Abstract
The molecular mechanisms that regulate megakaryocyte (MK) ploidization are poorly understood. Using MK differentiation from primary human CD34+ cells, we observed that p19INK4D expression was increased both at the mRNA and protein levels during ploidization. p19INK4D knockdown led to a moderate increase (31.7% ± 5%) in the mean ploidy of MKs suggesting a role of p19INK4D in the endomitotic arrest. This increase in ploidy was associated with a decrease in the more mature MK population (CD41highCD42high) at day 9 of culture, which was related to a delay in differentiation. Inversely, p19INK4D overexpression in CD34+ cells resulted in a decrease in mean ploidy level associated with an increase in CD41 and CD42 expression in each ploidy class. Confirming these in vitro results, bone marrow MKs from p19INK4D KO mice exhibited an increase in mean ploidy level from 18.7N (± 0.58N) to 52.7N (± 12.3N). Chromatin immunoprecipitation assays performed in human MKs revealed that AML-1 binds in vivo the p19INK4D promoter. Moreover, AML-1 inhibition led to the p19INK4D down-regulation in human MKs. These results may explain the molecular link at the transcriptional level between the arrest of endomitosis and the acceleration of MK differentiation.
Introduction
Megakaryocytes, cardiomyocytes, arterial smooth muscle cells, and hepatocytes are the rare mammalian cells undergoing polyploidization during their life span in the absence of any stimuli
Until now, the best-studied model of polyploidization concerns megakaryocytes (MKs) in which after several replicative rounds, the process of classic mitosis is replaced by an endomitotic process. MK endomitosis is very similar to mitosis, but MKs skip anaphase B, telophase, and cytokinesis giving rise to a polyploid cell with a single polylobulated nucleus.1,2 Although polyploidization is a part of the MK differentiation program, terminal differentiation that is associated with an increased platelet protein synthesis and the development of specific organelles requires an arrest of the cell cycle. Both the mitotic and endomitotic processes require an amplification of the DNA content. However, while mitosis leads to an increase in cell number, polyploidization results in an increase in the cell size and protein mass. In the case of MK, the platelet precursor, polyploidization increases platelet production.
DNA replication is a regulated mechanism that depends on a well-organized network of the cyclin-dependent kinases (CDKs) and cyclin-dependent inhibitors (CDKIs) of the cell cycle. Progression through G1 to S phase of cell cycle in mammalian cells requires the activity of G1-cyclins (D and E) associated with their catalytic subunits (CDK4 and CDK6). The mitogen-stimulated cyclin D and CDK4/6 complexes phosphorylate the retinoblastoma protein (Rb) allowing its dissociation from E2Fs transcription factors. Activated E2Fs then regulate the expression of genes necessary for DNA synthesis during the S phase of cell cycle. Cells exit cell cycle and accumulate in a quiescent G0/G1 state either in the absence of mitogenic stimuli when the cyclin D–dependent activity is lost or after CDK4/6 inhibition. CDKs are regulated by inhibitory phosphorylations mediated by the WEE1 and MYT1 kinases3 or by induction of specific inhibitors from the INK4 and CIP/KIP families. Till now, 4 members of the INK4 family (p19INK4D, p18INK4C, p16INK4A, and p15INK4B) are described to specifically block the activity of CDK4/6 either by forming inactive ternary INK4-CDK4/6-cyclin D or binary INK4-CDK4/6 complexes.4 The members of the CIP/KIP family (p21CIP1/WAF1, p27KIP1, and p57KIP2) inhibit cyclin A– or cyclin E–associated CDK2 activity, but stabilize cyclin D–associated CDK4/6 activity.5,6 In MKs, a high level of 2 CIP/KIP family members, p21CIP1 and p27KIP1,7,8 and 1 member of the INK4 family, p16INK4A,7 was detected at hypoploid (2N-4N) and polyploid (≥ 8N) stages. p21 overexpression decreased the ploidy level in human and murine MKs independently of the p21CIP1 status (p21−/− and p21+/+), suggesting that this protein could play an important role during the exit from the endomitotic cycle.7 During human MK differentiation, an up-regulation of p15INK4B 9 and a down-regulation of p16INK4A have been observed.10 Recently, it was reported that overexpression of p16INK4A resulted in the inhibition of MK polyploidization.11 By performing a transcriptional profile of genes regulated during MK ploidization, we have shown that, among the CDKI, only p19INK4D mRNA was linearly regulated through the 2N to 16N stages.12
p19INK4D is expressed at high levels in thymus and peripheral blood leukocytes, and at a moderate level in spleen, testis, skeletal muscle, and heart.13 Different in vitro studies indicate that p19INK4D is involved in myeloid differentiation14-17 as well as in erythroid differentiation from murine progenitors infected with the anemia-inducing strain of Friend virus.18 Mice lacking p19INK4D have a normal life span, do not develop tumors, and do not display hematologic abnormalities. However, in p19INK4D knockout mice, another CDKI could compensate p19INK4D. Indeed, double knockout mice provided evidence that collaboration between p18INK4C and p19INK4D is necessary to regulate spermatogenesis, notably during the normal meiotic maturation of spermatocytes.19 In addition, p19INK4D in cooperation with p27KIP1 is essential to the maintenance of the terminally differentiated postmitotic neurons20,21 and sensory hair cells22 in postnatal mice. Finally, in mice lacking Msx1, significantly elevated p19INK4D expression was linked to a reduction in cranial neural crest cell proliferation in dental mesenchyme, suggesting an important role of p19INK4D during tooth morphogenesis.23
To analyze the involvement of p19INK4D in MK development, we have used p19INK4D−/− mice and we have repressed or overexpressed this cell-cycle inhibitor in MK progenitors and have studied the impact of these 2 events on the ploidy level and differentiation in in vitro human MK differentiation.
Methods
Mice
Heterozygous p19INK4D+/− mice were a gift from Martine Roussel (Memphis, TN). p19INK4D+/+ and p19INK4D−/− mice were obtained by crossing the heterozygous mice.
In vitro growth of megakaryocytes from CD34+ cells
Cytapheresis samples were obtained after informed consent in accordance with the Declaration of Helsinki. Approval was obtained from the Assistance Publique des Hôpitaux de Paris. CD34+ cells were isolated using an immunomagnetic beads technique (Miltenyi Biotec, Paris, France)24 and grown in serum-free medium as previously reported.25 The culture medium was supplemented with recombinant human thrombopoietin (TPO, 10 ng/mL) or with a cytokine cocktail containing TPO (10 ng/mL), interleukin-3 (IL-3, 100 U/mL) (Immunex, Seattle, WA), interleukin-6 (IL-6, 10 ng/mL; Tebu, Perray-en-Yvelines, France), stem-cell factor (SCF; 25 ng/mL), and fetal liver tyrosine kinase 3 ligand (FLT3-L; 1 ng/mL) (both kindly provided by Amgen, Thousand Oaks, CA).
Lentiviral vector construction and production
Cloning the shRNA.
Small interfering RNAs (siRNAs) sequences were designed using siSearch software (Stockholm Bioinformatics Centre, Stockholm University, Sweden; http://sonnhammer.sbc.su.se/) and small hairpins sequences were created as previously described.26,27 Oligonucleotide p19INK4D and AML1 short hairpins (sh's) listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) were synthesized (Eurogentec, Angers, France) and inserted into a pBlue Script containing the human H1 promoter. The H1-shp19, H1-shAML-1, or H1-SCR (scramble control sequence) cassette was inserted into a lentiviral vector (pRRLsin-PGK-eGFP-WPRE; Genethon, Evry, France).
Cloning the p19INK4D cDNA.
A 502-bp human p19INK4D cDNA was amplified by polymerase chain reaction (PCR) from the plasmid pSG5p19 (kindly provided by E. T. Cánepa), and subcloned into a lentiviral vector pTRIPΔU3-EF1α-IRES-GFP (kindly provided by A. Dubart-Kupperschmitt).
Lentiviral production.
Lentiviral stocks were prepared as previously described.28 Viral stocks were stored at −80°C, and concentrations of viral particles were normalized according to the p24 (HIV-1 capsid protein) content in supernatants.
Cell transduction
CD34+ cells (105/mL) were prestimulated for 24 hours with TPO (100 ng/mL), IL-3 (1000 U/mL), IL-6 (100 ng/mL), SCF (250 ng/mL), and FLT3-L (100 ng/mL). Lentiviral particles were added at a concentration corresponding to 125 ng viral p24/100 μL for 12 hours followed by a second transduction, and then were cultured in the presence of TPO alone. To study the effect of p19INK4D silencing in polyploid MKs, CD34+ cells were cultured in the presence of TPO alone and transduced at days 5 and 6 of culture.
Cell sorting and flow cytometric and cytologic analyses
Cells were incubated for 30 minutes at 4°C with an APC-conjugated anti-CD41a MoAb and a PE-conjugated anti-CD42a MoAb (BD Pharmingen, San Diego, CA). After washing, cells were incubated for 2 hours in 0.01 mM Hoechst 33342 (Sigma-Aldrich, St Quentin Fallavier, France) at 37°C. To study endogenous p19INK4D expression in different ploidy classes, CD41+ cells were sorted as previously reported into 2N and 4N or 8N and 16N cell fractions.2 To study the ploidy level, CD41+CD42+ cells stained with Hoechst were analyzed on a LSRII (Becton Dickinson, Mountain View, CA). Approximately 50 000 cells were analyzed with the Cellquest software package (Becton Dickinson). The ploidy of murine bone marrow megakaryocytes was measured as described by Baccini et al.7
Real-time quantitative RT-PCR
Primers and internal probes for quantitative reverse-transcriptase (QRT)–PCR were designed using Primer Express Software (Perkin-Elmer Applied Biosystems, Foster City, CA) and are listed in Table S2. PCRs were carried out in the ABI Prism GeneAmp 5700 Sequence Detection System (Perkin-Elmer Applied Biosystems) using the TaqMan Universal PCR Master Mix (ABI) containing the specific primers (1.2 μM) and the specific probe (0.1 μM). The expression levels of all genes were expressed relatively to HPRT. To calculate the absolute quantity of p19INK4D in 100 ng total RNA, expression level was measured relatively to the plasmid harboring the p19INK4D cDNA corresponding to a defined number of plasmid copies per cell (from 102 to 106).
Western blot analysis
Western blot analyses were performed as previously reported.7 The following antibodies were used: anti-p19INK4D (M-167), anti–HDAC-1 (H-51) (Santa Cruz Biotechnology, Santa Cruz CA), and anti–phospho-Rb (Ser780; Cell Signaling, Beverly, MA). Coloration with ponceau S solution (Sigma) after transfer and immunolabeling with the HDAC-1 antibody was used to control the quantity of proteins.
Quantification of proplatelet MKs
GFP, CD41+, CD42+ MKs were sorted and plated in 96-well plates at a concentration of 2000 cells/well in serum-free medium containing TPO. Four days later, MKs displaying proplatelets were quantified by enumerating 500 cells per well using an inverted microscope at a magnification of ×200.
Luciferase assays
The p19INK4D promoter with 2 different lengths, 1.5 kb (plucF3) and 0.5 kb (plucF2), was amplified and subcloned into the pLuc-MCS vector (Stratagene, Charbonnières, France). Primers used for promoter amplification are listed in Table S3. AML1c cDNA was cloned into the vector MPI HAa,29 and CBFβ cDNA was cloned into the vector pEF6/V5-His-TOPO (Invitrogen, Cergy-Pontoise, France). HEL cells were transfected with pluc constructs, a renilla luciferase vector (for normalization), and AML-1c expression vector using Lipofectamine 2000 (Invitrogen). Cells were cultured in 12-well plates and transfected with a 0.5 to 2 μg DNA. The total amount of DNA was kept constant by the inclusion of appropriate amounts of empty vectors. Cells were lysed with Passive Lysis Buffer 48 hours after transfection, and the luciferase activity was evaluated using the Dual Luciferase Reporter Assay System (Promega, Madison, WI).
Chromatin immunoprecipitations
Chromatin immunoprecipitations (ChIP) assays were performed with a ChIP assay kit (Upstate-Millipore, Paris, France) using the H-65 anti–AML-1 antibody (Santa Cruz Biotechnology), and an anti–AML-1/RHD domain (Ab-2) antibody (Calbiochem, Mendon, France). These assays were performed using chromatin prepared from human MKs. Quantification of precipitated DNA fragments was carried out on an ABI PRISM 7000 sequence detection system using SYBR green (Eurogentec) in duplicate. Primer sequences for p19INK4D are described in Table S3. Relative occupancy of the immunoprecipitated factor at a locus was calculated using the following equation: 2(CtNegCtl − CtTarget), where CtNegCtl and CtTarget are mean threshold cycles of PCR done in duplicate on DNA samples from negative control ChIP (using non–immune IgG) and targeted ChIP (using specific antibody).
Results
Expression of p19INK4D in megakaryocytes
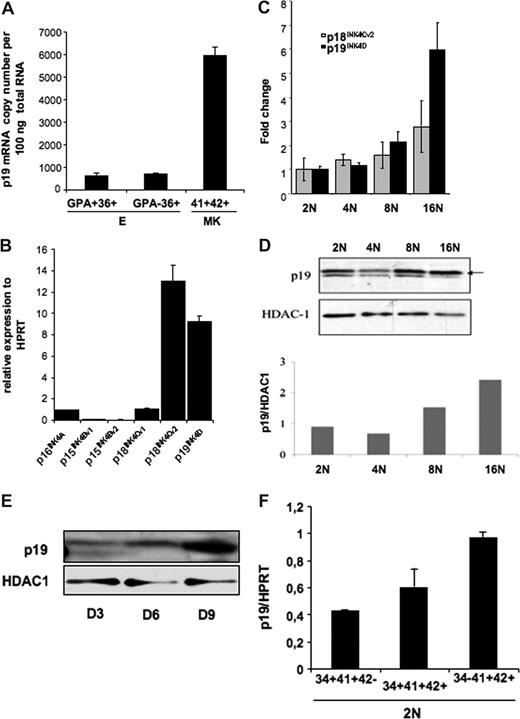
We studied the pattern of p19INK4D expression during MK differentiation. Since previous studies have shown that p19INK4D was expressed during erythroid differentiation, we compared the level of p19INK4D expression in mature MKs expressing CD41 and CD42 and in immature (CD36+GPA−) and mature (CD36+GPA+) erythroid cells. We found that p19INK4D mRNA was approximately 10-fold more abundant in MKs than in immature or mature erythroid cells (Figure 1A). Total RNA (100 ng) from CD41+CD42+ cells contained a mean of 5943 copies of p19INK4D mRNA, whereas approximately708 and 631 copies were detected in 100 ng total RNA extracted from CD36+GPA− and CD36+GPA+ cells, respectively.
p19INK4D expression in erythroid and megakaryocyte human cells. (A) The absolute expression level of p19INK4D per 100 ng total RNA was measured relatively to different dilutions of plasmids (corresponding to 102 to 106 copies) harboring a p19INK4D cDNA. The graph represents the expression level of p19INK4D in immature (CD36+GPA−) and mature (CD36+GPA+) erythroid cells and in mature (CD41+CD42+) MKs. The error bars represent the standard deviation of the mean of 3 experiments each performed in triplicate wells. (B-E) CD34+ cells were cultured with TPO for 9 days. Then, CD41- and CD42-positive cells stained with Hoechst were sorted on their ploidy level (from 2N to 16N). (B) mRNA levels of p15INK4B, p16INK4A, p18INK4C, and p19INK4D and their variants were quantified by real-time RT-PCR. Their relative expression was calculated in comparison with HPRT mRNA. (C) mRNA levels of p18INK4Cv2 and p19INK4D were quantified by RT-PCR in each ploidy class (from 2N to 16N) and normalized to their expression in diploid (2N) cell fraction. (D) The protein level of p19INK4D in different ploidy fractions was measured relatively to HDAC-1, a protein that we found stable during ploidization. The graph represents the ratio between p19INK4D and HDAC-1 levels for each ploidy class. The data illustrate 1 of 2 representative experiments that produced similar results. (E) To investigate the expression level of p19INK4D during differentiation, total cell population was harvested on day 3 (D3), day 6 (D6), and day 9 (D9) and submitted to Western blot analysis. (F) The expression level of p19INK4D mRNA was investigated by QRT-PCR in diploid (2N) cells sorted at day 6 in culture in 3 MK populations corresponding to very immature (CD34+CD41+CD42−), immature (CD34+CD41+CD42+), to more mature (CD34−CD41+CD42+) cells. The error bars represent the standard deviation of the mean of 3 repeated experiments each performed in triplicate wells.
p19INK4D expression in erythroid and megakaryocyte human cells. (A) The absolute expression level of p19INK4D per 100 ng total RNA was measured relatively to different dilutions of plasmids (corresponding to 102 to 106 copies) harboring a p19INK4D cDNA. The graph represents the expression level of p19INK4D in immature (CD36+GPA−) and mature (CD36+GPA+) erythroid cells and in mature (CD41+CD42+) MKs. The error bars represent the standard deviation of the mean of 3 experiments each performed in triplicate wells. (B-E) CD34+ cells were cultured with TPO for 9 days. Then, CD41- and CD42-positive cells stained with Hoechst were sorted on their ploidy level (from 2N to 16N). (B) mRNA levels of p15INK4B, p16INK4A, p18INK4C, and p19INK4D and their variants were quantified by real-time RT-PCR. Their relative expression was calculated in comparison with HPRT mRNA. (C) mRNA levels of p18INK4Cv2 and p19INK4D were quantified by RT-PCR in each ploidy class (from 2N to 16N) and normalized to their expression in diploid (2N) cell fraction. (D) The protein level of p19INK4D in different ploidy fractions was measured relatively to HDAC-1, a protein that we found stable during ploidization. The graph represents the ratio between p19INK4D and HDAC-1 levels for each ploidy class. The data illustrate 1 of 2 representative experiments that produced similar results. (E) To investigate the expression level of p19INK4D during differentiation, total cell population was harvested on day 3 (D3), day 6 (D6), and day 9 (D9) and submitted to Western blot analysis. (F) The expression level of p19INK4D mRNA was investigated by QRT-PCR in diploid (2N) cells sorted at day 6 in culture in 3 MK populations corresponding to very immature (CD34+CD41+CD42−), immature (CD34+CD41+CD42+), to more mature (CD34−CD41+CD42+) cells. The error bars represent the standard deviation of the mean of 3 repeated experiments each performed in triplicate wells.
Then we investigated the level of p19INK4D expression during MK ploidization because our previous data with microarrays have shown that p19INK4D was the only CDKI whose expression was increased during ploidization. First, we investigated the expression level of all transcription variants of p15INK4B, p16 INK4A, p18 INK4C, and p19 INK4D, each separately in MKs at day 9 of culture. Only p18INK4C variant 2 and p19INK4D were expressed at a high level (Figure 1B). Quantitative RT-PCR assays confirmed that mRNA levels of p19INK4D, but not p18INK4C variant 2, increased continuously from 2N to 16N cells (Figure 1C). Thus, we focused our study only on p19INK4D. An increase in p19INK4D protein was also observed with ploidy (Figure 1D). However, a slight decrease was reproducibly seen in the 4N class compared with 2N cells (Figure 1D) that could be explained by the degradation of p19INK4D occurring during mitosis exit. The increasing amount of p19INK4D during ploidization suggests a role of this protein in the cell-cycle arrest occurring at the end of endomitosis. As the exit from cell cycle is generally linked to the onset of maturation, p19INK4D expression may also be regulated along differentiation independently of ploidy. As a first approach, we studied the p19INK4D protein level at different days of culture (day 3, day 6, and day 9) in the presence of TPO and found that the amount of p19INK4D increased continuously (Figure 1E). However, as at days 6 and 9 the cell population already contains polyploid cells, we sorted diploid megakaryocytic cells at day 6 according to the expression of 3 differentiation markers and analyzed p19INK4D expression by quantitative RT-PCR. The data show that p19INK4D transcripts continuously increased with the progression of MK differentiation (from CD34+CD41+CD42− to CD34−CD41+CD42+ cells; Figure 1F).
Inhibition of p19INK4D induces a delay in MK maturation
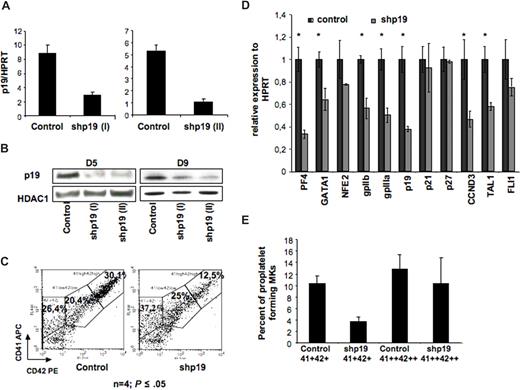
The fact that p19INK4D expression increases not only during ploidization but also during MK differentiation suggested that this protein was involved in the arrest of cell cycle that allows MK terminal differentiation. To confirm this hypothesis, we developed 2 lentiviruses encoding 2 different p19INK4D shRNA under the H1 promoter. The lentivirus vectors also expressed the GFP protein under the ubiquitous promoter PGK. To demonstrate that these vectors could achieve a stable p19INK4D knockdown in MKs, CD34+ cells were grown in the presence of IL-3, FLT3-L, SCF, and TPO; infected at days 1 and 2 of culture; and then cultured in the presence of TPO alone. At day 5 of culture (72 hours after transduction), cells were harvested and GFP-positive cells were sorted. The lentivirus encoding for shp19INK4D (I or II) reduced the p19INK4D transcripts approximately 5-fold in comparison with the control shRNA (Figure 2A). Similar results were observed at the protein level (at day 5), and the p19 inhibition was maintained at day 9 of culture (Figure 2B).
Effect of p19 INK4D knockdown on MK differentiation. CD34+ cells were transduced with a control lentivirus or a lentivirus encoding either shRNA p19 (I) or shRNA p19 (II) GFP positive (GFP+). (A) p19INK4D mRNAs were measured by QRT-PCR in GFP-positive cells at day 5 of culture. Error bars are standard deviations of the mean of 3 repeated experiments. (B) p19INK4D protein level was analyzed at day 5 and day 9 of culture by Western blot. HDAC-1 was used as internal control of a quantity. The data illustrate 1 of 2 representative experiments that produced similar results. (C) Flow cytometric analysis of mature megakaryocytes expressing a high level of CD41 and CD42 antigens in GFP+ cells at day 9 of culture. (D) Real-time RT-PCR was used to quantify PF4, NF-E2, GATA-1, GPIIb, GPIIIa, FLI-1, p19INK4D, p21, p27, cyclin D3 (CCND3), and TAL-1 mRNAs in GFP+ cells (at day 9 of culture). The relative expression of all these genes was calculated in comparison with HPRT mRNA. (E) GFP+ cells were sorted at day 9 either on the low coexpression of CD41 and CD42 (CD41+ CD42+ population) or on a high expression of both antigens (CD41++ CD42++ population). Cells were seeded at 2 × 103 cells/well in 96-well plate. At day 12, the percentage of MKs forming proplatelets was estimated by counting MKs exhibiting one or more cytoplasmic processes with areas of constriction among 500 cells per well (in 5 wells). Panel C shows 1 representative analysis of 4 repeated experiments with similar results. Error bars in histograms in panels D,E represent the standard deviation of 3 repeated experiments each performed in triplicate wells.
Effect of p19 INK4D knockdown on MK differentiation. CD34+ cells were transduced with a control lentivirus or a lentivirus encoding either shRNA p19 (I) or shRNA p19 (II) GFP positive (GFP+). (A) p19INK4D mRNAs were measured by QRT-PCR in GFP-positive cells at day 5 of culture. Error bars are standard deviations of the mean of 3 repeated experiments. (B) p19INK4D protein level was analyzed at day 5 and day 9 of culture by Western blot. HDAC-1 was used as internal control of a quantity. The data illustrate 1 of 2 representative experiments that produced similar results. (C) Flow cytometric analysis of mature megakaryocytes expressing a high level of CD41 and CD42 antigens in GFP+ cells at day 9 of culture. (D) Real-time RT-PCR was used to quantify PF4, NF-E2, GATA-1, GPIIb, GPIIIa, FLI-1, p19INK4D, p21, p27, cyclin D3 (CCND3), and TAL-1 mRNAs in GFP+ cells (at day 9 of culture). The relative expression of all these genes was calculated in comparison with HPRT mRNA. (E) GFP+ cells were sorted at day 9 either on the low coexpression of CD41 and CD42 (CD41+ CD42+ population) or on a high expression of both antigens (CD41++ CD42++ population). Cells were seeded at 2 × 103 cells/well in 96-well plate. At day 12, the percentage of MKs forming proplatelets was estimated by counting MKs exhibiting one or more cytoplasmic processes with areas of constriction among 500 cells per well (in 5 wells). Panel C shows 1 representative analysis of 4 repeated experiments with similar results. Error bars in histograms in panels D,E represent the standard deviation of 3 repeated experiments each performed in triplicate wells.
These 2 shRNAs (shp19INK4D I and II) were used to study the effect of p19INK4D knockdown on MK maturation. As shown in Figure 2C, p19INK4D knockdown inhibited MK maturation resulting in a 38.6% ± 17% diminution of mature CD41++CD42++ MKs at day 9 (n = 4, P ≤ .05).
To investigate further the effects of p19INK4D knockdown on genes important for MK lineage, the transcript levels of 4 MK transcription factors NF-E2, GATA-1, FLI-1, and TAL-1; 2 MK glycoproteins, GPIIb and GPIIIa; and 3 proteins involved in cell cycle p21CIP1, p27KIP1, and cyclin D3 (CCDN3) were then studied by quantitative RT-PCR. p19INK4D knockdown resulted in a significant decrease of all transcripts, except for the 2 cell-cycle inhibitors, p21CIP1and p27KIP1, and 2 transcription factors, NFE2 and Fli1 (Figure 2D).
Finally, we studied if p19INK4D knockdown would alter proplatelet formation. When the MK population was sorted on CD41+CD42+ expression, p19INK4D knockdown markedly decreased proplatelet formation. However, when the more mature MKs were purified on a high CD41 and CD42 expression (CD41++CD42++ cells), no difference in the percentage of proplatelet-forming MKs was seen between control and p19INK4D knockdown cells (Figure 2E). This result highly suggests that p19INK4D regulates more the rate of MK differentiation than the process of differentiation itself and, consequently, that p19INK4D is not directly involved in proplatelet formation.
p19INK4D is involved in the cell-cycle exit
To study the effect of p19INK4D knockdown on cell cycle, CD34+ cells were transduced at days 1 and 2 of culture as previously described and analyzed at day 9 of culture. The increase of Rb phosphorylation on serine 780 confirmed the effect of p19 inhibition on cyclin D–Cdk activity in shp19INK4D-transduced MK cells (Figure 3A). The ploidy level of transduced (GFP+) cells was analyzed in the gate that included the highest CD41++CD42++ expression. As illustrated for a representative experiment in Figure 3B, p19INK4D knockdown resulted in a 31.7% ± 5% increase in mean ploidy level (n = 3, P ≤ .015). When the mean fluorescence intensity of CD41 and CD42 was analyzed in each ploidy class, no change in CD41 expression was seen in shp19INK4D-transduced cells versus control. In contrast, the expression level of CD42 was slightly decreased in each ploidy class (n = 3, P ≤ .03, Figure 3C). When cells were shp19INK4D transduced at later days in culture (days 5 and 6), when some polyploid MKs are already present in the culture and analyzed at day 9, the mean ploidy level was also increased but to a lesser extent (22.3% ± 6%) (n = 3; P ≤ .05, Figure 3D). Moreover, p19INK4D silencing was associated with a decreased level of CD42 only in polyploid MKs (8N and 16N) but not in 2N and 4N MKs. In contrast, the expression level of CD41 was not modified (n = 3; P ≤ .05, Figure 3E), suggesting that p19INK4D is, in part, involved in the coupling between the arrest of the mitotic or endomitotic process and differentiation. Thus, in contrast to transduction performed at day 1 where CD42 was reduced in all ploidy class, p19INK4D knockdown started at day 5 affected only cells undergoing endomitosis but not the diploid and tetraploid cells that were already maturing. Together, these data suggest that when the expression level of p19INK4D is sufficient, the cell will exit cell cycle and will accelerate its differentiation.
Effect of p19INK4D knockdown on cell cycle and CD41 or CD42 expression. CD34+ cells were transduced at days 1 and 2 (A-C) or days 5 and 6 (D,E) of culture with a control lentivirus and the lentivirus encoding either shRNA p19 (I) or shRNA p19 (II). GFP+ cells were analyzed at day 9. (A) Immunoblot analysis showing that phosphorylation of Rb on Serine 780 (p-Rb) was increased after MK transduction by the 2 shRNA p19. (B,D) Only the CD41++CD42++ cell population (left panel), corresponding to mature MKs, was analyzed for ploidy level by Hoechst staining (right panel). The mean ploidy (N) was calculated from the number of cells at each ploidy level. (C,E) The mean fluorescence intensity (MFI) of CD41 (left panel) and of CD42 (right panel) was analyzed separately for each ploidy class (from 2N to 16N). Panels B-E illustrate 1 representative analysis of 3 repeated experiments with similar results, and P values refer to 3 independent experiments.
Effect of p19INK4D knockdown on cell cycle and CD41 or CD42 expression. CD34+ cells were transduced at days 1 and 2 (A-C) or days 5 and 6 (D,E) of culture with a control lentivirus and the lentivirus encoding either shRNA p19 (I) or shRNA p19 (II). GFP+ cells were analyzed at day 9. (A) Immunoblot analysis showing that phosphorylation of Rb on Serine 780 (p-Rb) was increased after MK transduction by the 2 shRNA p19. (B,D) Only the CD41++CD42++ cell population (left panel), corresponding to mature MKs, was analyzed for ploidy level by Hoechst staining (right panel). The mean ploidy (N) was calculated from the number of cells at each ploidy level. (C,E) The mean fluorescence intensity (MFI) of CD41 (left panel) and of CD42 (right panel) was analyzed separately for each ploidy class (from 2N to 16N). Panels B-E illustrate 1 representative analysis of 3 repeated experiments with similar results, and P values refer to 3 independent experiments.
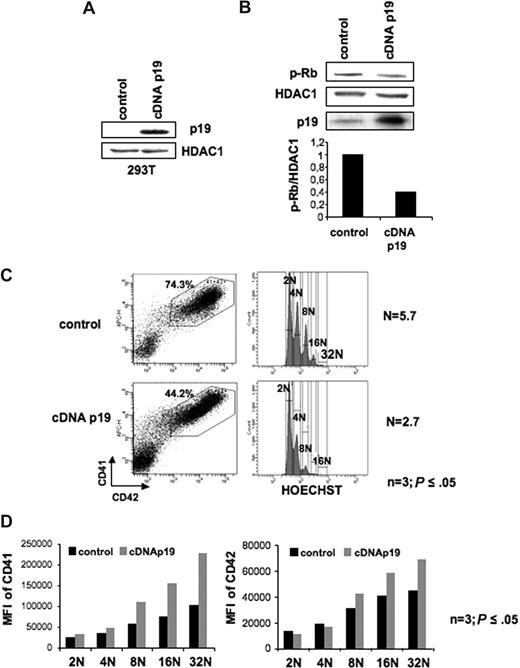
To confirm this hypothesis, we used a p19INK4D overexpression strategy using a lentiviral vector. Functionality of this lentivirus was first tested in transiently transfected 239T cells (Figure 4A). Then, CD34+ cells were transduced with the p19INK4D cDNA encoding lentivirus and the control lentivirus at days 1 and 2 of culture and cells were analyzed at day 9. In MKs overexpressing the p19INK4D, the decrease of Rb phosphorylation on serine 780 confirmed the effect of p19INK4D overexpression on cyclin D–Cdk activity (Figure 4B). As shown in Figure 4C, overexpression of p19INK4D induced a decrease in the percentage of cells expressing CD41 and CD42 from approximately 70% in control to 43%. Analysis of the mean ploidy level showed a marked decrease of approximately 60% in p19INK4D-overexpressing cells (n = 3; P ≤ .05, Figure 4C). Moreover, analysis of CD41 and CD42 expression in each ploidy class showed a marked increase of these 2 proteins in MKs overexpressing p19INK4D (n = 3; P ≤ .05, Figure 4D). Thus, in conclusion, p19INK4D is involved in cell cycle exit linked to the differentiation.
Effect of p19INK4D overexpression on cell cycle and CD41 or CD42 expression. (A) Validation of a lentiviral vector encoding a p19INK4D cDNA (cDNA p19). Immunoblot analysis of p19INK4D protein in 293T cell line transiently transfected with either the control vector or the p19 cDNA lentiviral vector. HDAC-1 was used as an internal control for protein quality and loading. (B) Immunoblot analysis shows that p19INK4D overexpression induced a 2-fold decrease in phosphorylation of Rb on serine 780 (p-Rb). (C-D) CD34+ cells were transduced at day 1 and 2 of culture with the control or the p19 cDNA lentiviral vectors and GFP+ cells were analyzed at day 9. The MK population positive for CD41 and CD42 (C left panel) was analyzed for ploidy level by Hoechst staining (C right panel). The mean ploidy (N) was calculated from the number of cells of each ploidy level. (D) The mean fluorescence intensity (MFI) of CD41 (left panel) and CD42 (right panel) was analyzed separately for each ploidy class (from 2N to 32N). Data in panels C,D represent analysis 1 of 3 repeated experiments giving similar results. P values refer to 3 independent experiments.
Effect of p19INK4D overexpression on cell cycle and CD41 or CD42 expression. (A) Validation of a lentiviral vector encoding a p19INK4D cDNA (cDNA p19). Immunoblot analysis of p19INK4D protein in 293T cell line transiently transfected with either the control vector or the p19 cDNA lentiviral vector. HDAC-1 was used as an internal control for protein quality and loading. (B) Immunoblot analysis shows that p19INK4D overexpression induced a 2-fold decrease in phosphorylation of Rb on serine 780 (p-Rb). (C-D) CD34+ cells were transduced at day 1 and 2 of culture with the control or the p19 cDNA lentiviral vectors and GFP+ cells were analyzed at day 9. The MK population positive for CD41 and CD42 (C left panel) was analyzed for ploidy level by Hoechst staining (C right panel). The mean ploidy (N) was calculated from the number of cells of each ploidy level. (D) The mean fluorescence intensity (MFI) of CD41 (left panel) and CD42 (right panel) was analyzed separately for each ploidy class (from 2N to 32N). Data in panels C,D represent analysis 1 of 3 repeated experiments giving similar results. P values refer to 3 independent experiments.
p19INK4D−/− murine bone marrow megakaryocytes have an increased ploidization process
To confirm that p19 is involved in the arrest of endomitosis in vivo, we analyzed the mean ploidy level in bone marrow megakaryocytes obtained from p19INK4D+/+, p19INK4D+/−, and p19INK4D−/− mice. As shown in Figure 5A, the modal peak in p19INK4D+/+ MKs is 16N; in p19INK4D+/− MKs, 32N; and in p19INK4D−/− MKs, 64N. As the CD41 antibody binds also to nonmegakaryocytic cells, the mean ploidy level was calculated only in polyploid cells (Figure 5A gate R3) and increased from 18.7N (± 0.58N) in p19INK4D+/+ MKs to 52.7N (± 12.3N) in p19INK4D−/− MKs (Figure 5B; n = 4).
Bone marrow p19INK4D knockout megakaryocytes have an increased ploidy. Bone marrow megakaryocyte ploidy from WT, p19INK4D+/−, and p19INK4D−/− was measured after labeling with an anti-CD41 antibody and propidium iodide (PI) staining using flow cytometry. More than 10 × 106 cells were recorded and the ploidy was measured in a CD41-high gate. (A) One representative experiment is shown. (B) The mean ploidy of 4 experiments is illustrated. In all experiments, the modal ploidy increased from 16N in wt MKs to 32N in p19INK4D+/− MKs and to 64N in p19INK4D−/− MKs as shown in panel A.
Bone marrow p19INK4D knockout megakaryocytes have an increased ploidy. Bone marrow megakaryocyte ploidy from WT, p19INK4D+/−, and p19INK4D−/− was measured after labeling with an anti-CD41 antibody and propidium iodide (PI) staining using flow cytometry. More than 10 × 106 cells were recorded and the ploidy was measured in a CD41-high gate. (A) One representative experiment is shown. (B) The mean ploidy of 4 experiments is illustrated. In all experiments, the modal ploidy increased from 16N in wt MKs to 32N in p19INK4D+/− MKs and to 64N in p19INK4D−/− MKs as shown in panel A.
p19INK4D is a direct target of AML-1 (RUNX-1) in megakaryocytes
To identify transcriptional regulators controlling p19INK4D expression, we used an in silico approach with the ChipMapper online software available at http://bio.chip.org/mapper.30 This analysis established that several AML-1 putative transcription factor–binding sites were present in the p19INK4D promoter (+2000 upstream region relative to the predicted transcriptional start). To determine whether p19INK4D was a direct target of AML-1, we first cloned 1.5 kb of the p19INK4D promoter (plucF3) upstream of the firefly luciferase gene (Figure 6A) and assayed the ability of AML-1 to induce expression in transient reporter assays in HEL cells. As shown in Figure 6B, the p19INK4D promoter displayed an induction in response to AML-1 that is increased in the presence of non—DNA-binding subunit of CBF, CBFβ. Then, a shorter promoter construct (500 bp) was generated (plucF2). Similar responses to AML-1 were obtained indicating that the most proximal 500 bases in the p19INK4D promoter are sufficient for full activity (Figure 6B).
p19INK4D is directly regulated by AML-1 (RUNX1). (A) Schematic representation of the p19INK4D human promoter region. The arrowhead represents the putative transduction start site. Black bars with diamonds designate genomic positions of the amplicons used in ChIP analysis (UCSC genomic localization http://genome.ucsc.edu/). plucF2 and plucF3 represent schematically the length of the promoter regions cloned in the pluc-mcs reporter. (B) Luciferase assay performed by transient transfection of HEL cells with the indicated micrograms of MPI vector containing AML1c and of pEF6/V5-His-TOPO vector containing CBFβ. Luciferase levels are shown as fold change relative to cells transfected with reporter construct alone. The total amount of transfected DNA was kept constant by transfection with an empty MPI vector. The histograms show 1 representative experiment of 2, each in triplicate. Error bars represent the standard deviation of triplicate. (C) Chromatin immunoprecipitation assay performed in CD34+-derived MKs (day 10 in culture) with primer sets (p19INK4DA, B, and C) directed toward AML-1–binding site predicted in silico, as well as control primer sets. Control rabbit IgG and 2 anti–AML-1 antibodies were used for immunoprecipitation. The histograms indicate relative occupancy of the AML-1–binding site in the p19INK4D promoter by AML-1. The figure illustrates 1 representative experiment using 2 different anti–AML-1 antibodies. Error bars represent the standard deviation of experiments performed on the same cell extracts with 2 different anti–AML-1 antibodies in duplicate. A similar result was obtained in a second independent experiment. (D,E) CD34+ cells were transduced at days 1 and 2 of culture with a control lentivirus and the lentivirus encoding either shRNA AML-1 or control shRNA. GFP+ cells were analyzed at day 9. (D) p19INK4D and AML-1 mRNAs were measured by QRT-PCR in GFP-positive cells. Error bars are standard deviations of the mean of 3 repeated experiments. (E) p19INK4D and AML-1 protein level was analyzed by Western blot. HDAC-1 was used as internal control of a quantity. The data illustrate 1 of 2 representative experiments that produced similar results.
p19INK4D is directly regulated by AML-1 (RUNX1). (A) Schematic representation of the p19INK4D human promoter region. The arrowhead represents the putative transduction start site. Black bars with diamonds designate genomic positions of the amplicons used in ChIP analysis (UCSC genomic localization http://genome.ucsc.edu/). plucF2 and plucF3 represent schematically the length of the promoter regions cloned in the pluc-mcs reporter. (B) Luciferase assay performed by transient transfection of HEL cells with the indicated micrograms of MPI vector containing AML1c and of pEF6/V5-His-TOPO vector containing CBFβ. Luciferase levels are shown as fold change relative to cells transfected with reporter construct alone. The total amount of transfected DNA was kept constant by transfection with an empty MPI vector. The histograms show 1 representative experiment of 2, each in triplicate. Error bars represent the standard deviation of triplicate. (C) Chromatin immunoprecipitation assay performed in CD34+-derived MKs (day 10 in culture) with primer sets (p19INK4DA, B, and C) directed toward AML-1–binding site predicted in silico, as well as control primer sets. Control rabbit IgG and 2 anti–AML-1 antibodies were used for immunoprecipitation. The histograms indicate relative occupancy of the AML-1–binding site in the p19INK4D promoter by AML-1. The figure illustrates 1 representative experiment using 2 different anti–AML-1 antibodies. Error bars represent the standard deviation of experiments performed on the same cell extracts with 2 different anti–AML-1 antibodies in duplicate. A similar result was obtained in a second independent experiment. (D,E) CD34+ cells were transduced at days 1 and 2 of culture with a control lentivirus and the lentivirus encoding either shRNA AML-1 or control shRNA. GFP+ cells were analyzed at day 9. (D) p19INK4D and AML-1 mRNAs were measured by QRT-PCR in GFP-positive cells. Error bars are standard deviations of the mean of 3 repeated experiments. (E) p19INK4D and AML-1 protein level was analyzed by Western blot. HDAC-1 was used as internal control of a quantity. The data illustrate 1 of 2 representative experiments that produced similar results.
To demonstrate that AML-1 is bound to the p19INK4D promoter in vivo, chromatin immunoprecipitation (ChIP) assays were performed using chromatin prepared from human MKs derived in culture from CD34+ cells. Quantitative PCR with primers amplifying the regions encompassing the predicted AML-1–binding sites identified (Figure 6A) revealed that AML-1 was indeed highly enriched on the p19INK4D promoter (Figure 6C).
To determine whether p19INK4D is positively or negatively regulated by AML-1, a shRNA of AML-1 was developed and vectorized in the lentivirus, and human CD34+ cells were transduced at days 1 and 2 of culture and analyzed at day 9. As shown in Figure 6E,F, the decrease in AML-1 level induced the diminution in p19INK4D level, suggesting that p19INK4D is regulated positively by AML-1 during MK differentiation.
Discussion
In the hematopoietic system, a link between G1 arrest and terminal differentiation was demonstrated. One of the best-studied lineage is the erythroid lineage in which the mitotic arrest is connected with several molecular events that lead to the inhibition of CDK2, CDK4, and CDK6 activities.31 Among these events, up-regulation of p18INK4C and p27KIP1 and down-regulation of CDK6 were reported.32 In addition, it has been demonstrated that GATA-1, the main transcription factor for the erythroid lineage, regulates not only differentiation but also the proliferation arrest by repressing c-myc and up-regulating p18INK4C and p27KIP1 probably through an indirect mechanism.32 A recent study shows that EKLF, another transcription factor also involved in erythroid differentiation, may directly regulate p18INK4C to induce mitotic arrest and differentiation.33
The megakaryocytic and erythroid differentiation programs have many similarities, including a common bipotent progenitor and the sharing of several transcription factors. However, these 2 cell lineages display marked differences during terminal differentiation. In particular, MK differentiation is characterized by a cell polyploidization that occurs through an endomitotic process. This process includes a succession of G1, S, and G2 phases followed by a mitotic phase that aborts at a late stage.1,2,34 Like in the erythroid lineage, terminal MK differentiation is also associated with a cell-cycle arrest in G1 phase and thus requires an inactivation of the CDK2, CDK4, and CDK6 activities.35 However, the different molecules and transcription factors that are involved in the cell-cycle arrest are poorly understood.
It was shown that p21CIP1 is expressed at a high level during MK differentiation,8 and it was initially suggested that p21CIP1 was necessary for the polyploidization process. Nevertheless, ectopically overexpressed p21CIP1 led to a decrease in MK ploidy suggesting that the main role of p21CIP1 was to induce an endomitotic arrest and to couple the end of polyploidization with maturation. However, MKs from p21CIP1 knockout mice display a normal ploidy.7 This observation was quite similar to what has been found in p27KIP1 knockout mice where no marked erythroid abnormalities were seen,36 although it was suggested that p27KIP1 had a key role in the erythroid lineage.18 Thus, both in the erythroid and the megakaryocytic lineages, it was likely that a redundant role was played by other molecules. During MK differentiation and among the different CDKIs, p27KIP1 is also expressed.7,8 When we studied p27KIP1 knockout mice or p21CIP1/p27KIP1 double mutant animals, we did not find any marked alteration in MK ploidization (Véronique Baccini, unpublished data, April 2003). This finding suggested that another CDKIs, especially of the INK4 family, might be involved in the regulation of the endomitotic process. We demonstrate here that, although p18INK4C variant 2 transcripts were expressed at a high level in MKs, p19INK4D plays a key role in coupling the endomitotic cycle arrest with maturation during normal MK differentiation.
The kinetics of p19INK4D expression was quite evocative of this function. We previously found by gene-profiling assays that p19INK4D expression was increasing linearly during ploidization.12 Such a result was not found for other CDKIs belonging to either the Cip/Kip or INK4 families, whether the analyses were performed by microarrays12 or by quantitative real-time RT-PCR (Figure 1C). In addition, our data (Figure 1D) show that the p19INK4D protein also increased with ploidization, except at the 4N stage. It is demonstrated that, in fibroblasts, the p19INK4D protein level oscillates during cell cycle due to an ubiquitin/proteasome mechanism.37 As the 4N MK cell population is a heterogeneous mixture of both precursor cells in mitosis and endomitotic cells, it remained possible that a degradation of the p19INK4D protein occurring during mitosis could explain the differences seen between the mRNA and protein levels. Notably, MKs can arrest at any ploidization stage during differentiation and can undergo maturation even at the 2N stage. Indeed, maturing 2N MKs are observed in human pathologies,38 during embryogenesis,39 or in in vitro cultures. We thus sorted only the 2N MKs at different maturation levels based on the expression of differentiation markers such as CD34, CD41, and CD42, and we measured the p19INK4D mRNA level in the various fractions. We observed that p19INK4D transcripts increased with differentiation and independently of ploidization. However, a plausible explanation might be that in the immature MKs we detect a low level of p19INK4D because these cells are still cycling, while when a high level p19INK4D is accumulated, cells exit cell cycle and enter terminal differentiation.
Using a knockdown strategy of p19INK4D with 2 different shRNAs and an ectopic p19INK4D overexpression, we have directly demonstrated that p19INK4D regulates the endomitotic cell-cycle arrest. Indeed, overexpression of p19INK4D led to a mitotic or endomitotic cell cycle arrest, as previously observed for p21CIP1,7 p16INK4A,11 and p27KIP1 (Véronique Baccini, unpublished data, December 2003). This is clearly related to the capacity of these molecules to inhibit CDK4, CDK6, and eventually CDK2 after their overexpression independently of the cell system. However, we have also shown that an inhibition of approximately 80% of p19INK4D mRNA by 2 shRNAs affecting the activity of cyclin D/CDK46 complex resulted in a moderate but significant increase in MK ploidy, even when the shRNAs were transduced during MK ploidization. The use of 2 different shRNAs producing similar consequences excludes the possibility of an off-target effect of the shRNA. Thus, the result indicates that p19INK4D plays an intrinsic role in the mechanism of the endomitotic arrest. Such a conclusion was not drawn for other CDKIs, since a decrease of their expression did not result in an increase in MK ploidy level. However, most approaches were performed in knockout mice but not by shRNA strategy, and it cannot be excluded that compensation by redundant CDKIs takes place when using a KO strategy. However, we could confirm our results obtained by shRNA in human MKs in p19INK4D KO mice. Indeed a marked and dose-dependent increase in MK ploidy was observed in p19INK4D KO mice: one modal ploidy class increase in p19INK4D+/− MKs and 2 modal ploidy class increases in p19INK4D−/− MKs.
Moreover, p19INK4D down-regulation or enforced expression leads to a change in maturation. p19INK4D knockdown was associated with a decrease in CD42 expression, but not in CD41 that is an earlier differentiation marker. This slight decrease was found in all ploidy classes when p19INK4D was down-regulated in MK precursors and only in polyploid MKs when p19INK4D knockdown was performed later on during differentiation. However, mature MKs in which p19INK4D was knocked down were capable of shedding platelets like control MKs, and the kinetics experiments (data not shown) confirmed that p19INK4D inhibition induced only a delay but not a blockage of maturation. The selective effect of p19INK4D inhibition on the kinetics of differentiation is consistent with the concept that p19INK4D has no direct effect on maturation, but couples mitotic arrest with differentiation. When p19INK4D is knocked down early during differentiation, the mitotic arrest is delayed in all ploidy classes, whereas when it is performed later, the delayed differentiation concerns only cells that have not yet finished their ploidization. This hypothesis is supported by the fact that p19INK4D knockout mice have normal platelet counts in basal conditions40 (and personal communication). In future experiments, it will be important to determine whether p19INK4D knockout would have an impact in stress conditions.
Knowing the important role of coupling cell-cycle arrest with terminal differentiation, it seems likely that the transcription factors regulating terminal differentiation may also induce the G1 arrest. In the erythroid lineage, GATA-1 and EKLF induce such a cell-cycle arrest by interacting directly with c-myc32 and p18INK4C,33 respectively. During megakaryopoiesis, knockout of GATA-141 or AML-142 or FLI-143 lead to a maturation blockage with a marked defect in MK polyploidization. Recently, it was reported that GATA-1–deficient murine MKs express a low level of cyclin D1 and a high level of p16INK4A, but it was also shown that only cyclin D1 was a direct target gene for GATA-1.11 These data may explain how GATA-1 regulates ploidization, but they do not indicate whether GATA-1 induces a cell-cycle arrest in MKs as described in erythroblasts. The p19INK4D promoter region contains multiple binding sites for different transcriptional factors, such Sp1, AP-2, CCAAT, SRY, GATA-3, and E2F.44 Whereas the GATA-binding site is not conserved between vertebrates, we identified by in silico analysis of the p19INK4D promoter region 2 conserved consensus binding sites for AML-1 and detected the in vivo binding of AML-1 to 1 of the 2 sites during MK differentiation. This specific positive regulation of p19INK4D transcription by AML-1 during MK differentiation could couple the endomitotic arrest with terminal differentiation. It is not excluded that a regulation loop exists between p19INK4D, cyclin D/CDK6, and AML-1 in megakaryocytes. Indeed it has been reported that CDK6 blocks the myeloid differentiation by inhibiting the activity of AML-1.45 In addition, CDK6 expression decreases during erythroid differentiation independently of the cell cycle.31 Thus, if a similar down-regulation of CDK6 occurs during megakaryopoiesis, it will be associated with an increase in AML-1 activity leading both to a cell-cycle arrest and MK differentiation. In fact, AML-1 plays an important role in megakaryocytopoiesis because its conditional knockout in the mouse42 or its haploinsufficiency in a human disease46 induce a thrombocytopenia. It has also been suggested that AML-1 and GATA-1 act in combination through a physical interaction during megakaryocytopoiesis.47 Megakaryocyte AML-1 target genes have not been well characterized, but p19INK4D appears to be one of the first AML-1 target genes directly implicated in megakaryocytopoiesis.
In summary, our study shows that p19INK4D plays an important role in inhibiting the endomitotic cell cycle. In megakaryocytes, expression of p19INK4D is positively regulated by AML-1, which could explain the coupling of cell-cycle arrest with maturation. Future characterization of other AML-1 target genes might provide important insights in megakaryocytopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to F. Wendling for critical reading of the paper and helpful suggestions; to K. Brewery (Tokyo, Japan) for the gift of human recombinant thrombopoietin; to A. Dubart-Kupperschmitt (Institut Cochin, Department of Hematology, Paris, France) for HIV-derived SIN lentiviral vector pTRIPΔU3-EF1α-IRES-GFP; to E. Cánepa (Facultad de Ciencias Exactas y Naturales-UBA, Ciudad Universitaria Pabellón II Piso, Ciudad de Buenos Aires, Argentina) for pSG5p19plasmid; and to O. Albagli (Inserm U790, France).
This work was supported by grants from Association de la recherche contre le Cancer (W.V.) and the Agence Nationale de la Recherche (ANR contrat blanc, and an ANR-maladies rares). R.G. was supported by a fellowship from the Académie Nationale de Médecine, and D.B. was supported by a postdoctoral fellowship from ANR.
Authorship
Contribution: L.G. and R.G. performed experiments and analyzed data; D.B., V.C.-L., C.L., R.F., and N.D. performed experiments; F.L. performed sorting experiments and analyzed data; W.V. designed the work and wrote the paper; H.R. designed and supervised the experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hana Raslova, PhD, Institut Gustave Roussy, Inserm U790, 39 rue Camille Desmoulins, 94805 Villejuif, France; e-mail: hraslova@igr.fr.
References
Author notes
*L.G. and R.G. contributed equally to this work.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal