7% (Table 2).
Characteristics of HLA-B7+ LRH-1+ patients who received a transplant from an LRH-1− donor
| Patient (Pt) . | Diagnosis . | Type of DLI . | Sample date, wk after DLI . | Presence of LRH-1–specific CD8+ T cells . | LRH-1–specific CD8+ T cells (%) in sample . | LRH-1–specific CD8+ T cells (%) after 2 weeks of culture . |
|---|---|---|---|---|---|---|
| 1 | CML | Therapeutic | 25 | + | 1.6 | 12.8 |
| 2 | CML | Therapeutic | 11 | + | 0.1 | 0.7 |
| 3 | AML | Preemptive | 10 | + | 6.9 | 15.4 |
| 4 | AML | Preemptive | 18 | − | < 0.01 | < 0.01 |
| 5 | B-ALL | Preemptive | 11 | − | < 0.01 | < 0.01 |
| 6 | T-ALL | Preemptive | 19 | − | < 0.01 | n.d. |
| 7 | MM | Preemptive | 12 | − | < 0.01 | < 0.01 |
| Patient (Pt) . | Diagnosis . | Type of DLI . | Sample date, wk after DLI . | Presence of LRH-1–specific CD8+ T cells . | LRH-1–specific CD8+ T cells (%) in sample . | LRH-1–specific CD8+ T cells (%) after 2 weeks of culture . |
|---|---|---|---|---|---|---|
| 1 | CML | Therapeutic | 25 | + | 1.6 | 12.8 |
| 2 | CML | Therapeutic | 11 | + | 0.1 | 0.7 |
| 3 | AML | Preemptive | 10 | + | 6.9 | 15.4 |
| 4 | AML | Preemptive | 18 | − | < 0.01 | < 0.01 |
| 5 | B-ALL | Preemptive | 11 | − | < 0.01 | < 0.01 |
| 6 | T-ALL | Preemptive | 19 | − | < 0.01 | n.d. |
| 7 | MM | Preemptive | 12 | − | < 0.01 | < 0.01 |
n.d. indicates not determined.
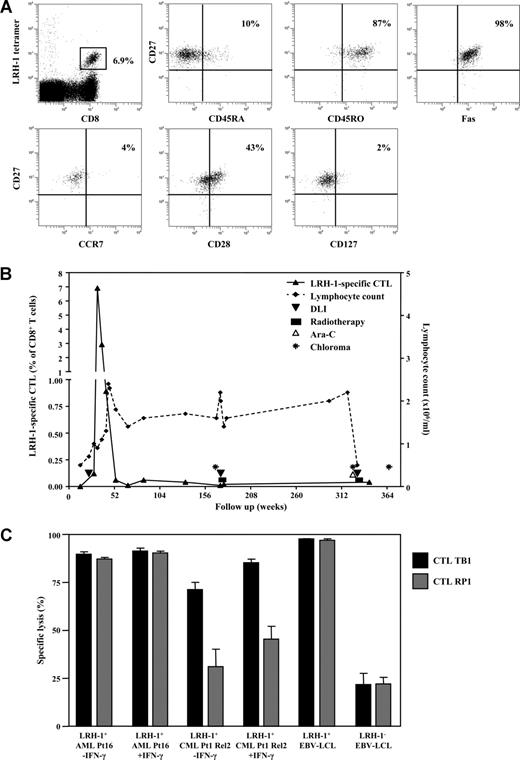
The other patient was an AML patient (Pt3) who was treated with preemptive DLI (0.1 × 108 CD3+ cells/kg body weight) at 5 months after allogeneic SCT. Strikingly, at 10 weeks after DLI a peak level of 6.9% LRH-1–specific CD8+ T cells could be detected in the PB (Table 2; Figure 7A). These in vivo–expanded LRH-1–specific CD8+ T cells had predominantly (∼90%) a CD45RA−CD45RO+CD27+CCR7−Fas+ effector/effector-memory phenotype (Figure 7A). A subfraction (43%) of the LRH-1 tetramer+ CD27+ T cells expressed CD28, which is indicative for the presence of early antigen-primed CD8+ T cells according to the differentiation model proposed by Appay et al.15 Most LRH-1–specific CD8+ T cells lacked IL-7Rα (CD127) expression at this time point. After the peak response, the number of circulating LRH-1 tetramer+ cells declined to 2.9% at 15 weeks and 0.9% at 20 weeks (Figure 7B). Thereafter, a small memory population of around 0.05% remained detectable in the PB for many years. Importantly, despite the presence of these very high numbers of LRH-1–specific CD8+ T cells the patient did not develop GVHD, which correlates with the hematopoietic-restricted expression of the P2X5 target gene. Furthermore, the patient remained in complete clinical and cytogenetic remission for years after preemptive DLI. However, 33 months after DLI, the patient developed a chloroma in the left upper eyelid. Remarkably, the patient remained in continuous cytogenetic remission in the BM. The patient was treated with local radiotherapy and DLI at a dose of 0.7 × 108 CD3+ cells/kg body weight. The chloroma disappeared and the DLI did not elicit GVHD. However, 3 years later (ie, 6 years after SCT) the patient developed a second chloroma paravertebrally, again without leukemic involvement in the BM. Despite successful treatment with high-dose cytarabine, DLI (1.4 × 108 CD38 cells/kg body weight), and local radiotherapy, a third chloroma in the gut was demonstrated 8 months later. She was given a second allogeneic SCT from an HLA-identical sister but developed GVHD grade 3 and died from infectious complications. During this period of extramedullary relapses and concomitant treatment, LRH-1–specific CD8+ memory T cells were present but did not respond (Figure 7B).
LRH-1–specific CTL TB1 of AML Pt3 efficiently recognizes LRH-1+ CD34+ AML and CML progenitor cells. (A) LRH-1–tetramer+ CD8+ T cells from Pt3 were phenotyped for their expression of CD45RA, CD27, Fas, CD127, CD28, CD45RO, and CCR7. (B) Longitudinal follow-up of LRH-1–tetramer+ CD8+ T cells in vivo from Pt3 in relation to clinical outcome. Depicted is the percentage of LRH-1–tetramer+ T cells in the CD8+ T-cell population (left y-axis) and the lymphocyte count (right y-axis) together with treatments and the occurrence of chloromas. (C) Specific lysis of CD34+ leukemic progenitor cells with or without preincubation with IFN-γ in the presence of LRH-1–specific CTL TB1 (■) or RP1 ( ) was determined in flow cytometry–based cytotoxicity assays. Lysis is calculated compared with medium after 2 days of coculture at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
) was determined in flow cytometry–based cytotoxicity assays. Lysis is calculated compared with medium after 2 days of coculture at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
LRH-1–specific CTL TB1 of AML Pt3 efficiently recognizes LRH-1+ CD34+ AML and CML progenitor cells. (A) LRH-1–tetramer+ CD8+ T cells from Pt3 were phenotyped for their expression of CD45RA, CD27, Fas, CD127, CD28, CD45RO, and CCR7. (B) Longitudinal follow-up of LRH-1–tetramer+ CD8+ T cells in vivo from Pt3 in relation to clinical outcome. Depicted is the percentage of LRH-1–tetramer+ T cells in the CD8+ T-cell population (left y-axis) and the lymphocyte count (right y-axis) together with treatments and the occurrence of chloromas. (C) Specific lysis of CD34+ leukemic progenitor cells with or without preincubation with IFN-γ in the presence of LRH-1–specific CTL TB1 (■) or RP1 ( ) was determined in flow cytometry–based cytotoxicity assays. Lysis is calculated compared with medium after 2 days of coculture at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
) was determined in flow cytometry–based cytotoxicity assays. Lysis is calculated compared with medium after 2 days of coculture at an E/T ratio of 0.5:1. Data are depicted as mean plus or minus SD of triplicate wells.
P2X5-expressing AML and CML cells are efficiently lysed by LRH-1–specific CTLs isolated from 2 different patients
To investigate whether LRH-1–specific CTLs present in AML Pt3 showed similar cytotoxicity as the LRH-1–specific CTLs generated from CML Pt1, we have isolated LRH-1 tetramer+ T cells from Pt3. After antigen-specific stimulation, tetramer analysis revealed that more than 95% of CD8+ T cells were LRH-1 tetramer positive (data not shown). This CD8+ CTL culture, termed TB1, mediated efficient lysis against LRH-1+ EBV-LCLs, but not against LRH-1− donor EBV-LCLs, demonstrating specific recognition of the LRH-1–epitope (Figure 7C). Targeting of HLA-B7+ LRH-1+ CD34+ AML cells from Pt16 by CTL TB1 at a low E/T ratio of 0.5:1 resulted in efficient lysis (Figure 7C). Similar strong lysis of AML cells is shown for the CML Pt1-derived CTL RP1. CD34+ CML cells from Pt1 were again less susceptible to LRH-1 CTL-mediated lysis, although CTL TB1 displayed somewhat more efficient lysis than CTL RP1. For both CTLs, lysis could be improved by pretreatment of the CD34+ CML cells with IFN-γ, similar to previous results. Furthermore, CTL TB1 as well as CTL RP1 did not produce IFN-γ upon recognition of AML cells or CML relapse cells from Pt1, irrespective of IFN-γ treatment, whereas IFN-γ was produced upon coculture with LRH-1+ EBV-LCLs (Figure S2B). Analysis of the phenotypical properties of both CTL cultures showed that RP1 and TB1 were both negative for CD27, CD28, CD62L, CCR7, and CD127 (IL-7Rα), and expressed similar levels of perforin (data not shown). Furthermore, we compared IFN-γ production by CTL RP1 and TB1 upon recognition of various concentrations of the LRH-1 peptide loaded on HLA-B*0702–transduced K562 cells. We observed that IFN-γ production by CTL TB1 was slightly higher than by CTL RP1, suggesting a somewhat higher avidity of CTL TB1 for LRH-1+ target cells (Figure S3). These observations are in agreement with the slightly better lysis of CD34+ CML relapse cells from Pt1 by CTL TB1 (Figure 7C).
Collectively, these results illustrate that LRH-1–specific T-cell responses can be induced in vivo in both CML and AML patients, and that these LRH-1–reactive CTLs are involved in targeting CD34+ AML and CML progenitor cells.
Discussion
Hematopoietic cell–restricted MiHAs are crucial targets for selective induction of a GVL response in the absence of GVHD after HLA-identical allogeneic SCT. In a previous study, we described the identification of a hematopoietic cell–restricted MiHA, designated LRH-1, which is derived from the polymorphic P2X5 protein.8 Interestingly, LRH-1 elicited an allogeneic CTL response in a patient with advanced CML treated with DLI, which was associated with clinical remission. Moreover, gene expression studies showed that the LRH-1–encoding P2X5 gene is selectively expressed by lymphoid cells and by normal and malignant CD34+ progenitor subpopulations. Importantly, no significant P2X5 gene expression was found in prominent GVHD target organs, such as skin, liver, colon, and small intestine. These results suggest that LRH-1 may contribute to induction of GVL immunity in leukemia patients treated with allogeneic SCT in the absence of GVHD. In this study, we investigated susceptibility of CD34+ myeloid leukemic progenitor cells to LRH-1 CTL-mediated lysis and the applicability of LRH-1 as target for immunotherapy of myeloid leukemia.
It has been recognized that for complete eradication of leukemia after SCT, induction of effective alloreactive T-cell responses against leukemic stem and progenitor cells is essential.16,17 Several studies have demonstrated a correlation between the presence of T cells recognizing leukemic CD34+ progenitor cells and the occurrence of complete remissions in vivo.18-21 Therefore, MiHAs that are expressed on CD34+ leukemic progenitor cells are highly relevant targets for immunotherapy. Previously, we reported that LRH-1 elicited a profound CTL response in a CML patient treated with DLI that was associated with the disappearance of Bcr-Abl–positive tumor cells (ie, Pt1 in this study). Although we observed that LRH-1–specific CTLs efficiently recognized and lysed lymphoid cells, bulk CML mononuclear cells of the patient from whom this CTL culture RP1 was originally isolated were not recognized.8 However, the majority of CML cells are mature myeloid cells that express very low levels of P2X5. In this study, we therefore investigated whether CD34+ leukemic progenitor cells are potential targets of LRH-1–specific CTLs. Significant P2X5 expression was detected in both CD34+CD38− and CD34+CD38+ subpopulations from most CML and AML patients.8 Based on the results of previous expression and CTL recognition studies, these expression levels are expected to be sufficient for LRH-1 CTL-mediated lysis.8 To determine whether primary leukemic progenitor cells are susceptible for LRH-1 CTL-mediated lysis, we performed cytotoxicity assays with CD34+ progenitor cells isolated from PB or BM of CML and AML patients. In these assays, we clearly demonstrated that leukemic CD34+ progenitor cells from chronic phase LRH-1+ CML patients as well as from LRH-1+ AML patients were efficiently lysed by LRH-1–specific CTLs. On the other hand, mature myeloid leukemic cells were not at all or only scarcely lysed, which was in concordance with a strong decrease in P2X5 gene expression levels after cytokine maturation. In addition to the LRH-1–specific CTLs from CML Pt1 (ie, RP1), we isolated a second LRH-1–specific CTL (ie, TB1) from AML Pt3, in whom a high level of LRH-1–specific CD8+ T cells was induced at 10 weeks after DLI. Interestingly, we observed that both CTLs displayed similar cytotoxicity against primary CD34+ CML and AML progenitor cells. These results show that LRH-1–specific T-cell responses can be induced in vivo in both CML and AML patients and that these LRH-1–specific CTLs can recognize and lyse both AML and CML progenitor cells.
Despite the effective cytotoxicity by LRH-1–specific CTLs observed in flow cytometry–based cytotoxicity assays, no IFN-γ production could be measured for both CTLs after stimulation with myeloid leukemic progenitor cells. By contrast, LRH-1–specific CTLs efficiently produced IFN-γ upon recognition of LRH-1+ EBV-LCLs. This may be explained by differences in antigen concentration at the cell surface and strength of CTL-target interactions between myeloid leukemic progenitor cells and EBV-LCLs. It has been well established that dual activation thresholds are present for cytotoxicity and cytokine production in CTLs upon antigen-specific interactions with its target.22,23 At low antigen concentrations a lytic immunologic synapse can be formed, whereas for production of IFN-γ higher antigen concentrations and the formation of a mature immunologic synapse are needed.22-24 Similar results were previously observed in the recognition of P2X5-expressing LRH-1+ lymphoid malignancies by LRH-1–specific CTLs.9
Although efficient lysis of most CD34+ CML and AML samples was observed, accelerated phase CD34+ relapse CML cells from Pt1 were less efficiently lysed by LRH-1–specific CTLs. However, lysis could be improved by increase of the E/T ratio and preincubation of CML cells with inflammatory cytokine IFN-γ. The observed lysis of CML relapse cells from Pt1 confirms the previously shown correlation between expansion of LRH-1–specific CD8+ T cells and the disappearance of CML.8 Preincubation of CD34+ cells with IFN-γ induced an increase of HLA-B7 and CD54, which probably resulted in more effective lytic immunologic synapses. A similar effect of IFN-γ pretreatment on the expression of HLA-B7 and CD54, resulting in improved CTL-mediated lysis, was previously observed for solid tumor cells.11 Therefore, the presence of IFN-γ may have a beneficial effect on the LRH-1–specific GVL response. However, we observed that LRH-1–specific CTLs did not produce IFN-γ in vitro upon recognition of myeloid leukemic progenitor cells. Nevertheless in vivo, IFN-γ could be supplied by coexisting CD8+ cytotoxic T cells for other MiHAs on leukemic cells.
Studies by Ravi et al showed that coexistence of specific mitochondrial signaling defects, such as decreased expression of Smac, together with expression of XIAP decreases the sensitivity of cancer cells to CTL-mediated apoptosis.25 To elucidate whether this mechanism could play a role in the relative resistance to lysis of CML relapse cells from Pt1, we compared expression levels of apoptosis-regulating genes between CD34+ cells from Pt1 and a panel of CML and AML samples from other patients. Interestingly, we observed elevated expression of the antiapoptotic gene XIAP in CD34+ cells from Pt1. Moreover, the levels of proapoptotic genes caspase-3 and Smac were relatively low, whereas expression levels of proapoptotic genes Bax and Bak were relatively high. However, XIAP-mediated inhibition of effector caspases may have a more dominant effect on CTL-mediated apoptosis than increased expression of Bax and Bak, which exert their function more upstream in the apoptosis pathway. In addition, we observed relatively low levels of XIAP-inhibitor Smac, which might result in more prominent resistance of XIAP-expressing leukemia cells of CML Pt1 following targeting by LRH-1 CTLs. We showed that pretreatment with IFN-γ and increased CTL-target ratios largely corrected the antiapoptotic balance and significantly improved lysis by LRH-1–specific CTLs. However, this mechanism could have played a role in evasion of leukemia cells in accelerated phase CML Pt1, eventually resulting in uncontrollable disease.8
At present, several MiHAs have been identified that can induce allogeneic T-cell responses in patients. Of these molecularly defined MiHAs, LRH-1,8 HB-1,26 HA-1,27 HA-2,28 PANE-1,29 LB-ECGF-1,30 ACC-1,31 ACC-2,31 and ACC-632 have a hematopoietic-restricted expression. The applicability of these MiHAs as targets for immunotherapy depends on the disparity of the MiHA+ allele in the recipient direction and on the population frequency of the HLA-restriction molecule in which the MiHA is presented.33 Here, we determined the population frequencies of the LRH-1+ and LRH-1− alleles by genotyping 90 unrelated individuals, which revealed a recipient disparity for the LRH-1+ allele of 13.6%. Consequently, the overall applicability of LRH-1–based immunotherapy is estimated around 2.3% based on the recipient disparity for the LRH-1+ allele (13.6%) and the HLA-B7 phenotype frequency (∼17% in the white population34 ). Therefore, LRH-1 belongs to the most suitable MiHAs for immunotherapy, together with MiHA HA-1, ACC-1, and ACC-2, which have an overall applicability of 10.6%, 5.8%, and 2.7%, respectively.33 Interestingly, the 9 amino acid disparity between donor and recipient cells caused by the frameshift polymorphism in the P2X5 protein may give rise to additional antigenic epitopes that are presented by other frequent HLA class I or class II alleles. Currently, we are investigating whether other putative peptides are derived from P2X5 that act as immunogenic T-cell epitopes. This may further increase the overall applicability of P2X5 as an immunotherapeutic target.
To determine the immunogenicity of LRH-1 in different patients, we investigated the occurrence of LRH-1–specific T-cell responses in LRH-1+ patients treated with DLI. Seven LRH-1+ patients who received DLI from an LRH-1− donor were analyzed for the presence of LRH-1–specific CD8+ T cells by tetramer staining. LRH-1–specific CD8+ T cells could be detected in 3 (43%) of 7 LRH-1+ recipients within 10 to 25 weeks after DLI. These results suggest that LRH-1 is a relatively immunogenic epitope. In the CML patient (Pt1) from whom the LRH-1–specific CD8+ CTL culture RP1 was originally isolated, we detected 1.6% LRH-1–tetramer+ CD8+ T cells at 25 weeks after therapeutic DLI. Although LRH-1 has a hematopoietic-restricted expression, this patient developed mild chronic GVHD. However, since it has been demonstrated that allogeneic T-cell responses against multiple MiHAs can be elicited after DLI,19 the mild GVHD may be due to the presence of T cells directed against MiHAs with ubiquitous expression. Remarkably, 6.9% LRH-1–specific CD8+ T cells in the total CD8+ T-cell population were present in an AML patient (Pt3) treated with preemptive DLI, who did not develop GVHD. This patient remained in complete cytogenetic remission for 33 months. However, years after the LRH-1 CTL response this patient developed relapses at sanctuary sites in the absence of leukemic involvement in the BM. Similar cases of extramedullary relapses in the presence of continuous BM remission have been described, suggesting disparate GVL effect at different sites in the body.35-37 In these patients, certain leukemic cells have migrated to immune-protected sites where they escaped from GVL immunity due to diminished T-cell homing to these tissues, evasion of immune recognition, and/or development of an immunosuppressive network at the tumor site. Finally, a lower amount of 0.1% LRH-1–specific CD8+ T cells was detected in a CML patient (Pt2) who was treated with imatinib mesylate (Glivec) in combination with low-dose DLI for relapsed disease. This low percentage of LRH-1–specific T cells may be caused by the presence of imatinib mesylate, which has been demonstrated to interfere with T-cell activation and proliferation.38 The absence of severe GVHD in these 3 patients together with the presence of significant numbers of LRH-1–specific T cells emphasizes the hematopoietic-restricted expression of LRH-1 and its potential to induce GVL without inducing GVHD. Together with our in vitro cytotoxicity data, the observed molecular remission in CML Pt1 after therapeutic DLI and the prolonged BM remission in AML Pt3 after preemptive DLI indicate that LRH-1–specific T-cell responses can be induced in vivo in LRH-1+ patients treated with an LRH-1− donor and that these LRH-1–specific CD8+ T cells can play an important role in specific lysis of CD34+ leukemic progenitor cells.
In conclusion, we demonstrate that MiHA LRH-1 is an attractive GVL-specific antigen, which induces CD8+ T-cell responses in multiple patients after DLI. We show that CD34+ leukemic progenitor cells, but not mature myeloid cells, are susceptible to lysis by in vivo induced LRH-1–specific CTLs. These results indicate that LRH-1–based immunotherapy following allogeneic SCT may be a potent treatment for CML and AML patients. Recently, we demonstrated that P2X5 is highly expressed in a broad range of lymphoid malignancies and several solid tumors, which results in susceptibility to LRH-1 CTL-mediated lysis.9,11 Collectively, these findings illustrate that LRH-1 represents a highly relevant target for posttransplantation vaccination or adoptive T-cell therapy to induce GVL-specific immunity in a broad range of hematologic malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rob Woestenenk and Jeroen van Velzen for assistance in flow cytometry, and Trix de Boer for technical assistance.
This work was supported by grants from the Radboud University Nijmegen (UMCN 2002-29; 2007-34), the Bekales Foundation (Brussels, Belgium), and the Ger Janssen Foundation (Nijmegen, The Netherlands).
Authorship
Contribution: W.J.N. performed research and analyzed data; I.M.O. analyzed data and wrote the paper; F.M., H.F., and J.C.M.V. performed research; R.v.d.V., A.V.S., and T.M.d.W. revised the paper; M.G.D.K., I.J., and J.H.F.F. provided vital material and revised the paper; and H.D. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: H. Dolstra, Central Hematology Laboratory, Radboud University Nijmegen Medical Centre, Geert Grooteplein 8, PO Box 9101, 6500 HB Nijmegen, The Netherlands; e-mail: h.dolstra@chl.umcn.nl.
References
Author notes
*W.J.N. and I.M.O. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal