Abstract
Urokinase-type plasminogen activator (uPA) participates in diverse (patho)physiological processes through intracellular signaling events that affect cell adhesion, migration, and proliferation, although the mechanisms by which these occur are only partially understood. Here we report that upon cell binding and internalization, single-chain uPA (scuPA) translocates to the nucleus within minutes. Nuclear translocation does not involve proteolytic activation or degradation of scuPA. Neither the urokinase receptor (uPAR) nor the low-density lipoprotein-related receptor (LRP) is required for nuclear targeting. Rather, translocation involves the binding of scuPA to the nucleocytoplasmic shuttle protein nucleolin through a region containing the kringle domain. RNA interference and mutational analysis demonstrate that nucleolin is required for the nuclear transport of scuPA. Furthermore, nucleolin is required for the induction smooth muscle α-actin (α-SMA) by scuPA. These data reveal a novel pathway by which uPA is rapidly translocated to the nucleus where it might participate in regulating gene expression.
Introduction
Urokinase-type plasminogen activator (uPA) is a multifunctional protein that has been implicated in several physiological and pathological processes, including cell proliferation and migration during angiogenesis, tissue regeneration, inflammatory responses, and tumor growth/metastases. These complex processes all involve intracellular signal transduction and regulation of gene transcription in addition to proteolysis (see Alfano et al1 for review). uPA is secreted as a single-chain protein (scuPA) that consists of an N-terminal EGF-like domain (GFD), a kringle domain (KD), and a serine protease domain. Binding of uPA to its high-affinity receptor CD87 (uPAR) is mediated by the GFD.2 Plasmin converts scuPA into a proteolytically active 2-chain enzyme (tcuPA)3 that is rapidly inhibited primarily by plasminogen activator inhibitor-1 (PAI-1). tcuPA-PAI-1 complexes are internalized with the aid of lipoprotein receptor–related protein (LRP)4 by clathrin-mediated endocytosis. The tcuPA-PAl-1 complexes traffic to lysosomes and are degraded, while unoccupied uPAR and LRP recycle back to the cell surface.5
uPA-induced signal transduction occurs via uPAR-dependent and uPAR-independent pathways (reviewed in Alfano et al1 ; Kjoller6 ; Blasi and Carmeliet7 ). Among the latter, we have shown that cleavage of scuPA by plasmin releases the GFD fragment, generating a form of uPA unable to bind to uPAR,8 but that stimulates migration of smooth muscle cells (SMCs).9 Signal transduction by this scuPA fragment may be mediated in part by LRP10 and certain integrins.11 However, there is limited information as to the mechanism by which uPA modifies gene transcription,12-15 and our previous studies have provided reason to hypothesize that cells express additional uPA-binding proteins that possess distinct signal-transducing activities involved in cell contractility, migration, and differentiation.9,10,16
We previously reported that uPAR forms a complex on the surface of human SMCs with nucleolin,17 a protein that regulates the organization of nucleolar chromatin, packaging of pre-RNA, rDNA transcription, and ribosome assembly (reviewed in Tuteja and Tuteja18 ). Nucleolin also regulates cell growth (reviewed in Srivastava and Pollard19 ), angiogenesis,20-22 and DNA replication,23,24 functions that overlap in part with those of uPA. Nucleolin is highly expressed in exponentially growing eukaryotic cells where it serves as a shuttle to import ribosomal proteins to the nucleus and to export ribosomal subunits to the cytoplasm.25-27 Although nucleolin is localized predominantly in the nucleolus, it has been identified in nucleoplasm and cytoplasm, as well as on the cell surface, where it is positioned to bind growth factors,28,29 lipoproteins,30 laminin-1,31 L-selectin,32 lactoferrin,33 endostatin,22 and certain viruses.34,35
Our finding that uPA/uPAR forms complexes with the nucleocytoplasmic shuttle protein nucleolin17 prompted us to follow the functional consequences of this interaction and to ask if uPA undergoes nuclear translocation. In this paper, we show that scuPA is rapidly translocated to the nucleus of cultured cells in a nucleolin-dependent manner. The interaction is mediated by the KD-containing region in scuPA and the C-terminal glycine-arginine–rich (GAR) domain in nucleolin. Nucleolin-mediated translocation of scuPA promotes up-regulation of smooth muscle α-actin (α-SMA) expression in human fibroblasts, a finding that might provide insight into tissue remodeling and vascular repair.
Methods
Plasmids and construction
Mouse (m) nucleolin cDNA (accession no. P09405; ATCC, Manassas, VA) was subcloned into PMT/BiP/V5 vector (Invitrogen, Frederick, MD) by introducing a FLAG epitope at the N-terminus. pcDNA3.1(+)-based constructs encoding C-terminus FLAG-tagged m-nucleolin mutants, in which the nuclear localization signal (NLS), N-terminus, or C-terminus was deleted (ΔNLS, ΔN, and ΔC, respectively), were kindly provided by Dr K. Kadomatsu (Nagoya University, Nagoya, Japan).28 N-terminus-FLAG-tagged m-nucleolin, ΔNLS-N-FLAG-m-nucleolin, and GAR-deficient N-FLAG-m-nucleolin (ΔGAR, nt 1-1938) were constructed and expressed in pcDNA3.1(+) (Invitrogen).
Proteins and purification
Recombinant human wild-type scuPA (WT-scuPA), scuPA lacking the GFD (ΔGFD-scuPA, amino acids 47-411), scuPA lacking the KD (lacking amino acids 47-135; ΔK-scuPA), ATF (GFD + KD, amino acids 1-143), low-molecular weight scuPA (LMW-scuPA, amino acids 144-411), and catalytically inactive scuPA (S356A-scuPA) were expressed using the Drosophila Expression System (Invitrogen) in Schneider S2 cells and purified as described.10,36 Recombinant N-FLAG-tagged m-nucleolin was expressed using the Drosophila system and purified from S2 cells using anti-FLAG agarose (Sigma-Aldrich, St Louis, MO) as per the manufacturer.
Cell culture and transfection
HeLa cells (ATCC) were cultured in DMEM (Invitrogen) containing 10% FBS (Hyclone, Logan, UT). Human coronary artery smooth muscle cells (CASMCs; Cascade Biologics, Portland, OR) were maintained in Medium 231 supplemented with SMGS supplement (Cascade Biologics) and used at passages 6 to 7. Human foreskin fibroblasts (BJ line), 293T-HEK, mouse embryonic fibroblasts MEF-2 (PEA13) (LRP−/−), and MEF-1 (LRP+/+)37 were maintained in DMEM/10% FBS. 293T cells were transfected using Fugene 6 (Roche, Indianapolis, IN); HeLa cells, using Lipofectamine 2000 (Invitrogen); and MEF-1 cells, using AMAXA nucleofector with the MEF-1 kit (Amaxa Biosystems, Gaithersburg, MD).
Subcellular fractionation
Iodination of scuPA variants was performed as described.9 HeLa, CASMC, BJ, MEF-1, and MEF-2 cells were incubated with 125I-WT-scuPA, 125I-ΔGFD-scuPA, or 125I-ΔK-scuPA (1–20 nM) for 5 to 60 minutes at 37°C in triplicate. The cells were washed with PBS; 100 mM glycine-HCl buffer (pH = 3.0) supplemented with 100 mM NaCl was added for 5 minutes to remove surface-associated ligand, and the cells were trypsinized and pelleted at 500g for 5 minutes at 4°C. To isolate nuclei, the cells were fractionated as described in Lin et al.38 All manipulations were performed on ice and centrifugations at 4°C. Briefly, the cells were homogenized in lysis buffer A (20 mM HEPES, pH 7.0, 10 mM KCl, 2 mM MgCl2, 0.5% NP40, and protease inhibitor cocktail [for use with mammalian cell and tissue extracts; Sigma-Aldrich]) by 30 strokes in a Dounce homogenizer. The homogenates were centrifuged at 1500g for 5 minutes. The nuclear pellets were washed twice and resuspended in buffer A containing 0.5 M NaCl to extract nuclear proteins. Nuclear extracts were separated from the nuclear pellet by centrifugation at 15 000g for 10 minutes. The nuclear pellets were dissolved in 1 × sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (Invitrogen). Separated subcellular fractions were counted in a γ-counter to quantify incorporated radioactive proteins or analyzed by SDS-PAGE and Western blotting (WB). Absolute amounts of cell-associated proteins in each fraction were normalized per 106 cells. In some experiments, cell lysis and homogenization procedures were followed by ultracentrifugation to separate the nuclei, as described in Blobel and Potter.39 Distribution of radioactive proteins in subcellular fractions, obtained with or without ultracentrifugation, was similar.
Cytoplasmic, wash, and nuclear fractions were analyzed by SDS-PAGE and WB using mouse α-GAPDH MAb and mouse α-histone H1 MAb (Chemicon, Temecula, CA), mouse α-transferrin receptor MAb (Zymed Labs, South San Francisco, CA), rat α-LAMP MAb (BD Biosciences, San Jose, CA), mouse α-calnexin MAb (BD Biosciences), α-vimentin MAb (Chemicon), and rabbit α-lamin A pAb (Santa Cruz Biotechnology, Santa Cruz, CA). Migration of radiolabeled proteins was assessed by SDS-PAGE and autoradiography under nonreducing and reducing conditions.
To analyze α-SMA content, BJ cells were starved for 24 hours, stimulated with scuPA variants for 24 hours, washed, and lysed in buffer containing 100 mM Tris-HCl, pH = 8.0, 150 mM NaCl, 1% Triton X-100, and the protease inhibitor cocktail (Sigma-Aldrich) described above. Lysates were analyzed by SDS-PAGE and WB using mouse anti–α-SMA MAb (Sigma-Aldrich) and mouse α-GAPDH MAb (Chemicon).
Lentivirus-based nucleolin RNA interference
The lentivirus packaging genome pCMV-dR8.74,40 the envelope pMD2G, and the transfer pLVTHM vectors were kindly provided by Dr D. Trono (University of Geneva, Geneva, Switzerland). The target sites in human nucleolin mRNA (accession no. NM005381) for RNAi were determined using the siRNA Selection Server (http://jura.wi.mit.edu/bioc/siRNAext/home.php)41 and designed as oligonucleotides encoding short hairpin RNAs (shRNAs): Nuc si64 forward: 5′-GATCCCCGGAGGTAGAAGAAGATAGTTTCAAGAGAACTATCTTCTTCTACCTCCTTTTTA-3′; Nuc si64 reverse: 5′-AGCTTAAAAAGGAGGTAGAAGAAGATAGTTCTCTTGAAACTATCTTCTTCTACCTCCGGG-3′; Nuc si719 forward: 5′-GATCCCCGACGCTAAAGAAGCTTTAATTCAAGAGATT-AAAGCTTCTTTAGCGTCTTTTTA-3′; Nuc si719 reverse: 5′-AGCTTAAAAAGACGCTAAAGAAGCTTTAATCTCTTGAATTAAAGCTTCT-TTAGCGTCGGG-3′.
Lentivirus particles were obtained as described in Frederick et al.42 HeLa or BJ cells were transduced with either control lentivirus (“empty” pLVTHM) or nucleolin shRNA-delivering lentivirus in the presence of 8 μg/mL polybrene (Sigma-Aldrich) for 24 hours. Fresh medium was added for an additional 48 hours. Nucleolin down-regulation in total cell extracts was monitored by WB and immunofluorescence using α-nucleolin MAb. Lentivirus-infected cells were incubated with either 125I-WT-scuPA or 125I-ΔGFD-scuPA (10 nM) for 1 hour and fractionated as described above to quantify intracellular protein trafficking. For immunofluorescence microscopy, lentivirus-infected cells were incubated with unlabeled ΔGFD-scuPA (10 nM) for 30 minutes, washed, fixed, and stained as described below.
Indirect immunofluorescence staining
To analyze the intracellular distribution of exogenously added uPAs in various cell lines, HeLa, BJ, or MEF-1 cells grown in 8-well chamber slides (LabTek, Campbell, CA) were incubated with 20 nM WT-scuPA, ΔK-scuPA, or ΔGFD-scuPA for the indicated times. The cells were fixed in ice-cold MeOH for 15 minutes at room temperature (RT) and blocked with 1% BSA in PBS. Rabbit α-uPA pAb (American Diagnostica, Greenwich, CT), mouse α-nucleolin MAb (Stressgen [Victoria, BC] and Santa Cruz Biotechnology), rabbit α-nucleolin pAb (Santa Cruz Biotechnology), mouse Cy3-α-FLAG MAb (M2; Sigma-Aldrich), and Alexa488- or Alexa555-conjugated preabsorbed goat α-rabbit and goat α-mouse pAbs (Molecular Probes, Eugene, OR) were used for immunostaining. Nuclei were counterstained with DAPI (0.5 μg/mL). Stained cells were mounted in ProLong Gold (Molecular Probes) and examined with a confocal laser-scanning microscope (Zeiss LSM 510; Carl Zeiss, Heidelberg, Germany). To detect α-SMA, BJ cells were fixed with 4% PFA in PBS and permeabilized with 0.5% Triton X-100. α-SMA was detected using mouse anti–α-SMA MAb (Sigma-Aldrich) and total fibrillar actin was detected using Alexa647-Phalloidin (Molecular Probes). Cells were analyzed using a Leica DMI 4000B fluorescence microscope equipped with a Leica DFC350FX camera (Leica Microsystems, Heidelberg, Germany).
Coimmunoprecipitation and immunoblotting
293HEK cells were transfected with pcDNA3.1(+)-based plasmids encoding FLAG-m-nucleolin mutants and lysed 48 hours later in 100 mM Tris-HCl, 1% Triton X-100, 150 mM NaCl (pH = 7.5), and a protease inhibitor cocktail (Sigma-Aldrich). 125I-WT-scuPA (5 nM) was added to the lysates, and the FLAG-tagged m-nucleolin-associated proteins were immunoprecipitated using α-FLAG agarose (Sigma-Aldrich). In a separate set of experiments, 125I-scuPA, 125I-ΔGFD-scuPA, or 125I-ΔK-scuPA (5 nM) were added to lysates of untransfected 293HEK cells. Endogenous nucleolin was immunoprecipitated using rabbit α-nucleolin pAb (Santa Cruz Biotechnology) bound to protein G-agarose (Invitrogen). HeLa cells were preincubated with 125I-WT-scuPA (20 nM) for 1 hour, washed, and lysed as above. Endogenous nucleolin was immunoprecipitated as above, and uPA was immunoprecipitated with rabbit α-uPA Ab. Immunoprecipitated proteins were analyzed by SDS-PAGE and WB. Mouse α-nucleolin MAb, rabbit α-nucleolin pAb, rabbit α-uPAR pAb, and mouse anti-FLAG HRP-conjugated MAb (Sigma-Aldrich) were used for WB.
Surface plasmon resonance analysis
The interaction between WT-scuPA and soluble uPAR or recombinant nucleolin was analyzed on a model 3000 Biosensor (Biacore, Uppsala, Sweden) as described.36 A detailed description of the methodology is found in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Real-time PCR
BJ cells were grown to 50% confluence, starved for 24 hours, and stimulated with 10 nM scuPA or mock-treated for an additional 24 hours. Experiments were performed in triplicates. Total RNA was isolated using RNAeasy Mini Kit and QIAshredder kit (Qiagen, Valencia, CA). Total RNA (5 μg) from each sample was reverse transcribed using the Superarray C-01 Reaction ready cDNA synthesis kit (SuperArray Bioscience Corp., Frederick, MD). Reverse-transcription–polymerase chain reaction (RT-PCR) with α-SMA and GAPDH primer pairs was performed in quadruplicate for each sample. α-SMA (ACTA2) and GAPDH mRNA levels were calculated from a standard curve derived from the same plate. RT-PCR was performed on the Icycler (BioRad, San Jose, CA) using the PA-011 SYBR green master mix (SuperArray). Differences in α-SMA mRNA expression between untreated and treated samples were calculated using the 2−Δ ΔCT method.43
Results
Nuclear translocation of uPA
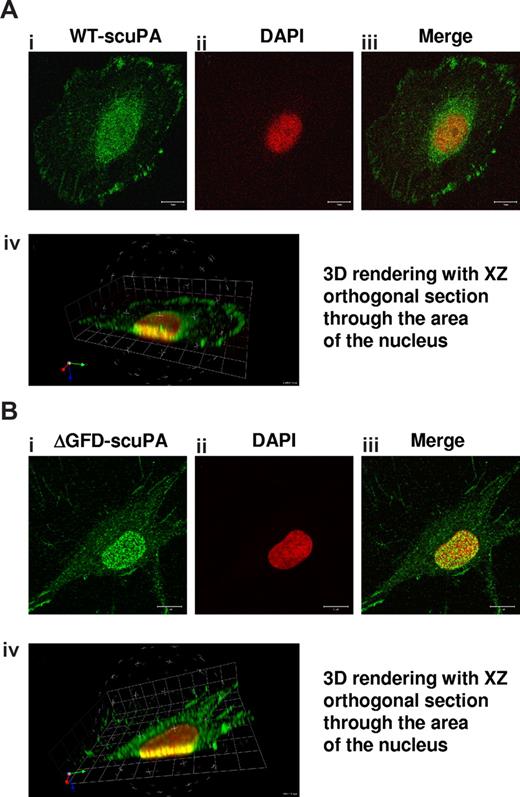
Our finding that uPA/uPAR forms complexes with the nucleocytoplasmic shuttle protein nucleolin17 prompted us to ask if uPA undergoes nuclear translocation. To examine this question, we first incubated HeLa cells and fibroblasts (BJ line) with 10 nM WT-scuPA for 60 minutes at 37°C. HeLa cells were chosen because of their low level of endogenous uPA. The cells were washed extensively and fixed in methanol to help unmask nuclear antigens, and uPA was localized using immunofluorescence and confocal microscopy. scuPA was found both within the nucleus and in the perinuclear region (Figure 1Ai-iii). 3D reconstruction of sequential images acquired at multiple focal planes (Z-stacks) affirmed the presence of scuPA throughout the nucleus (Figure 1Aiv; Video S1), where it was distributed in a punctate manner. The same results were seen in BJ cells (not shown).
uPA variants translocate to the nucleus. HeLa cells (A) or BJ cells (B) were incubated for 30 minutes with 10 nM WT-scuPA or ΔGFD-scuPA, respectively, in complete medium at 37°C, washed, and fixed in cold MeOH for 15 minutes. To visualize uPA, the fixed cells were incubated with α-uPA pAb and Alexa488-conjugated α-rabbit pAb. Nuclei were stained with DAPI. Images were taken using a Zeiss LSM 510 laser scanning confocal microscope with the z interval set at 0.3 μm. (i-iii) Confocal images taken through a single 0.3-μm optical slice. (i) Staining with α-uPA Ab is shown in green. (ii) Counterstain of the nucleus with DAPI is shown as pseudocolor in red. (iii) Merged images within one focal plane. Scale bar represents 10 μm. (iv) Sequences of merged images taken in various z sections at 0.3-μm intervals were subjected to 3-dimensional (3D) reconstitution using Volocity 4.1.0 software (Improvision, Lexington, MA). Virtual 3D image of the cell was sectioned along the XZ plane through the nucleus. The results reveal that a substantial fraction of scuPA and/or ΔGFD-scuPA is found within the nucleus.
uPA variants translocate to the nucleus. HeLa cells (A) or BJ cells (B) were incubated for 30 minutes with 10 nM WT-scuPA or ΔGFD-scuPA, respectively, in complete medium at 37°C, washed, and fixed in cold MeOH for 15 minutes. To visualize uPA, the fixed cells were incubated with α-uPA pAb and Alexa488-conjugated α-rabbit pAb. Nuclei were stained with DAPI. Images were taken using a Zeiss LSM 510 laser scanning confocal microscope with the z interval set at 0.3 μm. (i-iii) Confocal images taken through a single 0.3-μm optical slice. (i) Staining with α-uPA Ab is shown in green. (ii) Counterstain of the nucleus with DAPI is shown as pseudocolor in red. (iii) Merged images within one focal plane. Scale bar represents 10 μm. (iv) Sequences of merged images taken in various z sections at 0.3-μm intervals were subjected to 3-dimensional (3D) reconstitution using Volocity 4.1.0 software (Improvision, Lexington, MA). Virtual 3D image of the cell was sectioned along the XZ plane through the nucleus. The results reveal that a substantial fraction of scuPA and/or ΔGFD-scuPA is found within the nucleus.
Requirement for uPAR
scuPA binds with high affinity to cell surface uPAR/CD8744,45 through its GFD.2 To determine whether nuclear translocation of scuPA requires binding to uPAR, we used a GFD-deficient scuPA mutant (ΔGFD-scuPA) incapable of binding to uPAR.9,36 ΔGFD-scuPA translocated to the nucleus in BJ cells (Figure 1B) and HeLa cells (Figure S1). Although 125I-ΔGFD-scuPA binds with lower affinity to somewhat fewer total cell surface sites at 4°C (Kd: ∼22 nM; Bmax: 1.3 × 105 sites per cell) than 125I-WT-scuPA (Kd: ∼3.4 nM; Bmax: 1.7 × 105 sites per cell), nuclear accumulation was comparable (Figure 1Bi-iii) in all cell types studied. 3D reconstitution of sequential Z-stacks affirms that ΔGFD-scuPA is located in multiple planes through the nucleus (Figure 1Biv; Figure S1; Video S2). Neither untreated BJ nor HeLa cells showed intense nuclear staining for uPA under the same experimental conditions (Figure S2). Nuclear staining was not seen with control rabbit Ig (not shown). These results indicate that binding of scuPA to uPAR is not essential for nuclear translocation.
Quantification and kinetics of translocation
To assess translocation using an entirely independent approach and to determine the proportion of scuPA that translocates to the nucleus, HeLa and BJ cells were incubated with 125I-labeled WT-scuPA and scuPA variants lacking the GFD or the kringle over a range of concentrations. The cells were washed and the nuclei were isolated using cell homogenization combined with limited lysis in a low-salt buffer containing 0.5% NP40 detergent.38,46 To assess the “purity” of the nuclear fraction, we analyzed the cytoplasmic, wash, and nuclear fractions for specific compartmental markers (Figure 2A) using SDS-PAGE and WB. The cytoplasmic and initial wash fractions, but not the nuclear fraction, stained positively for transferrin receptor (plasma membrane and endosomal marker), LAMP-1 (lysosomal marker), calnexin (endoplasmic reticulum marker), vimentin (cytoskeleton), and GAPDH (cytosolic marker), whereas the nuclear fraction exclusively contained histone H1 and lamin A, as expected.
Subcellular distribution of cell-associated 125I-WT-scuPA and 125I-ΔGFD-scuPA. BJ cells were in-cubated with 125I-WT-scuPA (A-D) or 125I-ΔGFD-scuPA (E) in complete medium at 37°C at the indicated concentrations for 1 hour and then washed to remove unbound radioligand. Surface-bound uPA was eluted with glycine buffer, pH = 3.0 (surface). Subcellular fractions were then prepared as described in “Methods,” and the radioactivity in each fraction was counted. Radioactivity in the cytoplasmic extract and wash fractions were combined as a measure of total cytoplasmic content. Radioactivity in the nuclear extract and nuclear pellet were counted separately, but the values were combined as a measure of total nuclear content. (A) Distribution of nuclear and cytoplasmic cell markers. The cytoplasmic extract, the 2 wash fractions, and the nuclear extract were analyzed by SDS-PAGE under reducing conditions, electrotransferred to nitrocellulose membranes, and probed with antibodies to the indicated proteins. This figure indicates that the nuclear fraction does not contain detectable amounts of plasma membrane and cytoplasmic proteins or markers of organelles. (B) Subcellular distribution of 125I-WT-scuPA. BJ and HeLa cells were incubated with 125I-WT-scuPA, cell fractionation was performed as described in “Methods,” and the radioactivity in each fraction was measured. Total cell-associated radioactivity was designated as 100%. (C) 125I-WT-scuPA does not degrade/becomes cleaved upon translocation to the nucleus. BJ cells were incubated with 20 nM 125I-WT-scuPA for 1 hour. The subcellular fractions were isolated as above. The proteins were separated by SDS-PAGE and analyzed using autoradiography under nonreducing (NR) and reducing (1 mM DTT) (R) conditions. Input lane: 125I-WT-scuPA. This figure shows that cytoplasmic and nuclear scuPA migrates as a single band that corresponds to intact protein, and no additional low-molecular-weight bands were evident. (D,E) Dose-dependent accumulation and subcellular distribution of125I-WT-scuPA (D) and 125I-ΔGFD-scuPA (E). The experiments were performed as in panel B. All experiments were performed in triplicate and repeated 2 to 3 times. Data from a representative experiment are presented as mean plus or minus SE.
Subcellular distribution of cell-associated 125I-WT-scuPA and 125I-ΔGFD-scuPA. BJ cells were in-cubated with 125I-WT-scuPA (A-D) or 125I-ΔGFD-scuPA (E) in complete medium at 37°C at the indicated concentrations for 1 hour and then washed to remove unbound radioligand. Surface-bound uPA was eluted with glycine buffer, pH = 3.0 (surface). Subcellular fractions were then prepared as described in “Methods,” and the radioactivity in each fraction was counted. Radioactivity in the cytoplasmic extract and wash fractions were combined as a measure of total cytoplasmic content. Radioactivity in the nuclear extract and nuclear pellet were counted separately, but the values were combined as a measure of total nuclear content. (A) Distribution of nuclear and cytoplasmic cell markers. The cytoplasmic extract, the 2 wash fractions, and the nuclear extract were analyzed by SDS-PAGE under reducing conditions, electrotransferred to nitrocellulose membranes, and probed with antibodies to the indicated proteins. This figure indicates that the nuclear fraction does not contain detectable amounts of plasma membrane and cytoplasmic proteins or markers of organelles. (B) Subcellular distribution of 125I-WT-scuPA. BJ and HeLa cells were incubated with 125I-WT-scuPA, cell fractionation was performed as described in “Methods,” and the radioactivity in each fraction was measured. Total cell-associated radioactivity was designated as 100%. (C) 125I-WT-scuPA does not degrade/becomes cleaved upon translocation to the nucleus. BJ cells were incubated with 20 nM 125I-WT-scuPA for 1 hour. The subcellular fractions were isolated as above. The proteins were separated by SDS-PAGE and analyzed using autoradiography under nonreducing (NR) and reducing (1 mM DTT) (R) conditions. Input lane: 125I-WT-scuPA. This figure shows that cytoplasmic and nuclear scuPA migrates as a single band that corresponds to intact protein, and no additional low-molecular-weight bands were evident. (D,E) Dose-dependent accumulation and subcellular distribution of125I-WT-scuPA (D) and 125I-ΔGFD-scuPA (E). The experiments were performed as in panel B. All experiments were performed in triplicate and repeated 2 to 3 times. Data from a representative experiment are presented as mean plus or minus SE.
When BJ (and HeLa) cells were incubated with 10 nM 125I-WT-scuPA for 1 hour at 37°C, 27% (± 6%) of the radiolabeled protein remained associated with the cell surface as assessed by acid elution, 13% (± 1.5%) was found in the cytoplasmic fraction, and 60% (± 7%) was located within the nuclear fraction (Figure 2B). Nuclear and cytoplasmic 125I-WT-scuPA migrated as a full-length single-chain molecule as assessed by SDS-PAGE and autoradiography under nonreducing and reducing conditions (Figure 2C). Less than 0.1% of the radioactivity recovered from the nuclear extracts was TCA soluble, indicating that the internalized ligand had not undergone substantial proteolysis or degradation. Moreover, active 2-chain uPA (125I-tcuPA) was not incorporated into the nuclei in either BJ or HeLa cells under the same experimental conditions (< 0.5% ± 0.05% of the total cell-associated protein), indicating that the cells use a different intracellular trafficking/degradation pathway to handle active enzyme. Nuclear translocation of 125I-WT-scuPA and 125I-ΔGFD-scuPA at 37°C was evident within 5 minutes, reached a maximum by 30 minutes (not shown), and increased thereafter in a dose-dependent manner (Figure 2D,E). Comparable translocation of 125I-WT-scuPA and 125I-ΔGFD-scuPA into the nuclei was seen in human coronary artery SMCs (CASMCs) and human umbilical vein endothelial cells (HUVECs; not shown), as well as in mouse embryonic fibroblasts (below).
Mechanism of translocation: binding of uPA to nucleolin
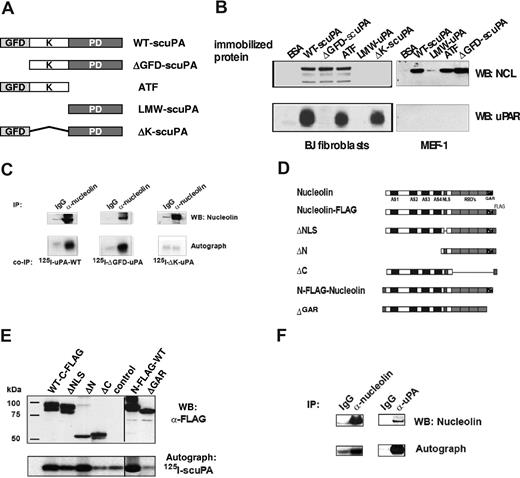
Guided by our observation that uPA/uPAR is found in complexes with nucleolin on the surface of CASMCs17 and with the knowledge that nucleolin mediates nucleocytoplasmic shuttling of several proteins,18,19,47 we asked whether scuPA is transported to the nucleus through a similar mechanism by binding to nucleolin. To begin to address this question, affinity matrices prepared with equimolar amounts of immobilized WT-scuPA or several scuPA deletion variants (Figure 3A) were added to lysates prepared from human CASMCs and the bound proteins were analyzed by WB. Affinity matrices containing immobilized WT-scuPA, ΔK-scuPA, and ATF bound suPAR (Figure 3B lower left panel). uPA variants containing the KD (including WT-scuPA, ATF, and ΔGFD-scuPA) bound nucleolin, whereas LMW-scuPA (proteolytic domain) and ΔK-scuPA did not (Figure 3B top left panel). These results show that amino acids 47 to 143, which include the KD, are required for binding to nucleolin and support our conclusion that uPAR is not essential for binding of scuPA to nucleolin. To further test this conclusion, lysates from MEF-1 cells that express mouse uPAR, which does not bind human uPA, were studied in the same way. Mouse nucleolin bound to human WT-scuPA and KD-containing scuPA mutants (Figure 3B top panels), whereas mouse uPAR did not bind to immobilized WT-scuPA or ATF, as expected (Figure 3B right bottom panel). Moreover, 125I-WT-scuPA and 125I-ΔGFD-scuPA added to lysates from uPAR-deficient 293 HEK cells coimmunoprecipitated with nucleolin (Figure 3C), whereas 125I-ΔK-scuPA did not, again indicating that the KD is necessary, but uPAR is not required, for scuPA to bind to nucleolin.
Binding of uPA to nucleolin. (A) Domain organization of uPA and its derivatives. GFD indicates growth factor-like domain; K, kringle-domain; and PD, protease domain. (B) Mapping of the nucleolin-binding domain in scuPA. Lysates from human CASMCs (designated as h-SMC on the figure) and mouse embryonic fibroblasts (MEF-1) were incubated with affinity matrices containing immobilized WT-scuPA, LMW-uPA, ATF, ΔGFD-scuPA, ΔK-scuPA, or BSA as a negative control. Bound proteins were detected by WB using α-nucleolin pAb (top panels), α-human uPAR pAb (lower left panel), and α-mouse uPAR pAb (bottom right panel). GFD-containing uPA mutants bound uPAR, whereas those containing the KD but not GFD bound nucleolin in uPAR-independent manner. (C) Pull-down of nucleolin with uPA deletion variants. 125I-WT-scuPA, 125I-ΔGFD-scuPA, or 125I-ΔK-scuPA (5 nM) was added to 293HEK lysates, and nucleolin was immunoprecipitated with an α-nucleolin pAb (lanes “α-nucleolin”). Rabbit Ig was used as a negative control (lanes “IgG”). The immune complexes were analyzed by WB. Nucleolin was detected using an α-nucleolin MAb (top panel). Co-IP: Bound scuPA deletion variants were detected by autoradiography (bottom panel). (D) Domain organization of nucleolin. AS indicates acidic stretch; GAR, glycine- and arginine-rich domain; RBD, RNA-binding domain; and NLS, nuclear localization signal. (E) Mapping the uPA-binding region in nucleolin. 293 HEK cells were transfected with pcDNA3.1 vectors encoding FLAG-tagged nucleolin mutants. Mutants are as described in panel D. The corresponding lysates were incubated with 125I-WT-scuPA for 1 hour at 20°C, and nucleolin mutants were immunoprecipitated with agarose-conjugated α-FLAG MAb. The immunoprecipitates were analyzed by WB, FLAG-nucleolin variants were visualized with an HRP-α-FLAG MAb (α-FLAG, top panel), and 125I-WT-scuPA was imaged by autoradiography (125I-scuPA, bottom panel). Vertical lines have been inserted to indicate repositioned gel lanes (“N-FLAG-WT” and “ΔGAR”). (F) Coimmunoprecipitation of nucleolin with 125I-WT-scuPA. HeLa cells were preincubated with 50 nM 125I-WT-scuPA for 1 hour at 37°C and then lysed. Nucleolin or scuPA was immunoprecipitated with rabbit polyclonal α-nucleolin Ab (lane “α-nucleolin”) or with rabbit polyclonal α-uPA Ab (lane “α-uPA”), respectively. Rabbit Ig was used as a negative control (lanes “IgG”). The immunoprecipitates were analyzed by WB using mouse α-nucleolin MAb (panel “WB: α-nucleolin”), and autoradiography to detect 125I-WT-scuPA (panel “Autograph” 125I-scuPA). An experiment representative of 3 so performed is shown.
Binding of uPA to nucleolin. (A) Domain organization of uPA and its derivatives. GFD indicates growth factor-like domain; K, kringle-domain; and PD, protease domain. (B) Mapping of the nucleolin-binding domain in scuPA. Lysates from human CASMCs (designated as h-SMC on the figure) and mouse embryonic fibroblasts (MEF-1) were incubated with affinity matrices containing immobilized WT-scuPA, LMW-uPA, ATF, ΔGFD-scuPA, ΔK-scuPA, or BSA as a negative control. Bound proteins were detected by WB using α-nucleolin pAb (top panels), α-human uPAR pAb (lower left panel), and α-mouse uPAR pAb (bottom right panel). GFD-containing uPA mutants bound uPAR, whereas those containing the KD but not GFD bound nucleolin in uPAR-independent manner. (C) Pull-down of nucleolin with uPA deletion variants. 125I-WT-scuPA, 125I-ΔGFD-scuPA, or 125I-ΔK-scuPA (5 nM) was added to 293HEK lysates, and nucleolin was immunoprecipitated with an α-nucleolin pAb (lanes “α-nucleolin”). Rabbit Ig was used as a negative control (lanes “IgG”). The immune complexes were analyzed by WB. Nucleolin was detected using an α-nucleolin MAb (top panel). Co-IP: Bound scuPA deletion variants were detected by autoradiography (bottom panel). (D) Domain organization of nucleolin. AS indicates acidic stretch; GAR, glycine- and arginine-rich domain; RBD, RNA-binding domain; and NLS, nuclear localization signal. (E) Mapping the uPA-binding region in nucleolin. 293 HEK cells were transfected with pcDNA3.1 vectors encoding FLAG-tagged nucleolin mutants. Mutants are as described in panel D. The corresponding lysates were incubated with 125I-WT-scuPA for 1 hour at 20°C, and nucleolin mutants were immunoprecipitated with agarose-conjugated α-FLAG MAb. The immunoprecipitates were analyzed by WB, FLAG-nucleolin variants were visualized with an HRP-α-FLAG MAb (α-FLAG, top panel), and 125I-WT-scuPA was imaged by autoradiography (125I-scuPA, bottom panel). Vertical lines have been inserted to indicate repositioned gel lanes (“N-FLAG-WT” and “ΔGAR”). (F) Coimmunoprecipitation of nucleolin with 125I-WT-scuPA. HeLa cells were preincubated with 50 nM 125I-WT-scuPA for 1 hour at 37°C and then lysed. Nucleolin or scuPA was immunoprecipitated with rabbit polyclonal α-nucleolin Ab (lane “α-nucleolin”) or with rabbit polyclonal α-uPA Ab (lane “α-uPA”), respectively. Rabbit Ig was used as a negative control (lanes “IgG”). The immunoprecipitates were analyzed by WB using mouse α-nucleolin MAb (panel “WB: α-nucleolin”), and autoradiography to detect 125I-WT-scuPA (panel “Autograph” 125I-scuPA). An experiment representative of 3 so performed is shown.
To determine the domain specificity within nucleolin for scuPA binding, cDNAs encoding C- and N-terminus FLAG-tagged mouse nucleolin deletion mutants (Figure 3D) were expressed in 293 HEK cells. Transfected cells were lysed and nucleolin mutants were immunoprecipitated in the presence of exogenous 125I-WT-scuPA. All nucleolin mutants except ΔC and ΔGAR bound 125I-WT-scuPA above background. The results show that the C-terminal glycine-arginine–rich domain (GAR) in nucleolin is required for scuPA binding (Figure 3E).
We next asked whether uPA and nucleolin associate within the cells and can be coimmunoprecipitated. HeLa cells were preincubated with 50 nM 125I-WT-scuPA for 1 hour at 37°C to permit internalization/nuclear translocation, washed, and lysed as described in “Methods.” Immunoprecipitates obtained with α-uPA or α-nucleolin Ab contained both proteins, whereas control rabbit IgG did not precipitate significant amounts of either protein (Figure 3F).
Requirement for LRP in nuclear translocation of uPA
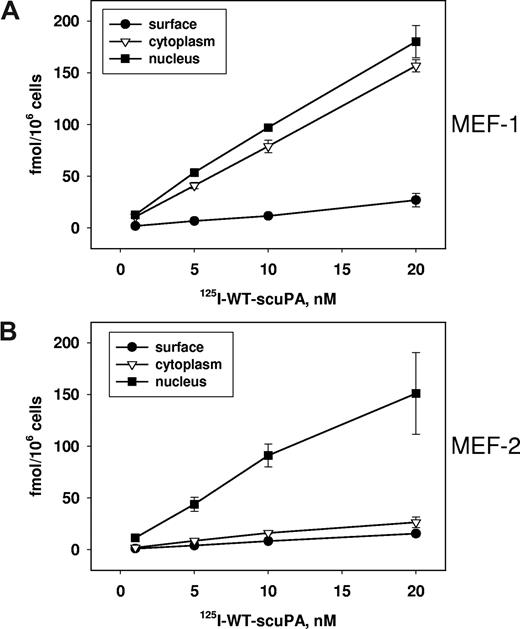
LRP is considered to be the predominant receptor through which uPA is endocytosed.48 Binding of uPA to uPAR promotes LRP-mediated lysosomal degradation of uPA.49 Therefore, we asked whether LRP is required for nuclear translocation of scuPA, a process in which ligand degradation is minimal (Figure 2B). LRP-expressing MEF-1 cells and their LRP−/− counterparts (MEF-2 cells) were incubated with 125I-WT-scuPA for 1 hour at the indicated concentrations, and the subcellular fractions were isolated as above. Binding of 125I-WT-scuPA to the surface of MEF-1 cells was significantly (P < .01) less than to BJ cells (Figure 2D), most likely due to the inability of human scuPA to bind mouse uPAR (Figure 4A). Bound 125I-WT-scuPA was distributed nearly equally between the cytoplasmic and nuclear fractions of MEF-1 cells. The cytoplasmic fraction of LRP−/− MEF-2 cells contained significantly less radiolabeled protein than MEF-1 cells at all scuPA concentrations (P < .01; Figure 4B). However, MEF-2 and MEF-1 cells transported 125I-scuPA to the nucleus with similar efficiency, indicating that nuclear translocation does not require LRP.
Requirement for LRP in the nuclear translocation of urokinase. (A,B) Subcellular distribution of 125I-WT-scuPA in MEF-1 (LRP+/+) and MEF-2 (LRP−/−) cells. (A) MEF-1 and (B) MEF-2 cells were incubated with 125I-WT-scuPA at the indicated concentrations for 1 hour. Subcellular fractions were prepared and analyzed as described in the legend to Figure 2.
Requirement for LRP in the nuclear translocation of urokinase. (A,B) Subcellular distribution of 125I-WT-scuPA in MEF-1 (LRP+/+) and MEF-2 (LRP−/−) cells. (A) MEF-1 and (B) MEF-2 cells were incubated with 125I-WT-scuPA at the indicated concentrations for 1 hour. Subcellular fractions were prepared and analyzed as described in the legend to Figure 2.
Transfer of uPA to nucleolin
Acidification reduces binding of uPA to uPAR. As a result, uPA dissociates from uPAR in endosomes leading to lysosomal degradation and return of unoccupied uPAR to the cell surface.5 uPAR is likely more abundant on the plasma membrane than nucleolin and uPA binds to uPAR with high affinity at neutral pH. Therefore, we asked whether the uPA-nucleolin interaction might be enhanced after internalization as a result of acidification that begins in early endosomes rather than occurring exclusively on the cell surface. To investigate this hypothesis, we measured the specific binding of 125I-WT-scuPA to immobilized recombinant mouse FLAG-nucleolin using a solid-phase binding assay. Binding of scuPA to nucleolin increased as the pH was lowered (Figure S3). Measurement of binding constants by surface plasmon resonance over a pH range of 5.5 to 7.4 revealed that the affinity of scuPA for suPAR fell from 2 nM at pH 7.436 to 17 nM at pH 6.5, whereas the affinity of scuPA for nucleolin increased from 150 nM to 28 nM over the same pH range (data not shown). The opposing effect of acidification on scuPA binding to suPAR and nucleolin supports a model in which the transfer of scuPA from uPAR to nucleolin is enhanced within endosomal compartments.
Nucleolin targets uPA to the nucleus
To elucidate the role of nucleolin in the nuclear translocation of uPA, we first examined the internalization and nuclear translocation of WT-scuPA and scuPA deletion mutants. 125I-WT-scuPA and 125I-ΔGFD-scuPA, but not 125I-ΔK-scuPA, were transported to the nucleus of BJ cells (Figure 5A), showing a correspondence between binding to nucleolin (Figure 3B,C) and nuclear translocation. These data were confirmed by immunofluorescence microscopy in both BJ and HeLa cells (Figure 1A,B; Figures S1,S4).
Transport of scuPA variants to the nucleus depends on their binding to nucleolin. (A) Subcellular distribution of 125I-WT-scuPA, 125I-ΔGFD-scuPA, and 125I-ΔK-scuPA in BJ cells. BJ cells were incubated with 10 nM 125I-WT-scuPA, 125I-ΔGFD-scuPA, or 125I-ΔK-scuPA for 1 hour at 37°C and washed, and the subcellular fractions were isolated as in Figure 2. The radioactivity in each fraction was measured and normalized per 106 cells to compare the absolute amounts of proteins incorporated into each fraction. The results of 1 experiment, representative of 3 so performed, are shown. (B) Nucleolin mediates nuclear translocation of scuPA in the absence of LRP and uPAR. MEF-2 cells were preincubated for 1 hour in medium alone (control) or media supplemented with either 8 μM human lactoferrin, or 2.5 μM recombinant FLAG-tagged C-terminal fragment of m-nucleolin (GAR), or 5 μM recombinant full-length m-nucleolin. 125I-WT-scuPA (10 nM) was added in the continued presence of potential competitors. The incubation was continued for an additional hour, and the radioactivity in the subcellular fractions was determined as in Figure 2. Nuclear transport of 125I-WT-scuPA in control cells was taken as 100%. Nuclear contents of 125I-WT-scuPA in cells pretreated with these potential inhibitors were expressed as percentage of control. Experiments were performed in triplicates, and data are presented as mean plus or minus SE. *indicates a result that is statistically significantly different (P < .05) from control.
Transport of scuPA variants to the nucleus depends on their binding to nucleolin. (A) Subcellular distribution of 125I-WT-scuPA, 125I-ΔGFD-scuPA, and 125I-ΔK-scuPA in BJ cells. BJ cells were incubated with 10 nM 125I-WT-scuPA, 125I-ΔGFD-scuPA, or 125I-ΔK-scuPA for 1 hour at 37°C and washed, and the subcellular fractions were isolated as in Figure 2. The radioactivity in each fraction was measured and normalized per 106 cells to compare the absolute amounts of proteins incorporated into each fraction. The results of 1 experiment, representative of 3 so performed, are shown. (B) Nucleolin mediates nuclear translocation of scuPA in the absence of LRP and uPAR. MEF-2 cells were preincubated for 1 hour in medium alone (control) or media supplemented with either 8 μM human lactoferrin, or 2.5 μM recombinant FLAG-tagged C-terminal fragment of m-nucleolin (GAR), or 5 μM recombinant full-length m-nucleolin. 125I-WT-scuPA (10 nM) was added in the continued presence of potential competitors. The incubation was continued for an additional hour, and the radioactivity in the subcellular fractions was determined as in Figure 2. Nuclear transport of 125I-WT-scuPA in control cells was taken as 100%. Nuclear contents of 125I-WT-scuPA in cells pretreated with these potential inhibitors were expressed as percentage of control. Experiments were performed in triplicates, and data are presented as mean plus or minus SE. *indicates a result that is statistically significantly different (P < .05) from control.
Nucleolin shuttles specific proteins from the cell surface22,28,33 and from the cytoplasm26 to the nucleus. To further elucidate the role of nucleolin in nuclear translocation of scuPA, we studied MEF-2 cells and human WT-scuPA to obviate the potential contribution of uPAR and LRP. MEF-2 cells were preincubated with either recombinant mouse FLAG-nucleolin or with a recombinant FLAG-GAR fragment of nucleolin, which is required for uPA binding (Figure 3D,E). Lactoferrin, which translocates to the nucleus in a nucleolin-dependent manner,33 was used as an auxiliary nucleolin competitor. The cells were then incubated with 10 nM 125I-WT-scuPA for an additional hour at 37°C in the presence of potential competitors. Lactoferrin, nucleolin, and the GAR fragment of nucleolin reduced nuclear transport of 125I-WT-scuPA by 72%, 66%, and 28%, respectively (Figure 5B) compared with cells preincubated with serum-containing medium alone. These observations suggest that nucleolin might act as a carrier for nuclear translocation of WT-scuPA.
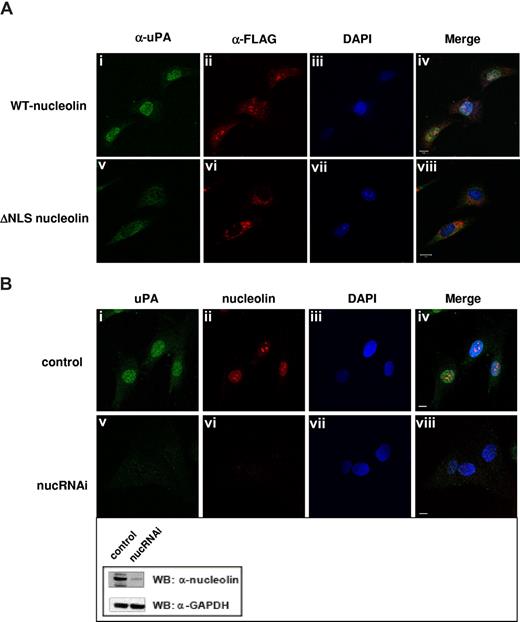
Translocation of karyophilic proteins to the nucleus requires a bipartite NLS, which lies between the N-terminal and central domains. NLS-containing proteins bind specific carriers belonging to the importin/karyopherin superfamily of import proteins that permeate the nuclear pore complex.50,51 To determine whether nucleolin uses a similar mechanism to import uPA into the nucleus, we transfected MEF-1 cells with vectors encoding either N-terminal FLAG-tagged full-length m-nucleolin or a NLS-deficient nucleolin mutant (ΔNLS, Figure 3D) that is unable to enter the nucleus.28 We then used immunofluorescence microscopy to follow the intracellular trafficking of ΔGFD-scuPA, which binds to nucleolin (Figure 3B) and translocates into the nucleus (Figures 1B,2D). Efficient nuclear translocation of ΔGFD-scuPA was observed in MEF-1 cells expressing full-length FLAG-m-nucleolin (Figure 6Ai-iv), but not in FLAG-ΔNLS-m-nucleolin–expressing cells where ΔGFD-scuPA and FLAG-ΔNLS-nucleolin were found predominantly in the cytoplasm (Figure 6Av-viii). The small amount of uPA found in the nucleus may reflect endogenous WT-nucleolin. We then used lentivirus-based RNA interference (shRNA) to suppress expression of nucleolin in BJ cells. Western blot analysis of cell lysates using α-nucleolin MAb showed reduction in the expression of nucleolin of approximately 80% (Figure 6B inset), which was consistent with the results of immunofluorescence microscopy (Figure 6Bvi). Nucleolin shRNA dramatically reduced transport of ΔGFD-scuPA into the nucleus in BJ cells as visualized by immunofluorescence microscopy (Figure 6Bv). These results were confirmed by cell fractionation studies, which demonstrated that nucleolin shRNA reduced the nuclear transport of 125I-WT-scuPA and 125I-ΔGFD-scuPA (10 nM) into nuclei isolated from BJ cells by 51% (± 2.8%) and 67% (± 1.7%), respectively. These results demonstrate a role for nucleolin in nuclear targeting by scuPA.
Nucleolin mediates transport of scuPA to the nucleus. (A) ΔNLS-nucleolin abrogates nuclear translocation of ΔGFD-scuPA. MEF-1 cells, transfected with vectors encoding either mouse WT-nucleolin-FLAG (top panel) or ΔNLS-nucleolin-FLAG (bottom panel), were incubated with ΔGFD-scuPA (20 nM) for 30 minutes at 37°C and then fixed in MeOH as in Figure 1. ΔGFD-scuPA was visualized using polyclonal α-uPA Ab and Alexa488-conjugated secondary Ab (green; i,v). FLAG-tagged nuceolin variants were visualized with Cy3-conjugated mouse α-FLAG MAb (red; ii,vi). Nuclei were stained with DAPI (blue; iii,vii). Panels iv and viii show merged images. (B) Effect of nucleolin down-regulation on nuclear translocation of ΔGFD-scuPA. BJ cells were transduced with “empty” lentivirus (top panel) or lentivirus delivering a cassette expressing a nucleolin-targeting shRNA (bottom panel, nucRNAi). Cells were incubated with 20 nM ΔGFD-uPA for 30 minutes, fixed, and stained as above, except that endogenous nucleolin was detected using mouse α-nucleolin MAb and Alexa546-conjugated α-mouse Ab (red, ii,vi). Panels iv and viii show merged images. Scale bar represents 10 μm. (Inset) WB of lysates from cells transduced with lentivirus variants as in panel B using mouse α-nucleolin MAb to analyze nucleolin content and α-GAPDH MAb for the control of total protein loading. A decrease in expression of nucleolin, but not GAPDH, is seen.
Nucleolin mediates transport of scuPA to the nucleus. (A) ΔNLS-nucleolin abrogates nuclear translocation of ΔGFD-scuPA. MEF-1 cells, transfected with vectors encoding either mouse WT-nucleolin-FLAG (top panel) or ΔNLS-nucleolin-FLAG (bottom panel), were incubated with ΔGFD-scuPA (20 nM) for 30 minutes at 37°C and then fixed in MeOH as in Figure 1. ΔGFD-scuPA was visualized using polyclonal α-uPA Ab and Alexa488-conjugated secondary Ab (green; i,v). FLAG-tagged nuceolin variants were visualized with Cy3-conjugated mouse α-FLAG MAb (red; ii,vi). Nuclei were stained with DAPI (blue; iii,vii). Panels iv and viii show merged images. (B) Effect of nucleolin down-regulation on nuclear translocation of ΔGFD-scuPA. BJ cells were transduced with “empty” lentivirus (top panel) or lentivirus delivering a cassette expressing a nucleolin-targeting shRNA (bottom panel, nucRNAi). Cells were incubated with 20 nM ΔGFD-uPA for 30 minutes, fixed, and stained as above, except that endogenous nucleolin was detected using mouse α-nucleolin MAb and Alexa546-conjugated α-mouse Ab (red, ii,vi). Panels iv and viii show merged images. Scale bar represents 10 μm. (Inset) WB of lysates from cells transduced with lentivirus variants as in panel B using mouse α-nucleolin MAb to analyze nucleolin content and α-GAPDH MAb for the control of total protein loading. A decrease in expression of nucleolin, but not GAPDH, is seen.
Regulation of α-SMA expression by nuclear uPA
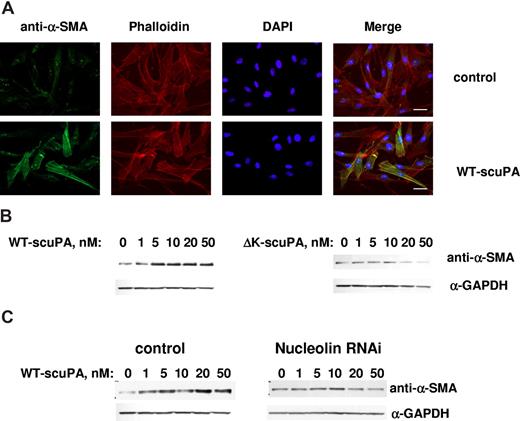
We recently reported that scuPA accelerates the transformation of adventitial fibroblasts into myofibroblasts, which is characterized by up-regulation in the expression of α-SMA.52 WT-scuPA (10 nM) stimulated the expression of α-SMA in BJ cells as assessed by immunofluorescence microscopy (Figure 7A) and caused a 3- to 5-fold increase in α-SMA levels as detected by WB (Figure 7B left panel, n = 3, P < .005). WT-scuPA (10 nM) also stimulated α-SMA mRNA expression by 1.3-fold (P < .02) as measured by real-time PCR. We then asked whether the up-regulation of α-SMA in human fibroblasts in response to scuPA was mediated through nucleolin. To do so, we first examined whether ΔK-scuPA, which does not translocate to the nucleus, stimulates expression of α-SMA. BJ cells were incubated with WT-scuPA or ΔK-scuPA (5-50 nM) for 24 hours, and the cell lysates were analyzed by WB. WT-scuPA induced α-SMA expression under these conditions, whereas ΔK-scuPA did not (Figure 7B). Furthermore, suppression of nucleolin expression by lentivirus-shRNA (Figure 6B inset) abrogated the stimulatory effect of WT-scuPA on α-SMA expression (Figure 7C). These results suggest that nucleolin-dependent nuclear translocation is required for scuPA-induced α-SMA expression. Importantly, catalytically inactive scuPA (S356A-scuPA) did not stimulate expression of α-SMA notwithstanding undergoing translocation to the nucleus (Figure S5), indicating that nuclear translocation alone is not sufficient for scuPA-dependent induction of fibroblast-to-myofibroblast transformation.
Nuclear uPA up-regulates expression of α-SMA in BJ cells. (A) BJ cells were serum-starved for 24 hours and serum-free medium containing 10 nM WT-scuPA (bottom panel) was then added for 24 hours. Control cells were replenished with fresh serum-free medium alone (top panel). The cells were washed, fixed in 4% PFA for 15 minutes, and permeabilized in 0.1% TritonX-100/PBS. α-SMA was visualized using mouse anti–α-SMA MAb and goat α-mouse Alexa 488-conjugated pAb (green). F-actin was detected with Alexa 647-conjugated phalloidin (red). Nuclei were stained with DAPI (blue). Images were taken using a Leica DMI 4000B microscope at 40× magnification equipped with Leica DFC350FX camera and Leica Application Suite (version 2.3.4R2) software (Leica Microsystems CMS). Scale bar represents 20 μm. (B) WT-scuPA, but not ΔK-scuPA, up-regulates α-SMA. BJ cells were serum-starved for 24 hours and serum-free medium containing either 1 to 50 nM WT-scuPA (left panel) or 1 to 50 nM ΔK-scuPA (right panel) was added for 24 hours. Cell lysates were prepared as in “Methods,” analyzed by SDS-PAGE and WB using mouse anti–α-SMA MAb and α-GAPDH MAb. The results shown are from 1 experiment representative of 5 so performed. (C) Nucleolin mediates scuPA-induced up-regulation of α-SMA. BJ cells were transduced with either control lentivirus (control) or lentivirus carrying nucleolin-targeting shRNA as described in “Methods.” Cells were starved as above and stimulated with WT-scuPA as in panel B. Expression of α-SMA and GAPDH was analyzed as in panel B. The results shown are from 1 experiment representative of 3 so performed.
Nuclear uPA up-regulates expression of α-SMA in BJ cells. (A) BJ cells were serum-starved for 24 hours and serum-free medium containing 10 nM WT-scuPA (bottom panel) was then added for 24 hours. Control cells were replenished with fresh serum-free medium alone (top panel). The cells were washed, fixed in 4% PFA for 15 minutes, and permeabilized in 0.1% TritonX-100/PBS. α-SMA was visualized using mouse anti–α-SMA MAb and goat α-mouse Alexa 488-conjugated pAb (green). F-actin was detected with Alexa 647-conjugated phalloidin (red). Nuclei were stained with DAPI (blue). Images were taken using a Leica DMI 4000B microscope at 40× magnification equipped with Leica DFC350FX camera and Leica Application Suite (version 2.3.4R2) software (Leica Microsystems CMS). Scale bar represents 20 μm. (B) WT-scuPA, but not ΔK-scuPA, up-regulates α-SMA. BJ cells were serum-starved for 24 hours and serum-free medium containing either 1 to 50 nM WT-scuPA (left panel) or 1 to 50 nM ΔK-scuPA (right panel) was added for 24 hours. Cell lysates were prepared as in “Methods,” analyzed by SDS-PAGE and WB using mouse anti–α-SMA MAb and α-GAPDH MAb. The results shown are from 1 experiment representative of 5 so performed. (C) Nucleolin mediates scuPA-induced up-regulation of α-SMA. BJ cells were transduced with either control lentivirus (control) or lentivirus carrying nucleolin-targeting shRNA as described in “Methods.” Cells were starved as above and stimulated with WT-scuPA as in panel B. Expression of α-SMA and GAPDH was analyzed as in panel B. The results shown are from 1 experiment representative of 3 so performed.
Discussion
We report that scuPA translocates from the extracellular space to the nucleus in the absence of proteolytic activation or degradation. Translocation is mediated by the nucleocytoplasmic shuttle protein nucleolin. Nuclear translocation of scuPA is followed by increased expression of α-SMA in fibroblasts.
scuPA lacks a canonical nuclear localization signal, and it is unlikely that it diffuses passively through nuclear pores due to its size. Rather, several lines of evidence support the concept that transport of scuPA to the nucleus is mediated by nucleolin. Immobilized scuPA binds nucleolin in cell lysates, and nucleolin coimmunoprecipitates from lysates of cells preincubated with scuPA. Only those scuPA variants that bind to nucleolin reach the nucleus. Soluble nucleolin, its uPA-binding segment GAR, and its known ligand lactoferrin33 inhibit nuclear transport of scuPA. Nuclear translocation of uPA is seen only in cells expressing full-length nucleolin, but not in cells expressing a dominant negative NLS-deficient nucleolin variant where uPA is trapped in cytoplasm due to inability of ΔNLS-nucleolin to enter the nucleus.28 Down-regulation of nucleolin by shRNA inhibits nuclear transport of scuPA. Taken together, these results indicate that nucleolin is required for efficient nuclear trans-location of scuPA.
The kringle domain of scuPA is required for nuclear transport by nucleolin. WT-scuPA and only those uPA deletion variants that contain the kringle domain bind to nucleolin and undergo nuclear translocation, whereas ΔK-scuPA fails to reach the nucleus. These findings should be interpreted in the context of previous studies that show that the kringle domain participates in uPA-mediated signal transduction,16,53,54 enhancement of cell migration,9 adhesion,11 and inhibition of angiogenesis,55 although the role of nuclear translocation in these processes and more precise dissection of the active motif within the kringle domain remain to be determined.
The ability of ΔGFD-scuPA to bind nucleolin directly and to translocate to the nucleus taken together with the observation that the importation of human scuPA to the nucleus takes place in mouse uPAR-expressing MEF-1 cells (as well as in human uPAR-expressing HeLa and BJ cells) indicates that uPAR is not required. The finding that nuclear transport of scuPA is comparable in LRP-deficient MEF-2 cells and LRP-expressing MEF-1 cells, while cytoplasmic accumulation is dramatically reduced, indicates that binding to LRP is also not essential for nuclear translocation of scuPA. However, expression of both receptors promotes binding and endocytosis of scuPA, and may thereby facilitate nuclear transport. The finding that scuPA is transported to the nucleus and does not undergo substantial degradation, whereas tcuPA does not translocate to the nucleus, suggests that conformational changes in tcuPA resulting from proteolytic activation may help guide uPAR/LRP-mediated intracellular molecular trafficking of active enzyme to lysosomes. However, a detailed characterization of how these and other surface uPA-binding receptors impact nuclear translocation and the structural changes in urokinase that govern intracellular trafficking requires further study.
Nucleolin is found on cell surfaces in some biologic contexts,17,20,21,32,35,56-58 for example, on endothelial cells during angiogenesis20,21 and on tumor vessels where it has recently been shown to act as a cell surface receptor for endostatin.22 We have previously reported that uPAR and nucleolin coimmunoprecipitate,17,59 colocalize on the cell surface, and cointernalize.17 Binding of GFD to uPAR may facilitate scuPA binding to nucleolin through its kringle. Alternatively, the uPA-nucleolin interaction might take place after endocytosis rather than on the cell surface as a result of progressive acidification within early endosomes that lowers uPA's affinity for uPAR5 while increasing its affinity for nucleolin. A similar postendocytic itinerary has been suggested for the interaction of midkine with nucleolin, which subsequently translocates to the nucleus in a nucleolin-dependent manner.28
Nucleolin is a highly motile shuttle protein with short dwell times in subcellular compartments, including the plasma membrane,26,28,34 which would amplify the nuclear import process. In settings (eg, in tumors or during wound repair7,60-63 ) where uPA secretion is sufficient to saturate uPAR, and cell surface expression of nucleolin is up-regulated, direct binding of uPA to cell surface nucleolin may contribute to nuclear transport. The finding that scuPA is translocated to the nucleus in MEF-2 cells, which do not express LRP or species-compatible uPAR, suggests that scuPA may bind directly to cell surface nucleolin. The observation that nuclear transport of scuPA is quantitatively similar in MEF-1 and MEF-2 cells suggests that upon binding LRP directs scuPA toward lysosomes when uPAR is not present, while the fraction bound to cell surface nucleolin is translocated to the nucleus.
Alternatively, scuPA might permeate the membrane of endocytic vesicles with the aid of proton and Na+/K+ pumps, as imputed for FGF-2,64,65 encountering nucleolin in the cytosol. Thus, there are several, nonmutually exclusive mechanisms by which scuPA may encounter nucleolin, and the relative contribution of each pathway is likely to depend on context. Studies are in progress to elucidate the signals and subcellular pathway(s) that regulate nuclear trafficking of scuPA.
We recently reported that uPA promotes the transformation of vascular adventitial fibroblasts to myofibroblasts after carotid artery injury, which is evidenced by increased expression of α-SMA.52 Expression of α-SMC by fibroblasts is usually associated with their differentiation into myofibroblasts. The mechanism by which uPA promotes fibroblast differentiation has not been well characterized. The extent of α-SMA induction in response to scuPA (3- to 5-fold) is comparable with the effect of TGFβ-1,66 a known mediator of fibroblast-to-myofibroblast differentiation.66,67 Our observations that uPA up-regulates α-SMA expression in fibroblasts in a nucleolin-dependent manner opens the possibility that scuPA may modulate gene transcription. For example, it has been reported that uPA binds selectively to conserved purine-rich single-stranded DNA (ssDNA) sequences in vitro.68 Our pilot data raise the possibility that uPA associates with chromatin by binding to DNA directly or in complex with transcription factors and/or with nucleolin, which itself possesses helicase69 and ssDNA- and dsDNA-binding and transcriptional activities.70,71 Whether uPA/nucleolin complexes exert transcriptional activity is currently under study. The finding that catalytically inactive S356A-scuPA translocates to the nucleus but does not induce α-SMA could indicate either that the low intrinsic proteolytic activity of scuPA is involved in its nuclear function or that there are conformational differences between the heavy chains of WT and inactive scuPA, and interactions between scuPA's heavy chain with other mediators (DNA or transcription factors) are of functional importance.
In summary, the studies presented here identify a novel pathway by which uPA is translocated to the nucleus where it might regulate gene expression alone or in complex with nucleolin. This pathway might identify a new platform for pharmacological modulation once its (patho)physiological implications are understood in greater detail.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr K. Kadomatsu for pcDNA3.1 constructs encoding mouse nucleolin C-terminus-FLAG-tagged mutants, Dr T. Willnow for providing MEF-1 and MEF-2 cells, Dr A. Shevelev for providing pcDNA3.1 construct encoding human uPA, Dr D. Trono for providing lentiviral vectors, M. W. Izzo (Improvision) for assistance with 3D data visualization and presentation, and Y. Bdeir for technical assistance.
The work was supported in part by National Institutes of Health grants (HL076406, CA83121, HL076206, and 1 R21 HL81864-01 [D.B.C and A.A.-R.H.]); Philip Morris USA and Philip Morris International grants (D.B.C., K.B., and A.A.-R.H.); University of Pennsylvania Research Foundation awards (D.B.C., and K.B.); AHA Scientist Development Grants (0430209N [V.S.] and 0535258N [S.Z.]); National Heart, Lung, and Blood Institute funds for exchanges of scientists under the auspices of the US-Russia Joint Agreement in Cardiopulmonary Research; Fogarty International Research Collaboration Award 5-RO3-TW01468-03 (D.B.C. and V.A.T.); Volkswagen Research Foundation I/76 889 (V.S. and I.D.); Deutsche Forschungsgemeinschaft Grant DU 344/2-4 (I.D.); and Russian Foundation for Basic Research (RFBR) grants 04-04-49481 (V.S.), 05-04-49108a to (Y.P.), and 05-04-49278a (V.A.T.).
National Institutes of Health
Authorship
Contribution: V.S. and D.B.C. designed the research; V.S., T.L., A.K., S.Y., S.Z., K.B., and Y.P. performed the research and collected data; V.S., V.A.T., A.A.-R.H., M.S.M., and D.B.C. analyzed the data; S.T. and I.D. developed vital new reagents; V.S., A.A.-R.H., and D.B.C. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Victoria Stepanova, Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, 512 Stellar-Chance Bldg, 422 Curie Blvd, Philadelphia, PA 19104; e-mail: vstepano@mail.med.upenn.edu.