Abstract
A small population of CD55−CD59− blood cells was detected in a patient who developed donor-type late graft failure after allogeneic stem cell transplantation (SCT) for treatment of aplastic anemia (AA). Chimerism and PIGA gene analyses showed the paroxysmal nocturnal hemoglobinuria (PNH)–type granulocytes to be of a donor-derived stem cell with a thymine insertion in PIGA exon 2. A sensitive mutation-specific polymerase chain reaction (PCR)–based analysis detected the mutation exclusively in DNA derived from the donor bone marrow (BM) cells. The patient responded to immunosuppressive therapy and achieved transfusion independence. The small population of PNH-type cells was undetectable in any of the 50 SCT recipients showing stable engraftment. The de novo development of donor cell–derived AA with a small population of PNH-type cells in this patient supports the concept that glycosyl phosphatidylinositol–anchored protein–deficient stem cells have a survival advantage in the setting of immune-mediated BM injury.
Introduction
Although small populations of CD55−CD59− blood cells are often detectable in patients with aplastic anemia (AA), it remains unclear how such paroxysmal nocturnal hemoglobinuria (PNH)-type cells arise.1 We recently encountered a patient with immune-mediated late graft failure (LGF) following allogeneic stem cell transplantation (SCT) for treatment of AA. Analyses of the patient's peripheral blood (PB) and bone marrow (BM) showed hematopoietic stem cells (HSCs) of donor origin with mutant PIGA, supporting the concept that glycosyl phosphatidylinositol–anchored protein (GPI-AP)–deficient stem cells have a survival advantage in the setting of immune mediated BM injury.
Methods
Patients
A 59-year-old man underwent allogeneic PBSCT from a human leukocyte antigen (HLA)–matched sibling donor after conditioning with fludarabin (120 mg/m2), cyclophosphamide (1200 mg/m2), and antithymocyte globulin (60 mg/kg) for treatment of very severe AA in April 2002 (Table 1) and achieved complete donor chimerism with normal blood cell counts. In January 2006, he developed pancytopenia and was diagnosed as having LGF without residual recipient cells. The patient underwent a second PBSCT from the original donor without preconditioning on February 8, 2006. Pancytopenia resolved completely by day 16 after PBSCT. However, at approximately day 60, the blood counts decreased gradually, and the patient became transfusion-dependent. On day 196 after the second PBSCT, the white blood cell (WBC) count was 5.3 × 109/L with 17% neutrophils, the hemoglobin concentration was 75 g/L, and the platelet count was 22 × 109/L. Treatment with horse antithymocyte globulin (ATG) and cyclosporine was started on day 205 after the second PBSCT. Transfusions were terminated after 88 days of the immunosuppressive therapy. Although the patient presently receives low-dose tacrolimus for treatment of chronic graft-versus-host disease, which developed 1 year after the second PBSCT, his pancytopenia has markedly improved as shown in Table 1. PB and BM of the patient were subjected to analyses of chimerism and flow cytometry to detect CD55−CD59− cells and PIGA gene analysis.
Hematologic parameters of donor and recipient
| Date . | Donor . | Recipient . | ||||
|---|---|---|---|---|---|---|
| Before 1st SCT . | Before 2nd SCT . | At ATG therapy . | After 20 mo of ATG therapy . | |||
| Apr 2002 . | May 2008 . | Apr 2002 . | Jan 2006 . | Aug 2006 . | Apr 2008 . | |
| WBC count, × 109/L | 7.0 | 5.1 | 1.2 | 1.7 | 5.3 | 4.0 |
| Neutrophil proportions, % | 77 | 65 | 0 | 0 | 17 | 62 |
| RBC count, × 1012/L | 4.21 | 4.43 | 2.20 | 2.75 | 2.07 | 3.04 |
| Reticulocytes, × 109/L | not tested | 35 | 2 | 3 | 26 | 61 |
| Hemoglobin, g/L | 146 | 150 | 72 | 89 | 75 | 120 |
| Platelet count, × 109/L | 261 | 230 | 19 | 52 | 22 | 54 |
| Date . | Donor . | Recipient . | ||||
|---|---|---|---|---|---|---|
| Before 1st SCT . | Before 2nd SCT . | At ATG therapy . | After 20 mo of ATG therapy . | |||
| Apr 2002 . | May 2008 . | Apr 2002 . | Jan 2006 . | Aug 2006 . | Apr 2008 . | |
| WBC count, × 109/L | 7.0 | 5.1 | 1.2 | 1.7 | 5.3 | 4.0 |
| Neutrophil proportions, % | 77 | 65 | 0 | 0 | 17 | 62 |
| RBC count, × 1012/L | 4.21 | 4.43 | 2.20 | 2.75 | 2.07 | 3.04 |
| Reticulocytes, × 109/L | not tested | 35 | 2 | 3 | 26 | 61 |
| Hemoglobin, g/L | 146 | 150 | 72 | 89 | 75 | 120 |
| Platelet count, × 109/L | 261 | 230 | 19 | 52 | 22 | 54 |
As controls, the PB from 51 SCT recipients (48 with hematologic malignancies and 3 with AA) who achieved a complete recovery of donor-derived hematopoiesis were subjected to flow cytometric analysis for the screening of CD55−CD59− cells. Of the 51 patients, 4 and 23, respectively, had acute graft-versus-host disease (GVHD) of grade II or higher and chronic GVHD at sampling.
BM aspirates were obtained from the patient's donor and 10 healthy individuals for PIGA gene analysis. Informed consent was obtained from all patients and healthy individuals in accordance with the Declaration of Helsinki for blood examination, and the experimental protocol for PIGA gene analysis was approved by our participating institutional ethics committee (No.157).
Detection of PNH-type cells
To detect GPI-AP deficient (GPI-AP−), PNH-type cells, we performed high-sensitivity 2-color flow cytometry of granulocytes and red blood cells (RBCs), as described previously.1 The presence of 0.003% or more CD55−CD59−CD11b+ granulocytes and 0.005% or more CD55−CD59−glycophorin-A+ RBCs was defined as an abnormal increase based on the results in 183 healthy individuals.2
Cell sorting and chimerism analysis
CD3+cells were isolated from the PB mononuclear cells of the patient using magnetic-activated cell sorting (MACS) CD3 Microbeads (Miltenyi Biotec, Auburn, CA). The CD55−CD59−CD11b+ granulocytes were separated from the CD55+CD59+CD11b+ granulocytes with a cell sorter (JSAN; Bay Bioscience, Yokohama, Japan). More than 95% of the sorted cells were CD55−CD59−CD11b+. The D1S80 locus was amplified from DNA of different cell populations with an AmpliFLP D1S80 PCR Amplification Kit (Perkin-Elmer Cetus, Norwalk, CT).
PIGA gene analysis
The coding regions of PIGA were amplified by seminested PCR or nested PCR from DNA extracted from the sorted PNH-type cells using 12 primer sets3,4 (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), and 6 ligation reactions were used to transform competent Escherichia coli JM109 cells (Nippon Gene, Tokyo, Japan). Five clones were selected randomly from each group of transfectants and subjected to sequencing with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Amplification refractory mutation system PCR
On the basis of a mutant sequence detected in PIGA of the patient, a nested amplification refractory mutation system (ARMS) forward primer with a 3′-terminal nucleotide sequence complementary to the mutant sequence was prepared5 (Table S1). To enhance the specificity, a mismatch at the penultimate nucleotide position of the mutation site was incorporated in the ARMS forward primer (P1).6,7 P1 and a reverse primer (P3) were used to amplify a 127 bp fragment containing the mutant sequence from the exon 2 amplified product. PCR was conducted under the following conditions; denaturation for 30 seconds at 94°C, annealing for 60 seconds at 64°C and extension for 90 seconds at 72°C for 20 cycles. Another forward primer (P2), complementary to the wild-type PIGA sequence upstream of the mutation site, was used in combination with P3 to amplify an internal control according to the same condition of ARMS-PCR.
Results and discussion
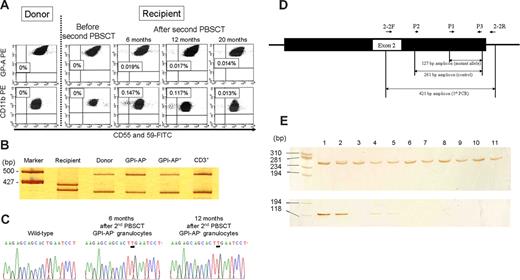
PNH-type cells were not detected in the donor or the patient at the time of development of the first LGF, whereas 0.147% PNH-type granulocytes and 0.019% PNH-type RBCs were detected in the PB obtained at the time of development of the second LGF (Figure 1A). Similar percentages of PNH-type blood cells were detectable in the PB of the patient 6 and 14 months later. When PB from 51 SCT recipients was examined, none of the patients were found to have detectable PNH-type cells (data not shown). PNH-type blood cells were also undetectable in a donor PB sample obtained 21 months later.
Analysis of PNH-type cells after the second PBSCT. (A) High-sensitivity flow cytometry detected small populations of CD55−CD59−cells in both granulocytes and red blood cells at the development of the second LGF as well as in those obtained 6 and 12 months later, but did not detect PNH-type cells in the donor or in the recipient before the second PBSCT. The numbers denote the proportion of PNH-type cells in CD11b+ granulocytes or glycophorin A+ RBCs. (B) D1S80 allelic patterns of sorted GPI-AP− granulocytes, GPI-AP+ granulocytes, and CD3+ lymphocytes. The polymerase chain reaction (PCR) products were subjected to 8% polyacrylamide gel electrophoresis and visualized by silver staining. (C) Nucleotide sequences of PIGA exon 2 in DNA from PNH-type granulocytes obtained 6 and 12 months after the second PBSCT. (D) A schematic illustration for ARMS-PCR is shown. Primer positions for the first, second are shown by short arrows. A black box and adjacent lines represent exon 2 and introns, respectively. (E) Amplified products of control PCR (the upper gel) and ARMS-PCR (the lower gel) were electrophoresed in 12.5% polyacrylamide gel and visualized by the silver staining. A pMD20-T vector containing the mutated exon 2 fragment was used as a positive control for ARMS-PCR. The template DNA derives from a plasmid containing the mutated exon 2 in lane 1, donor BM in lane 2, donor PB in lane 3, recipient BM in lane 4, recipient PB in lane 5, and BM from healthy individuals in lanes 6 to 11. PCR with a 5′ primer specific to the nucleotide sequence upstream of the mutated sequence amplified a 261 bp fragment from DNA of the donor and all healthy individuals.
Analysis of PNH-type cells after the second PBSCT. (A) High-sensitivity flow cytometry detected small populations of CD55−CD59−cells in both granulocytes and red blood cells at the development of the second LGF as well as in those obtained 6 and 12 months later, but did not detect PNH-type cells in the donor or in the recipient before the second PBSCT. The numbers denote the proportion of PNH-type cells in CD11b+ granulocytes or glycophorin A+ RBCs. (B) D1S80 allelic patterns of sorted GPI-AP− granulocytes, GPI-AP+ granulocytes, and CD3+ lymphocytes. The polymerase chain reaction (PCR) products were subjected to 8% polyacrylamide gel electrophoresis and visualized by silver staining. (C) Nucleotide sequences of PIGA exon 2 in DNA from PNH-type granulocytes obtained 6 and 12 months after the second PBSCT. (D) A schematic illustration for ARMS-PCR is shown. Primer positions for the first, second are shown by short arrows. A black box and adjacent lines represent exon 2 and introns, respectively. (E) Amplified products of control PCR (the upper gel) and ARMS-PCR (the lower gel) were electrophoresed in 12.5% polyacrylamide gel and visualized by the silver staining. A pMD20-T vector containing the mutated exon 2 fragment was used as a positive control for ARMS-PCR. The template DNA derives from a plasmid containing the mutated exon 2 in lane 1, donor BM in lane 2, donor PB in lane 3, recipient BM in lane 4, recipient PB in lane 5, and BM from healthy individuals in lanes 6 to 11. PCR with a 5′ primer specific to the nucleotide sequence upstream of the mutated sequence amplified a 261 bp fragment from DNA of the donor and all healthy individuals.
The D1S80 locus allelic pattern of the PNH-type granulocytes in the patient was compatible to that of the donor (Figure 1B). The emergence of donor-derived PNH-type cells and hematologic improvement after immunosuppressive therapy suggest that LGF arises as a result of de novo development of AA which affects the donor-derived hematopoietic stem cells (HSCs).
PIGA gene analysis of the DNA prepared from the sorted PNH-type cells of the patient obtained at the development of LGF and 6 months later showed an insertion of thymine at position 593 (codon 198) in 3 of 5 clones and 5 of 5 clones examined, respectively (Figure 1C). Mutations in other exons were not identified. The presence of a single PIGA mutation in PNH-type granulocytes and its persistence over 6 months suggest that these PNH-type cells are derived from a mutant HSC rather than from a committed granulocyte progenitor cell. Moreover, an ARMS-PCR with a 5′ primer specific to the mutated sequence amplified a 127 bp fragment from DNA of the donor BM as well as of the recipient BM and PB while it failed to amplify the same fragment in donor PB and in BM of all 10 healthy individuals (Figure 1D).
These experiments demonstrate that PIGA-mutant HSCs were present in the BM of the donor in a dormant state and were transplanted into the recipient and provide, for the first time, in vivo evidence that PIGA mutant, GPI-AP–deficient HSCs have a survival advantage in the setting of immune mediated BM injury. Similarly, relative resistance to immune injury likely accounts for the high incidence of PNH observed in association with acquired AA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ms Shizuka Yasue, Ms Megumi Yoshii, and Ms Rie Oumi for their excellent technical assistance. We also thank Dr Charles Parker for his critical reading of this manuscript.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No.15390298), a grant from the Ministry of Health, Labour and Welfare of Japan, and a grant from the Japan Intractable Diseases Research Foundation.
Authorship
Contribution: K.M. and C.S. participated in designing and performing the research. Z.Q. and X.L. performed experiments. K.M., C.S., and S.N. wrote the paper. C.S., A.T., K.I., Y.K., H.Y., and H.O. provided patient care. All authors have approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shinji Nakao, Cellular Transplantation Biology, Division of Cancer Medicine, Kanazawa University Graduate School of Medical Science, 13-1 Takaramachi, Kanazawa, Ishikawa 920-8641, Japan; e-mail: snakao@med3.m.kanazawa-u.ac.jp.
References
Author notes
K.M. and C.S. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal