Abstract
Entry into the cell cycle is mediated by cyclin-dependent kinase 4/6 (CDK4/6) activation, followed by CDK2 activation. We found that pharmacologic inhibition of the Flt3 internal tandem duplication (ITD), a mutated receptor tyrosine kinase commonly found in patients with acute myelogenous leukemia (AML), led to the down-regulation of cyclin D2 and D3 followed by retinoblastoma protein (pRb) dephosphorylation and G1 cell-cycle arrest. This implicated the D-cyclin-CDK4/6 complex as a downstream effector of Flt3 ITD signaling. Indeed, single-agent PD0332991, a selective CDK4/6 inhibitor, caused sustained cell-cycle arrest in Flt3 ITD AML cell lines and prolonged survival in an in vivo model of Flt3 ITD AML. PD0332991 caused an initial cell-cycle arrest in well-established Flt3 wild-type (wt) AML cell lines, but this was overcome by down-regulation of p27Kip and reactivation of CDK2. This acquired resistance was not observed in a Flt3 ITD and a Flt3 wt sample from a patient with primary AML. In summary, the mechanism of cell-cycle arrest after treatment of Flt3 ITD AML with a Flt3 inhibitor involves down-regulation of cyclin D2 and D3. As such, CDK4/6 can be a therapeutic target in Flt3 ITD AML but also in primary Flt3 wt AML. Finally, acquired resistance to CDK4/6 inhibition can arise through activation CDK2.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease at both the cytogenetic and molecular levels. Many of these alterations have prognostic impact on clinical outcome, and some also dictate therapeutic approaches.1 Approximately 25% of patients express a constitutively active Flt3 internal tandem duplication (ITD), a mutant form of the receptor tyrosine kinase Flt3. The Flt3 ITD is a marker for poor prognosis.2,3 The availability of cell lines expressing Flt3 ITD as well as Flt3 inhibitors has made it possible to study the signaling pathways activated by mutant Flt3. Flt3 ITD signaling regulates a multitude of proteins involved in proliferation, differentiation, and survival, reflecting a complex signaling network that promotes oncogenesis.4–10 It has also been reported that the pharmacologic inhibition of Flt3 ITD leads to cell-cycle arrest, but the underlying molecular mechanism and functional significance of this observation remains unclear.11,12
Progression through the cell cycle from G1/G0 to S, G2, and M phases is initiated by cyclin-dependent kinase 4 (CDK4) and the highly homologous enzyme CDK6. CDK4 and CDK6 form a complex with one of their activating subunits, which are the cyclins D1, D2, and D3.13–15 The activity of CDK4/6 is negatively regulated by the INK4 proteins.16 The D-cyclin/CDK4/6 complex phosphorylates the retinoblastoma protein (pRb) and the related proteins p107 and p130,13,15,17 as well as Smad3.18 In addition to being catalytically active, the cyclin D-CDK4/6 complexes can also sequester the cell-cycle inhibitors p21Cip1 and p27Kip1. This promotes the activation of the Cyclin E-CDK2 complex,19 which further phosphorylates pRb. Hyperphosphorylated pRb loses its inhibitory effect on the E2F transcription factor family. As a result of E2F activation, the transcription of genes promoting entry into S phase is initiated.20 Here we show that Flt3 ITD activates the cyclin D, CDK4/6, INK4, pRb pathway by up-regulating cyclin D2 and D3 gene expression and that this pathway may constitute a drug target in AML.
Materials and methods
Compounds
THRX-165724 was obtained from Theravance (South San Francisco, CA). PD0332991 was obtained from Pfizer (La Jolla, CA). For in vitro studies, both compounds were dissolved in dimethyl sulfoxide (DMSO; 10 mM stock solutions) and stored at −20°C.
Ribonuclease protection assay
The ribonuclease protection assay was performed with the RiboQuant hCYC-1 Multi-Probe Template Set (BD Pharmingen, San Diego, CA) according to the manufacturer's instructions.
Cell culture
The THP-1 and U937 cell lines were obtained from the American Type Culture Collection (Manassas, VA). The MV4-11 and MOLM13 cell lines were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). The cell lines were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 units/mL streptomycin. Frozen samples from patients with AML were obtained from the Leukemia Tissue Bank at The Ohio State University. Fresh CD34(;) cells were purchased from AllCells (Emeryville, CA). All studies with human specimens were performed with the approval from The Ohio State University Institutional Review Board.
Immunoprecipitation and Western blot
MV4-11 cells (1.0 × 107) were incubated in 5 mL RPMI 1640 medium plus the indicated concentration (Figure 1B) of THRX-165724 or PD0332991 (30-minute incubation). The cells were lysed in 0.75 mL of modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, and protease inhibitor mixture [Roche Diagnostics, Indianapolis, IN]). The lysates were incubated with a Flt3 antibody (S-18; Santa Cruz Biotechnology, Santa Cruz, CA) and protein G beads (Sigma) overnight at 4°C. The immunocomplexes were recovered by centrifugation, washed with RIPA buffer, boiled in Laemmli sample buffer, and resolved by sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis (PAGE). The proteins were transferred to a nitrocellulose membrane (Santa Cruz Biotechnology), blocked with phosphate-buffered saline (PBS), 0.1% Tween 20, and 3% bovine serum albumin, and probed with an anti-phosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY) overnight at 4°C. Subsequently, the blot was washed with PBS and 0.1% Tween 20, and specific antibody binding was detected with a horseradish peroxidase-coupled secondary antibody, followed by enhanced chemiluminescence (ECL; GE Healthcare, Chalfont St. Giles, United Kingdom) and exposure to film. To reprobe the blot, the primary antibody was stripped with ImmunoPure IgG Elution Buffer (Pierce, Rockford, IL).
For Western blots not involving immunoprecipitation, cells were centrifuged, the pellet was resuspended in PBS (10 μL/1 × 105 cells), and an equal volume of 2× Laemmli sample buffer was added to lyse the cells. The lysate was heated at 95°C for 10 minutes followed by centrifugation at 13 200 rpm. Typically, 20 μl of lysate were loaded on a gel for SDS/PAGE. The proteins were transferred to a nitrocellulose membrane as described above. For immunoblotting, the following antibodies were used: γ-tubulin (Santa Cruz Biotechnology), β-actin (Sigma, St Louis, MO), cyclin D2 (Cell Signaling Technology, Danvers, MA), cyclin D3 (Cell Signaling Technology), pRb (4H1; Cell Signaling Technology), CDK2 (Santa Cruz Biotechnology), CDK4 (Cell Signaling Technology), CDK6 (Cell Signaling Technology), cyclin E (M-20; Santa Cruz Biotechnology), p27Kip1 (Cell Signaling Technology), Grb2 (Cell Signaling Technology), Foxo3a (Cell Signaling Technology), and phospho-Foxo3a (Thr32) (Cell Signaling Technology). By using the pRb signal on the film of a Western blot, densitometry was used to quantify the percentage of dephosphorylated pRb versus total pRb.
Kinase assay
Cells were treated with DMSO or PD0332991 (500 nM) for 24 or 96 hours. The cells were centrifuged, washed with ice-cold PBS, and then lysed for 10 minutes in ice-cold lysis buffer (20 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, and 0.5% Nonidet P-40) containing a protease inhibitor cocktail (Roche Diagnostics) and 1 mM sodium orthovanadate, 10 mM sodium fluoride, and 10 mM β-glycerophosphate. Lysates were cleared by centrifugation at 13 000g for 5 minutes at 4°C. The protein concentration of the cleared lysates was determined by the bicinchoninic acid protein assay (Pierce). For immunoprecipitation, 200 μg of total protein from each lysate was brought to 600 μL with lysis buffer. Antibodies were added to each lysate at a concentration of 1 μg. The immunoprecipitation was performed at 4°C for 1 hour. Immunocomplexes were captured by protein G Agarose beads (Invitrogen). Agarose beads were collected and washed 3 times with kinase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, and 1 mM dithiothreitol). CDK2 immunoprecipitates were incubated in 30 μL of kinase buffer with 5 μg histone H1 and 10 μCi of [γ-32P]ATP (3000 Ci/mmol; GE Healthcare) for 30 minutes at 30°C. After addition of Laemmli sample buffer, samples were analyzed by SDS-PAGE followed by Western Blot transfer. Phosphorylated histone H1 was visualized by autoradiography.
Cell proliferation and viability
Single agent effects on cell proliferation and viability were assessed by using the 3-(4,5-dimethylthaizol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Roche Diagnostics). The drug combination studies were also performed using the MTT assay. The analysis was executed with an in-house spreadsheet according to the median-effect method introduced by Chou and Talalay21 and as described previously.22
Cell cycle analysis
Logarithmically growing cells were incubated with THRX-165724, PD0223991, or DMSO (vehicle control) for the indicated periods of time. The cells were washed in ice-cold PBS and fixed in 70% ethanol. Subsequently, the cells were stained with propidium iodide staining buffer (0.05 mg/mL propidium iodide [Sigma-Aldrich] and 0.1% RNase [Invitrogen]) for 30 minutes at room temperature. The analysis was performed by flow cytometry.
Real-time PCR
cDNA was synthesized from 500 ng of total RNA using Superscript III reverse transcriptase (Invitrogen) with oligo dT and p27Kip1 gene-specific primers. cDNA products were quantitated by using SYBR green fluorescence on an AB7000 thermal cycler (Applied Biosystems, Foster City, CA). Primer sets for mRNA quantitation included 2 different exon spanning primer sets. Cycle threshold results were normalized to β-Actin gene expression. p27Kip1 primer pair A: 913 forward, GGAATAAGGAAGCGACCTGCA; 1026 reverse, CGTCTGCTCCACAGAACCG. Primer pair B: 1029 forward, CAAGAAGCCTGGCCTCAGAAG; 1176 reverse, CCATTCCATGAAGTCAGCGAT. Actin primer pair: forward, CCTGGCACCCAGCACAAT; reverse, GCCGATCCACACGGAGTACT.
Phospho-pRb analysis in primary bone marrow cells
Bone marrow cells (5 × 106) per mouse were treated with Fix Buffer I (BD Biosciences) for 10 minutes at 37°C and permeabilized with Perm Buffer III (BD Biosciences) for 30 minutes on ice. After washing the cells twice with staining buffer, they were stained with phycoerythrin (PE)-mouse anti-phospho pRb (pSer780) (BD Biosciences) and allophycocyanin (APC)–mouse antihuman CD45 (BD Biosciences). PE- and APC-conjugated mouse isotype antibodies served as negative controls in the subsequent analysis by flow cytometry.
Immunohistochemistry
Sections of formalin-fixed, decalcified, paraffin-embedded tissues were mounted on Plus slides (Fisher Scientific, Pittsburgh, PA). Slides were deparaffinized and antigen retrieval (TRS solution [DAKO North America, Carpinteria, CA], 20 minutes in steamer, 20 minutes cooling to room temperature) was performed. Peroxidase and protein blocking was carried out using blocking solutions from DAKO as directed. Primary antibodies were applied for 30 minutes at dilutions of 1:100 for phospho-pRb (pSer807/11) and 1:250 for human CD45. Appropriate secondary antibodies were from Vector Elite kits. They were incubated with the sections for 30 minutes. The chromagen was 3,3[prime[-diaminobenzidine (DAB), and sections were lightly counterstained with hematoxylin. After dehydration, coverslips were applied using a xylene-based mounting medium.
Animal survival studies
All mouse experiments were performed with the approval of the Institutional Animal Care and Use Committees at The Ohio State University. Twenty nonobese diabetic (NOD)/severely compromised immunodeficient (SCID) mice (age, 12 weeks) were conditioned with 300 rads of total body irradiation and immediately injected with 6 × 106 log-phase MOLM-13 cells via the lateral tail vein. The mice were divided randomly into 2 groups and dosed once daily with PD0332991 (150 mg/kg, 10 μL/g) or an equivalent volume of vehicle (lactic acid buffer, pH 4.0). Dosing was performed by oral gavage starting on day 6 after engraftment. Survival times were compared with the logrank test using Prism (GraphPad Software, San Diego, CA).
Results
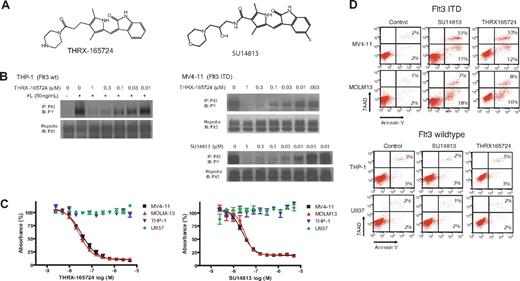
THRX-165724 and SU14813 Were Potent Inhibitors of Flt3
To study Flt3 ITD signaling, we chose 4 AML cell lines as a model system: THP-1 and U937, which express Flt3 wild type (wt), and MV4-11 and MOLM13, which express Flt3 ITD. THRX-16572423 and SU14813,24 inhibitors of the platelet-derived growth factor receptor family, which includes Flt3, were used as pharmacologic tools (Figure 1A).
THRX-165724 and SU14813 are potent inhibitors of Flt3 autophosphorylation and induce apoptosis in AML cells with Flt3 ITD. (A) Chemical structure of THRX-165724 and SU14813. (B) Inhibition of Flt3 autophosphorylation with THRX-165724 and SU14813. MV4-11 cells were incubated with increasing concentrations of THRX-165724 and SU14813 for 30 minutes. THP-1 cells were incubated with increasing concentrations of THRX-165724 and Flt3 wild-type autophosphorylation was stimulated with FL (50 ng/mL for 5 minutes). Flt3 phosphorylation status was determined by Flt3 immunoprecipitation followed by SDS-PAGE and Western blot with an anti-phosphotyrosine antibody. (C) MTT assay. MV4-11, MOLM13, THP-1, and U937 cells were incubated with increasing concentrations of THRX-165724 and SU14813 for 72 hours. Cell viability and proliferation was assessed using the MTT assay. (D) Induction of apoptosis. MV4-11, MOLM13, THP-1, and U937 cells were incubated for 72 hours with THRX-165724 (300 nM) or SU14813 (100 nM). Apoptosis was assessed by staining with Annexin V-PE and 7-AAD. The percentage of cells in the upper right quadrant denote cells that stain positive for Annexin V and 7-AAD. The cells in the lower right quadrant stain positive for Annexin V only.
THRX-165724 and SU14813 are potent inhibitors of Flt3 autophosphorylation and induce apoptosis in AML cells with Flt3 ITD. (A) Chemical structure of THRX-165724 and SU14813. (B) Inhibition of Flt3 autophosphorylation with THRX-165724 and SU14813. MV4-11 cells were incubated with increasing concentrations of THRX-165724 and SU14813 for 30 minutes. THP-1 cells were incubated with increasing concentrations of THRX-165724 and Flt3 wild-type autophosphorylation was stimulated with FL (50 ng/mL for 5 minutes). Flt3 phosphorylation status was determined by Flt3 immunoprecipitation followed by SDS-PAGE and Western blot with an anti-phosphotyrosine antibody. (C) MTT assay. MV4-11, MOLM13, THP-1, and U937 cells were incubated with increasing concentrations of THRX-165724 and SU14813 for 72 hours. Cell viability and proliferation was assessed using the MTT assay. (D) Induction of apoptosis. MV4-11, MOLM13, THP-1, and U937 cells were incubated for 72 hours with THRX-165724 (300 nM) or SU14813 (100 nM). Apoptosis was assessed by staining with Annexin V-PE and 7-AAD. The percentage of cells in the upper right quadrant denote cells that stain positive for Annexin V and 7-AAD. The cells in the lower right quadrant stain positive for Annexin V only.
To characterize the activity and potency of THRX-165724, we first investigated the compound's effect on Flt3 autophosphorylation. AML cell lines expressing Flt3 wt (THP-1) and Flt3 ITD (MV4-11) were incubated with increasing concentrations of THRX-165724. In THP-1 cells, Flt3 autophosphorylation was stimulated by the addition of Flt3 ligand (FL). In MV4-11, Flt3 ITD is constitutively phosphorylated and therefore did not require activation with FL. Flt3 was immunoprecipitated, and an immunoblot with an anti-phosphotyrosine antibody was performed. The experiment revealed that THRX-165724 is an equipotent inhibitor of Flt3 wt and Flt3 ITD autophosphorylation with an half-maximal inhibitory concentration (IC50) of approximately 30 nM (Figure 1B). Using the MV4-11 cell line, we found that the IC50 of SU14813-mediated inhibition of Flt3 autophosphorylation in this assay was approximately 10 nM (Figure 1B). An MTT assay was performed to investigate the effect of THRX-165724 and SU14813 on proliferation and cell viability. Both compounds affected MV4-11 and MOLM13, the 2 cell lines with Flt3 ITD. The IC50 in this assay was 50-100 nM for THRX-165724 and 10-20 nM for SU14813. In contrast, THP-1 and U937 cells, which express Flt3 wt, were not affected by either THRX-165724 or SU14813 up to 5 μM (Figure 1C). The effect caused by THRX-165724 and SU14813 is a result of apoptosis because MOLM13 and MV4-11 cells (Flt3 ITD) became positive for the apoptosis marker Annexin V, whereas U937 and THP-1 cells (Flt3 wt) did not (Figure 1D). To learn whether the induction of apoptosis by THRX-165724 and SU14813 is reversible, we performed a drug wash-out experiment. This experiment revealed that the removal of THRX-165724 or SU14813 after a 24-hour incubation period rescues the cells from cell death (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In summary, these experiments demonstrate that THRX-165724 and SU14813 are potent inhibitors of Flt3 autophosphorylation that can induce programmed cell death in AML cells with Flt3 ITD.
The inhibition of Flt3 ITD led to the down-regulation of cyclin D2 and D3
It has been reported that the inhibition of Flt3 ITD signaling affects the cell cycle before inducing apoptosis.11,12 We investigated the effect of THRX-165724 and SU14813 on the cell cycle by incubating MOLM13, MV4-11, THP-1, and U937 cells with the inhibitors for 24 hours. THRX-165724 as well as SU14813 induced a specific G1 cell-cycle arrest in MOLM13 and MV4-11 (Flt3 ITD) but not in THP-1 and U937 (Flt3 wt) (Figure 2A). The cell-cycle arrest was reversible. MOLM13 and MV4-11 cells were able to re-enter the cell cycle after an incubation with THRX-165724 and SU14813 for 24 hours followed by a wash-out of the inhibitors (Figure S2). To gain insight into the cell-cycle genes controlled by Flt3 ITD signaling, we isolated total RNA from MV4-11 cells that had been treated with THRX-165724 or DMSO (vehicle control) for 3 hours. This relatively short incubation time is expected to identify changes in the expression of genes that are directly downstream of Flt3 ITD signaling. A ribonuclease protection assay was performed using various sets of probes for genes involved in proliferation and the cell cycle. Using this approach we observed significant down-regulation of cyclin D2 and D3 transcript levels after treating the cells with THRX-165724 (Figure 2B).
The inhibition of Flt3 ITD induces a G1 cell-cycle arrest and down-regulation of D-cyclins. (A) MV4-11, MOLM13, THP-1, and U937 cells were treated with THRX-165724 (300 nM) or SU14813 (100 nM) for 24 hours. After propidium iodide staining, cell-cycle analysis was performed by flow cytometry. (B) Ribonuclease protection assay. MV4-11 cells were treated with THRX-165724 (300 nM) or with vehicle (DMSO) for 3 hours. Changes in mRNA levels of a set of cyclin genes was determined by ribonuclease protection assay (RPA) P indicates probe). (C) Western blot. MV4-11 cells were treated with THRX-165724 for 0, 1, 2, 4, 8, and 12 hours. Lysates were immunoblotted with the indicated antibodies. Probing with anti-tubulin was used to ensure equal loading. (D) Western blot. MOLM13, MV4-11, U937, and THP-1 cells were treated with THRX-165724 (300 nM) or DMSO (vehicle control) as well as SU14813 (100 nM) or DMSO (vehicle control) for 16 hours. A Western blot was performed with cell lysates probing for cyclin D2, D3, and pRb. Actin served as marker for equal loading.
The inhibition of Flt3 ITD induces a G1 cell-cycle arrest and down-regulation of D-cyclins. (A) MV4-11, MOLM13, THP-1, and U937 cells were treated with THRX-165724 (300 nM) or SU14813 (100 nM) for 24 hours. After propidium iodide staining, cell-cycle analysis was performed by flow cytometry. (B) Ribonuclease protection assay. MV4-11 cells were treated with THRX-165724 (300 nM) or with vehicle (DMSO) for 3 hours. Changes in mRNA levels of a set of cyclin genes was determined by ribonuclease protection assay (RPA) P indicates probe). (C) Western blot. MV4-11 cells were treated with THRX-165724 for 0, 1, 2, 4, 8, and 12 hours. Lysates were immunoblotted with the indicated antibodies. Probing with anti-tubulin was used to ensure equal loading. (D) Western blot. MOLM13, MV4-11, U937, and THP-1 cells were treated with THRX-165724 (300 nM) or DMSO (vehicle control) as well as SU14813 (100 nM) or DMSO (vehicle control) for 16 hours. A Western blot was performed with cell lysates probing for cyclin D2, D3, and pRb. Actin served as marker for equal loading.
As shown by Western blotting, the change in gene expression level correlates with a change in cyclin D2 and D3 protein level (Figure 2C). After 4 hours of treatment with THRX-165724, cyclin D2 and D3 are greatly reduced. Cyclin D1 protein could not be detected by Western blot. It follows that the reduction of cyclin D2 and D3 protein leads to the reduction of CDK4/6 activity.25 pRb is known to be hyperphosphorylated when CDK4/6 is active, so the loss of CDK4/6 activity leads to the dephosphorylation of pRb,25 which is reflected in a shift of the apparent molecular weight of the protein. We found that at 4 hours after the start of THRX-165724 treatment, as cyclin D2 and D3 levels decrease, pRb phosphorylation also decreases. At 12 hours, when cyclin D2 and D3 levels are very low, pRb is almost completely dephosphorylated. The THRX-165724 mediated down-regulation of cyclin D2 and D3 as well as the dephosphorylation of pRb was also observed in Flt3 ITD MOLM13 cells (Figure S3). THRX-165724 had no effect on the D-cyclin protein level and pRb phosphorylation status in the Flt3 wt cell lines THP-1 and U937 (Figure 2D). Like THRX-165724, the second Flt3 inhibitor, SU14813, induced the down-regulation of cyclin D2 and D3 as well as pRb dephosphorylation in MV4-11 and MOLM13 (Flt3 ITD) but not in THP-1 and U937 (Flt3 wt) (Figure 2D). These data suggest that CDK4/6 is activated by Flt3 ITD signaling through the up-regulation of cyclin D2 and D3 and that inhibition of the Flt3 ITD inhibits activation of CDK4/6.
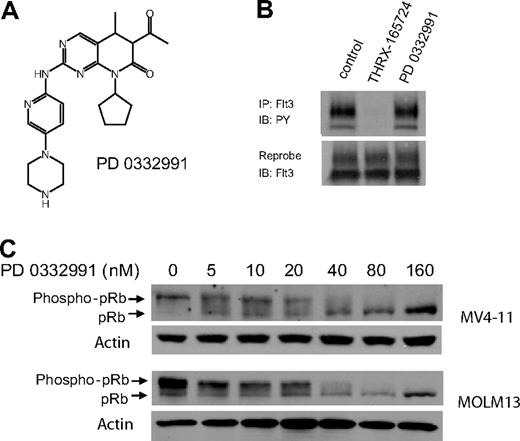
PD0332991 was a potent CDK4/6 inhibitor
To evaluate the role of CDK4/6 in the AML cell lines, we used PD0332991, a small molecule inhibitor of CDK4/6 that is currently in phase II clinical trials (Figure 3A). PD0332991 has little or no activity against a panel of 30 kinases, including CDK2, suggesting that this inhibitor is highly selective for CDK4/6.26,27 It is noteworthy that we found 500 nM PD0332991 to have no effect on Flt3 ITD autophosphorylation, which makes this compound a suitable tool to study CDK4/6 in our AML model system (Figure 3B). We incubated Flt3 ITD+, MV4-11, and MOLM13 cells with increasing concentrations of PD0332991. A Western blot of cell lysates showed decreasing pRb phosphorylation, confirming CDK4/6 inhibition by PD0332991 (Figure 3C). The IC50 of PD0332991 in this assay in both cell lines was 30 nM.
PD0332991 is a potent inhibitor of CDK4/6 but does not inhibit Flt3 ITD. (A) Structure of PD0332991. (B) MV4-11 cells were incubated with DMSO (vehicle control), THRX-165724 (500 nM), and PD0332991 (500 nM) for 30 minutes. The cells were lysed, Flt3 ITD was immunoprecipitated, and an immunoblot was performed with an anti-phosphotyrosine antibody. The blot was subsequently stripped and reprobed with anti-Flt3. (C) Western blot. MV4-11 and MOLM13 cells were incubated with increasing concentrations of PD0332991 for 16 hours. Lysates were resolved by SDS-PAGE followed by immunoblotting with anti-pRb.
PD0332991 is a potent inhibitor of CDK4/6 but does not inhibit Flt3 ITD. (A) Structure of PD0332991. (B) MV4-11 cells were incubated with DMSO (vehicle control), THRX-165724 (500 nM), and PD0332991 (500 nM) for 30 minutes. The cells were lysed, Flt3 ITD was immunoprecipitated, and an immunoblot was performed with an anti-phosphotyrosine antibody. The blot was subsequently stripped and reprobed with anti-Flt3. (C) Western blot. MV4-11 and MOLM13 cells were incubated with increasing concentrations of PD0332991 for 16 hours. Lysates were resolved by SDS-PAGE followed by immunoblotting with anti-pRb.
We next tested the effect of PD0332991 on the cell cycle of our model cell lines after incubation periods of 24 and 120 hours. Figure 4A demonstrates that MOLM13 cells (Flt3 ITD positive) are strongly arrested in G1 at 24 and 120 hours. Furthermore, a sub-G1 peak at 120 hours suggests that some cells are undergoing apoptosis. In contrast, U937 cells (Flt3 wt) were arrested in G1 at the 24-hour time point, but these cells had re-entered the cell cycle after 120 hours. Like MOLM13, the Flt3 ITD positive MV4-11 cell line was arrested in G1 at 24 and 120 hours whereas Flt3 wt THP-1 cells were arrested in G1 at the 24-hour time point but had re-entered the cell cycle at 120 hours (Figure 4B). However, despite re-entering the cell cycle, PD0332991 treated THP-1 and U937 were still showing significantly fewer cells in S and G2 of the cell cycle than the respective control cells. As expected, the effect of PD0332991 on the cell cycle also led to an effect on proliferation (Figure 4C). MOLM13 and MV4-11 cells incubated with PD0332991 as a single agent for 8 days decreased in cell number, and although THP-1 and U937 cell numbers increased, the increase was considerably smaller than for the same cells incubated with the vehicle control. Hence, the inhibition of entry into the cell cycle and proliferation in these well-established Flt3 wt AML cell lines is only partial, consistent with an acquired mechanism of resistance to PD0332991 after prolonged exposure as a single agent.
PD0332991 affects entry into the cell cycle and proliferation of AML model cell lines. (A) Cell-cycle analysis. Flow cytometric analysis of MOLM13 and U937 cells treated with DMSO (vehicle control) for 120 hours and PD0332991 (500 nM) for 24 and 120 hours and stained with propidium iodide. (B) Bar graph representation of cell-cycle analysis of MOLM13, MV4-11, THP-1, and U937 cells treated for 24 (□) and 120 hours (▒) with PD0332991 (500 nM). The cells were stained with propidium iodide and analyzed by flow cytometry. The percentage of cells in S/G2 compared with the control cells, which were normalized to 100% (treated with DMSO for 24 and 120 hours, respectively), was plotted (mean ± SE of 3 independent experiments). (C) Proliferation assay. MV4-11, MOLM13, THP-1, and U937 cells were plated in the presence of DMSO (vehicle control) or PD0332991 (500 nM) at a density of 0.5 × 106 / mL. The cells were counted every 2 days, and the cell number was adjusted to 0.5 × 106/ mL (mean ± SE of 3 independent experiments). (D) MTT assay. MV4-11, MOLM13, U937, THP-1 cells, as well as primary patient blasts from 2 patients (Pt 17 [Flt3 ITD] and Pt 75 [Flt3 wt]) were incubated in triplicate for 3 days with DMSO (control), PD0332991 (500 nM) (░), SU14813 (30 nM for cell lines and 100 nM for patient samples) (▒), or PD0332991 (500 nM) plus SU14813 (30 nM for cell lines and 100 nM for patient samples) (▓). The DMSO control is normalized to 100% (mean ± SE of 3 independent experiments for the cell lines and one experiment with triplicate data points for the patient samples). (E) Apoptosis assay. MV4-11, MOLM13, THP-1, and U937 cells were incubated for 3 days with DMSO (control), PD0332911 (250 nM) (░), SU14813 (100 nM) (▒), or PD0332991 (250 nM) plus SU14813 (100 nM) (▓). The DMSO control is normalized to 100%. The cells were stained with Annexin V-PE and 7-AAD followed by flow cytometric analysis (see Figure S4 for primary data).
PD0332991 affects entry into the cell cycle and proliferation of AML model cell lines. (A) Cell-cycle analysis. Flow cytometric analysis of MOLM13 and U937 cells treated with DMSO (vehicle control) for 120 hours and PD0332991 (500 nM) for 24 and 120 hours and stained with propidium iodide. (B) Bar graph representation of cell-cycle analysis of MOLM13, MV4-11, THP-1, and U937 cells treated for 24 (□) and 120 hours (▒) with PD0332991 (500 nM). The cells were stained with propidium iodide and analyzed by flow cytometry. The percentage of cells in S/G2 compared with the control cells, which were normalized to 100% (treated with DMSO for 24 and 120 hours, respectively), was plotted (mean ± SE of 3 independent experiments). (C) Proliferation assay. MV4-11, MOLM13, THP-1, and U937 cells were plated in the presence of DMSO (vehicle control) or PD0332991 (500 nM) at a density of 0.5 × 106 / mL. The cells were counted every 2 days, and the cell number was adjusted to 0.5 × 106/ mL (mean ± SE of 3 independent experiments). (D) MTT assay. MV4-11, MOLM13, U937, THP-1 cells, as well as primary patient blasts from 2 patients (Pt 17 [Flt3 ITD] and Pt 75 [Flt3 wt]) were incubated in triplicate for 3 days with DMSO (control), PD0332991 (500 nM) (░), SU14813 (30 nM for cell lines and 100 nM for patient samples) (▒), or PD0332991 (500 nM) plus SU14813 (30 nM for cell lines and 100 nM for patient samples) (▓). The DMSO control is normalized to 100% (mean ± SE of 3 independent experiments for the cell lines and one experiment with triplicate data points for the patient samples). (E) Apoptosis assay. MV4-11, MOLM13, THP-1, and U937 cells were incubated for 3 days with DMSO (control), PD0332911 (250 nM) (░), SU14813 (100 nM) (▒), or PD0332991 (250 nM) plus SU14813 (100 nM) (▓). The DMSO control is normalized to 100%. The cells were stained with Annexin V-PE and 7-AAD followed by flow cytometric analysis (see Figure S4 for primary data).
We next tested whether treatment of the 4 AML model cell lines with PD0332991 can enhance the effect of the Flt3 inhibitor SU14813. In an MTT assay, the 4 AML cell lines as well as primary blasts from patient 17 (Flt3 ITD) and patient 75 (Flt3 wt) were incubated for 3 days with DMSO (vehicle control), PD0332991, SU14813, or a combination of the compounds. PD0332991 strongly enhanced the effect of SU14813 in MOLM13 and MV4-11 (both Flt3 ITD) and to a lesser degree in cells from patient 17 (Flt3 ITD). Neither SU14813 nor PD0332991 alone or in combination had a significant effect on THP-1 and U937 (Flt3 wt) cells or cells from patient 75 (Flt3 wt) (Figure 4D). A median effect analysis according to the method of Chou and Talalay21 was performed to determine whether the enhanced activity of the drug combination in MV4-11 and MOLM13 was synergistic. We found that at 50 nM PD0332991 plus 25 nM SU14813, concentrations close to the IC50 values of these compounds in the 2 cell lines, the resulting combination index (CI) is 0.78 for MV4-11 and 0.67 for MOLM13. A CI value of less than 0.9 is defined as synergistic activity; hence, PD0332991 and SU14813 act synergistically in this assay.
PD0332991 also strongly enhanced the proapoptotic activity of SU14813 in MOLM13 and MV4-11 as assayed by Annexin V and 7-AAD staining. In contrast to the cell lines, in primary blasts from patient 17 (Flt3 ITD), the combination of PD0332991 and SU14813 did not increase in Annexin V staining compared with SU14813 alone. In fact, Annexin V staining was somewhat reduced in the combination treatment. THP-1 and U937 cells, as well as cells from patient 75, showed no enhanced Annexin V staining in response to any of the compound treatments. The level of induction of apoptosis is depicted in a bar graph representation in Figure 4E and the primary flow cytometric data are presented in Figure S4.
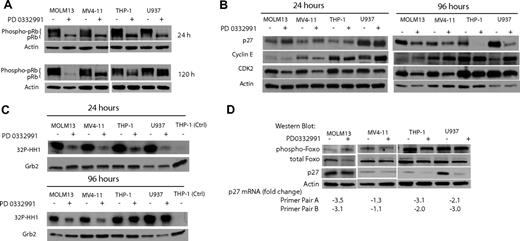
CDK2 is activated by p27Kip1 down-regulation
To investigate whether the differential effect of PD0332991 on the cell cycle of MV4-11 and MOLM13 (both Flt3 ITD) versus THP-1 and U937 (both Flt3 wt) is reflected in pRb phosphorylation status, we performed a Western blot. After 24 hours of exposure to PD0332991, pRb is completely dephosphorylated in all cell lines (Figure 5A). The complete dephosphorylation of pRb at 24 hours correlates well with the strong inhibitory effect on the cell cycle at this time point in all AML cell lines (Figure 4B). After 5 days of incubation with PD0332991, pRb is still completely dephosphorylated in the MOLM13 and MV4-11 Flt3 ITD AML cell lines. In contrast, pRb is partly phosphorylated again in the Flt3 wt U937 and THP-1 cell lines (Figure 5A), reflecting the reentry into the cell cycle as was detected by flow cytometry (Figure 4B).
THP-1 and U937 cells can rephosphorylate pRb after multiple days of PD0332991 treatment by reactivating CDK2. (A) Western blot. The indicated AML cell lines were treated with DMSO (vehicle control) or PD0332991 (500 nM) for 24 or 120 hours. Lysates were resolved by SDS-PAGE followed by immunoblotting with anti-pRb. (B) Western blot. Cells were treated for 24 or 96 hours with PD0332991 (500 nM) or DMSO (vehicle control). Lysates were subjected to SDS-PAGE followed by Western blot analysis with the indicated antibodies. An antibody against actin was used as a loading control. (C) CDK2 in vitro kinase assay: Cells were incubated for 96 hours with DMSO (vehicle control) or PD0332991 (500 nM). Cell lysates were prepared and normalized by protein concentration. CDK2 was immunoprecipitated to perform the in vitro kinase assay. Incorporation of 32P in the CDK2 substrate histone H1 was assessed by SDS-PAGE followed by autoradiography. As a control for the kinase assay, a lysate from THP-1 cells was incubated with an isotype-matched antibody to form an immunocomplex. To confirm equal protein amounts in each normalized extract, aliquots were separated by SDS-PAGE and analyzed by Western blot with an anti-Grb-2 antibody. (D) Western blot and real-time PCR. The indicated AML cell lines were treated with DMSO (vehicle control) or PD0332991 (500 nM) for 96 hours. Lysates were prepared and resolved by SDS-PAGE followed by immunoblotting with phospho-Foxo3a (Thr32), Foxo3a, p27, and actin as a loading control. The cells were also used to prepare mRNA to determine by real-time PCR the pairwise changes in p27 mRNA levels between DMSO- and PD0332991-treated cells. Two primer pairs were used for the analysis.
THP-1 and U937 cells can rephosphorylate pRb after multiple days of PD0332991 treatment by reactivating CDK2. (A) Western blot. The indicated AML cell lines were treated with DMSO (vehicle control) or PD0332991 (500 nM) for 24 or 120 hours. Lysates were resolved by SDS-PAGE followed by immunoblotting with anti-pRb. (B) Western blot. Cells were treated for 24 or 96 hours with PD0332991 (500 nM) or DMSO (vehicle control). Lysates were subjected to SDS-PAGE followed by Western blot analysis with the indicated antibodies. An antibody against actin was used as a loading control. (C) CDK2 in vitro kinase assay: Cells were incubated for 96 hours with DMSO (vehicle control) or PD0332991 (500 nM). Cell lysates were prepared and normalized by protein concentration. CDK2 was immunoprecipitated to perform the in vitro kinase assay. Incorporation of 32P in the CDK2 substrate histone H1 was assessed by SDS-PAGE followed by autoradiography. As a control for the kinase assay, a lysate from THP-1 cells was incubated with an isotype-matched antibody to form an immunocomplex. To confirm equal protein amounts in each normalized extract, aliquots were separated by SDS-PAGE and analyzed by Western blot with an anti-Grb-2 antibody. (D) Western blot and real-time PCR. The indicated AML cell lines were treated with DMSO (vehicle control) or PD0332991 (500 nM) for 96 hours. Lysates were prepared and resolved by SDS-PAGE followed by immunoblotting with phospho-Foxo3a (Thr32), Foxo3a, p27, and actin as a loading control. The cells were also used to prepare mRNA to determine by real-time PCR the pairwise changes in p27 mRNA levels between DMSO- and PD0332991-treated cells. Two primer pairs were used for the analysis.
To explore the possibility that CDK2 activity can compensate for the loss of CDK4/6 activity in THP-1 and U937 cells, we investigated the expression level of proteins involved in CDK2 regulation. After a 24-hour incubation period with PD0332991, neither p27Kip1 nor Cyclin E protein levels changed significantly (Figure 5B). However, CDK2 protein was slightly down-regulated in all 4 cell lines. It is noteworthy that after 4 days of incubation with PD0332991, the CDK2 inhibitor p27Kip1 was strongly down-regulated in Flt3 wt THP-1 and U937. Furthermore, in U937 Cyclin E was up-regulated. In contrast, no significant changes in the protein level of p27Kip1 and Cyclin E were observed in Flt3 ITD MOLM13 and MV4-11 (Figure 5B). These data led us to hypothesize that after treatment with PD0332991 for multiple days, the selective down-regulation of p27Kip1 in Flt3 wild-type THP-1 and U937 cells is responsible for higher CDK2 activity than is seen in MOLM13 and MV4-11 cells. To test this hypothesis, we performed a CDK2 in vitro kinase assay with cells treated for 24 and 96 hours with PD0332991 (Figure 5C). After treatment for 24 hours, CDK2 kinase activity was reduced in all cell lines, suggesting that CDK2 activation is dependent on CDK4/6 activity at this early time point. At the 96-hour time point, however, CDK2 activity in THP-1 and U937 cells was restored.
p27Kip1 transcription can be promoted by the transcription factor Foxo3a and the phosphorylation of Foxo3a leads to its inactivation.28–31 We found, however, no changes in Foxo3a phosphorylation status in any of the 4 AML cell lines treated for 96 hours with PD0332991 (Figure 5D). We used mRNA prepared from cells of the same experiment to perform a real-time PCR analysis to detect changes in p27Kip1 transcript levels. We applied 2 different sets of p27Kip1-specific primers for this experiment. This experiment showed that p27Kip1 mRNA is modestly reduced between 2- and 3-fold in THP-1, U937, and MOLM13 cells treated with PD0332991. There was no significant change of p27Kip1 transcript levels in MV4-11 cells.
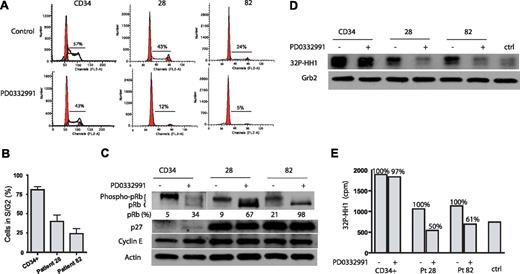
Primary AML cells are sensitive to PD0332991
To study the effect of CDK4/6 inhibition on primary cells, we used 2 AML patient samples and CD34+ hematopoietic progenitor cells from a healthy donor. Patient 28 expressed Flt3 wt, whereas patient 82 expressed Flt3 ITD. Figure 6A,B demonstrates that PD0332991 treatment for 4 days significantly reduced patient cells that are in S or G2 of the cell cycle. The effect of PD0332991 on the cell cycle of CD34+ cells is less pronounced. PD0332991 treatment for 4 days leads to the almost complete dephosphorylation of pRb in cells of patient 82 (Flt3 ITD) (98% of pRb is dephosphorylated) and strong dephosphorylation of pRb in cells from patient 28 (Flt3 wt) (68% of pRb dephosphorylated). In contrast, CD34+ cells show only 34% of pRb in its dephosphorylated form after treatment with PD0332991 (Figure 6C). A CDK2 in vitro kinase assay revealed that the treatment with PD0332991 for 4 days greatly reduces CDK2 activity in the 2 patient samples but not in the CD34+ cells (Figure 6D). The reduction in 32P incorporation into the CDK2 substrate histone H1 in this assay was 50% for patient 28 and 39% for patient 82, but only 3% in CD34+ cells (Figure 6E). Hence, in contrast to the 2 well-established Flt3 wt cell lines THP-1 and U937, which display autonomous growth ex vivo, the 2 primary AML patient samples, regardless of the Flt3 status, displayed strong sensitivity to PD0332991.
Effect of PD0332991 on primary AML cells. CD34+ and primary AML cells from patients 28 (Flt3 wt) and 82 (Flt3 ITD) were plated in methylcellulose and treated with PD0332991 (500 nM) or DMSO (vehicle control) for 4 days. The cells were fixed and stained with propidium iodide, and cell-cycle analysis was performed by flow cytometry. (A) Representative flow cytometric cell-cycle analysis of 1 experiment with CD34+ cells as well as cells from patients 28 and 82. (B) Bar graph analysis of the percentage of PD0332991-treated cells in S and G2 compared with control cells using data from 3 independent experiments (mean ± SE). (C) Western blot. CD34+ and primary AML cells from patients 28 and 82 were treated as in (A). Cell lysates were prepared and resolved by SDS/PAGE, and an immunoblot was performed with the indicated antibodies. (D) Cells were treated as in (panel A). Cell lysates were prepared and normalized by protein concentration. CDK2 was immunoprecipitated to perform the in vitro kinase assay. Incorporation of 32P in the CDK2 substrate histone H1 was assessed by SDS-PAGE followed by autoradiography. As a control for the specificity of the kinase assay, a lysate from CD34+ cells was incubated with an isotype-matched antibody to form an immunocomplex. To confirm equal protein amounts in each normalized extract, aliquots were separated by SDS-PAGE and analyzed by Western blot with an anti-Grb-2 antibody. (E) Quantitative representation of 32P-histone H1(cpm) of the experiment depicted in (panel D).
Effect of PD0332991 on primary AML cells. CD34+ and primary AML cells from patients 28 (Flt3 wt) and 82 (Flt3 ITD) were plated in methylcellulose and treated with PD0332991 (500 nM) or DMSO (vehicle control) for 4 days. The cells were fixed and stained with propidium iodide, and cell-cycle analysis was performed by flow cytometry. (A) Representative flow cytometric cell-cycle analysis of 1 experiment with CD34+ cells as well as cells from patients 28 and 82. (B) Bar graph analysis of the percentage of PD0332991-treated cells in S and G2 compared with control cells using data from 3 independent experiments (mean ± SE). (C) Western blot. CD34+ and primary AML cells from patients 28 and 82 were treated as in (A). Cell lysates were prepared and resolved by SDS/PAGE, and an immunoblot was performed with the indicated antibodies. (D) Cells were treated as in (panel A). Cell lysates were prepared and normalized by protein concentration. CDK2 was immunoprecipitated to perform the in vitro kinase assay. Incorporation of 32P in the CDK2 substrate histone H1 was assessed by SDS-PAGE followed by autoradiography. As a control for the specificity of the kinase assay, a lysate from CD34+ cells was incubated with an isotype-matched antibody to form an immunocomplex. To confirm equal protein amounts in each normalized extract, aliquots were separated by SDS-PAGE and analyzed by Western blot with an anti-Grb-2 antibody. (E) Quantitative representation of 32P-histone H1(cpm) of the experiment depicted in (panel D).
PD0332991 is active in a chimeric mouse-human model of AML
In the first mouse experiment we tested whether PD0332991 could induce the dephosphorylation of pRb in vivo. NOD/SCID mice engrafted with MOLM13 were treated 24 and 4 hours before sacrificing with PD0332991 or buffer (vehicle control) by oral gavage. The bone marrow cells were isolated and costained with anti-human CD45 to identify the leukemic MOLM13 cells and with human specific anti-phospho-pRb. The engraftment with human CD45 cells was at least 50% in all mice tested. Figure 7A demonstrates that CD45 cells from mice treated with PD0332991 showed a significant reduction in pRb phosphorylation compared with CD45 cells from mice treated with buffer as a control. Hence, the compound is active in the desired cell population in vivo. In the subsequent aggressive survival experiment we found that single agent PD0332991 could prolong the survival of NOD/SCID mice engrafted with MOLM13 by 4 to 5 days (P < .001) (Figure 7B). In the final experiment, we engrafted mice with MOLM13, treated the mice for 5 days starting on day 5 after engraftment and performed immunohistochemistry on the bone marrow. Staining with H&E and anti-human CD45 demonstrated the presence of MOLM13 cells in the bone marrow of control mice but to a lesser extent in the bone marrow of mice treated with PD0332991. Furthermore, the MOLM13 cells in the control mice showed strong phospho-pRb staining (Figure 7C).
AML mouse model to study the efficacy of PD0332991 in vivo. (A) Phospho-pRb staining in human CD45 cells. 6 NOD/SCID mice were inoculated with MOLM13 cells. On day 9 after inoculation, 3 mice were dosed with PD0332991 (150 mg/kg), and 3 mice were dosed with vehicle (lactic acid buffer). On day 10, the mice were dosed a second time with PD0332991 (150 mg/kg) or vehicle 4 hours before sacrificing them. The bone marrow was isolated, stained with anti-human CD45 plus anti-phospho-pRb, and analyzed by flow cytometry. The cells detected in the upper right and lower right quadrant are human leukemia cells because they stain positive for human CD45. The cells in the upper right quadrant stain positive for phospho-pRb. The cells in the lower right are negative for phospho-pRb. (B) Kaplan-Meyer survival plot. NOD/SCID mice were inoculated with MOLM13 cells. Starting on day 6 after inoculation, one group (10 mice) was dosed with PD0332991 (150 mg/kg), the other group (10 mice) was dosed with vehicle (lactic acid buffer). The survival of both groups was analyzed by Kaplan-Meyer plot. The survival benefit of mice treated with PD0332991 was statistically significant (P=.001, logrank test). (C) Immunohistochemistry. 10 NOD/SCID mice were engrafted as above. Five mice were each dosed with PD0332991 (150 mg/kg) or lactic acid buffer (control) starting on day 5 after inoculation. On day 10, all mice were killed. Sections of the sterna were prepared and stained as indicated with H&E, anti-human CD45, and anti-phospho pRb. The arrows point at the human leukemia cells. Images were visualized using an Olympus BX41 microscope equipped with an Olympus 3040 camera and a Plan 40×/0.65 NA objective lens (Olympus America, Melville, NY). The image medium was air. Adobe Photoshop CS version 8 (Adobe Systems, San Jose, CA) was used for image acquisition and processing.
AML mouse model to study the efficacy of PD0332991 in vivo. (A) Phospho-pRb staining in human CD45 cells. 6 NOD/SCID mice were inoculated with MOLM13 cells. On day 9 after inoculation, 3 mice were dosed with PD0332991 (150 mg/kg), and 3 mice were dosed with vehicle (lactic acid buffer). On day 10, the mice were dosed a second time with PD0332991 (150 mg/kg) or vehicle 4 hours before sacrificing them. The bone marrow was isolated, stained with anti-human CD45 plus anti-phospho-pRb, and analyzed by flow cytometry. The cells detected in the upper right and lower right quadrant are human leukemia cells because they stain positive for human CD45. The cells in the upper right quadrant stain positive for phospho-pRb. The cells in the lower right are negative for phospho-pRb. (B) Kaplan-Meyer survival plot. NOD/SCID mice were inoculated with MOLM13 cells. Starting on day 6 after inoculation, one group (10 mice) was dosed with PD0332991 (150 mg/kg), the other group (10 mice) was dosed with vehicle (lactic acid buffer). The survival of both groups was analyzed by Kaplan-Meyer plot. The survival benefit of mice treated with PD0332991 was statistically significant (P=.001, logrank test). (C) Immunohistochemistry. 10 NOD/SCID mice were engrafted as above. Five mice were each dosed with PD0332991 (150 mg/kg) or lactic acid buffer (control) starting on day 5 after inoculation. On day 10, all mice were killed. Sections of the sterna were prepared and stained as indicated with H&E, anti-human CD45, and anti-phospho pRb. The arrows point at the human leukemia cells. Images were visualized using an Olympus BX41 microscope equipped with an Olympus 3040 camera and a Plan 40×/0.65 NA objective lens (Olympus America, Melville, NY). The image medium was air. Adobe Photoshop CS version 8 (Adobe Systems, San Jose, CA) was used for image acquisition and processing.
Discussion
This study began with a focus on the molecular basis by which pharmacologic inhibition of Flt3 ITD autophosphorylation antagonizes the entry into the cell cycle. We found that the inhibition of Flt3 ITD signaling leads to a significant down-regulation of cyclin D2 and D3 gene and protein expression levels, thus affecting CDK4/6 activity as reflected in the dephosphorylation of pRb. Hence, CDK4/6 is a downstream effector of Flt3 ITD-mediated oncogenic pathways.
To study the role of CDK4/6 in AML with Flt3 ITD as well as in AML with Flt3 wt, we obtained PD0332991, a selective CDK4/6 inhibitor with activity in model systems for multiple myeloma, mantle-cell lymphoma, and rhabdomyosarcoma.32–34 In our AML model system, PD0332991 established a tight and sustained cell-cycle block in the Flt3 ITD MV4-11 and MOLM13 cell lines but only a transient cell-cycle block in Flt3 wt THP-1 and U937. In contrast to MV4-11 and MOLM13 cells, THP-1 and U937 cells were able to rephosphorylate pRb. The molecular basis for the rephosphorylation of pRb in THP-1 and U937 is probably the result of the observed reactivation of CDK2 activity. This reactivation may be caused by p27Kip1 down-regulation, which we found to be significant in U937 and THP-1 cells after PD0332991 treatment for 4 days. The down-regulation of p27Kip1 could occur post-translationally or at the transcriptional level. p27Kip1 gene transcription can be controlled by the Forkhead transcription factor Foxo3a (FKHR-L1), which is inactivated by phosphorylation.28–31 We observed no increase in Foxo3a phosphorylation in THP-1 or U937 in response to PD0332991, which suggests that a decrease in Foxo3a transcriptional activity is not involved in the down-regulation of p27Kip1 protein. It is noteworthy, however, that in measuring p27Kip1 mRNA levels by real-time RT-PCR, we observed a reduction of p27Kip1 gene expression in MOLM13, THP-1, and U937 in response to PD0332991. The changes in p27Kip1 transcript levels were modest, approximately 2- to 3-fold. The reduction in p27Kip1 gene transcription in THP-1 and U937 cells could account for the reduction in p27Kip1 protein levels. However, because a similar reduction in p27Kip1 gene expression without the concomitant drop in protein is observed in MOLM13, additional post translational effects in THP-1 and U937 may contribute to the PD0332991-mediated reduction in p27Kip1 levels.
The ability to compensate at least partially for the pharmacologic inhibition of CDK4/6 by using CDK2 may be one way for established AML cell lines to escape the antiproliferative effect of an inhibitor such as PD0332991. We found it encouraging, however, that both the Flt3 ITD+ and the Flt3 wt primary AML samples that we tested were sensitive to PD0332991 and did not up-regulate CDK2 activity after prolonged exposure. In the Flt3 ITD primary AML sample, the observed cell-cycle arrest, the pronounced dephosphorylation of pRb, and CDK2 down-regulation in response to CDK4/6 inhibition are consistent with the observations noted in Flt3 ITD AML cell lines. An identical pattern of cell-cycle arrest and CDK2 down-regulation observed in the Flt3 wt primary AML patient sample was in contrast to that observed in the well-established Flt3 wt AML cell lines. However, the Flt3 wt patient sample and Flt3 wt AML cell lines were similar with respect to the observation that pRb dephosphorylation was less complete than in Flt3 ITD cells. The dephosphorylation of pRb in the Flt3 wt patient cells was 68% versus 98% in the Flt3 ITD patient cells.
The partial dephosphorylation of pRb in CD34+ cells (only 34% of pRb is dephosphorylated) could be due to incomplete inhibition of CDK4/6 by PD0332991. However, because we treated the CD34+ cells in parallel and under the same conditions as the strongly responding primary AML cells, we believe that the basis for the remaining pRb phosphorylation in CD34+ cells is a result of compensatory CDK2 activity that we were able to measure by an in vitro kinase assay.
Finally, an MTT assay and Annexin V staining demonstrated that the inhibition of CDK4/6 by PD0332991 strongly enhanced the activity of the Flt3 inhibitor SU14813 in the AML cell lines MV4-11 and MOLM13 (both are Flt3 ITD positive). In contrast to SU14813, PD0332991 as a single agent did not induce Annexin V staining in the 3-day incubation period of this experiment. The enhancement of SU14813 activity by PD0332991 was also observed in blasts from a patient with Flt3 ITD in the MTT assay but not by Annexin V staining. In fact, Annexin V staining was slightly reduced in cells treated with the drug combination compared with cells treated with SU14813 alone. The improved efficacy of the drug combination in the MTT proliferation/viability assay but not in the Annexin V apoptosis assay may indicate that in primary cells with Flt3 ITD, the advantage of a PD0332991/SU14813 combination treatment may result from a tighter cell-cycle block and not necessarily from enhanced apoptosis as observed in cell lines.
In summary, we provide evidence that CDK4/6 is a downstream effector of Flt3 ITD signaling. The inhibition of CDK4/6 alone or in combination with inhibition of Flt3 ITD may have a therapeutic effect in AML. However, resistance to CDK4/6 inhibition may arise through down-regulation of p27Kip1 and CDK2 activation.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Cristina Lewis and Gerrit Los at Pfizer for providing PD0332991 and SU14813, and Mathai Mammen and Patrick Humphrey at Theravance for providing THRX-165724. Thanks to Audrey Papp and Zunyan Dai for technical help with the real-time PCR experiments.
This work was supported by a grant from the Leukemia & Lymphoma society (R.B.).
Authorship
Contribution: L.W. conducted research and analyzed data; J.W. conducted research and analyzed data; B.W.B. designed and conducted research and analyzed data; A.-M.D. conducted research and analyzed data; D.K. conducted research and analyzed data; T.L. conducted research and analyzed data; M.A.C. analyzed data and wrote the manuscript; R.B. designed research, analyzed data, and wrote the manuscript. L.W. and J.W. contributed equally to this work.
Conflict-of-interest disclosure: R.B. has declared a financial interest in Theravance, whose potential product was studied in the present work. The remaining authors declare no competing financial interests.
Correspondence: Roger Briesewitz, Department of Pharmacology, Ohio State University, 5065 Graves Hall, 333 W. 10th Ave, Columbus, OH 43210; e-mail: roger.briesewitz@osumc.edu.


![Figure 4. PD0332991 affects entry into the cell cycle and proliferation of AML model cell lines. (A) Cell-cycle analysis. Flow cytometric analysis of MOLM13 and U937 cells treated with DMSO (vehicle control) for 120 hours and PD0332991 (500 nM) for 24 and 120 hours and stained with propidium iodide. (B) Bar graph representation of cell-cycle analysis of MOLM13, MV4-11, THP-1, and U937 cells treated for 24 (□) and 120 hours (▒) with PD0332991 (500 nM). The cells were stained with propidium iodide and analyzed by flow cytometry. The percentage of cells in S/G2 compared with the control cells, which were normalized to 100% (treated with DMSO for 24 and 120 hours, respectively), was plotted (mean ± SE of 3 independent experiments). (C) Proliferation assay. MV4-11, MOLM13, THP-1, and U937 cells were plated in the presence of DMSO (vehicle control) or PD0332991 (500 nM) at a density of 0.5 × 106 / mL. The cells were counted every 2 days, and the cell number was adjusted to 0.5 × 106/ mL (mean ± SE of 3 independent experiments). (D) MTT assay. MV4-11, MOLM13, U937, THP-1 cells, as well as primary patient blasts from 2 patients (Pt 17 [Flt3 ITD] and Pt 75 [Flt3 wt]) were incubated in triplicate for 3 days with DMSO (control), PD0332991 (500 nM) (░), SU14813 (30 nM for cell lines and 100 nM for patient samples) (▒), or PD0332991 (500 nM) plus SU14813 (30 nM for cell lines and 100 nM for patient samples) (▓). The DMSO control is normalized to 100% (mean ± SE of 3 independent experiments for the cell lines and one experiment with triplicate data points for the patient samples). (E) Apoptosis assay. MV4-11, MOLM13, THP-1, and U937 cells were incubated for 3 days with DMSO (control), PD0332911 (250 nM) (░), SU14813 (100 nM) (▒), or PD0332991 (250 nM) plus SU14813 (100 nM) (▓). The DMSO control is normalized to 100%. The cells were stained with Annexin V-PE and 7-AAD followed by flow cytometric analysis (see Figure S4 for primary data).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/6/10.1182_blood-2007-02-071266/7/m_zh80180706750004.jpeg?Expires=1765903618&Signature=pgS~~DmwMvEcWpBx9-EIUv1MdyoPA4RtAXZgyh68APKbWzEr7tyNUzymYwdyQaq~pHCPbg1CU5zEiPWZfGK9qM0ybBYcNU2PyWYOeubQW7Yf6SgA1rR4SqazfDm4F9JuQTKZ0Zvvt5EHBRgaWKNd67ubJNwft1hExm3GlgoDJRFLXPyUiuC-FANqbEDxqqb47CxzIq9SuPDVa0MUJceszz8RgVpaeycovqUSLR-vJbvY8ssJcfv5YLLInrAsk3PHRUo8eXV3yM2odBggi5678bWu8q7IXX7SX-jDJHWs6O~M7ECauFT2CShYMAjKk5YTTnjMDoE5lHL3RCbTPHg4Gw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal