Abstract
Little is known about the behavior of hematopoietic stem cells (HSCs) in primates because direct observations and competitive-repopulation assays are not feasible. Therefore, we used 2 different and independent experimental strategies, the tracking of transgene expression after retroviral-mediated gene transfer (N = 11 baboons; N = 7 rhesus macaques) and quantitation of the average telomere length of granulocytes (N = 132 baboons; N = 14 macaques), together with stochastic methods, to study HSC kinetics in vivo. The average replication rate for baboon HSCs is once per 36 weeks according to gene-marking analyses and once per 23 weeks according to telomere-shortening analyses. Comparable results were derived from the macaque data. These rates are substantially slower than the average replication rates previously reported for HSCs in mice (once per 2.5 weeks) and cats (once per 8.3 weeks). Because baboons and macaques live for 25 to 45 years, much longer than mice (∼2 years) and cats (12-18 years), we can compute that HSCs undergo a relatively constant number (∼80-200) of lifetime replications. Thus, our data suggest that the self-renewal capacity of mammalian stem cells in vivo is defined and evolutionarily conserved.
Introduction
Hematopoiesis is the ordered process by which hematopoietic stem cells (HSCs) proliferate and differentiate into mature blood cells. Through replication and differentiation, HSCs generate clones of progenitors, precursors, and mature cells, which support hematopoiesis throughout an animal's life. HSCs cannot be directly observed and are generally defined and identified by their function (their ability to reconstitute and maintain hematopoiesis).
The most informative experimental approach for studying HSC kinetics is limiting-dilution competitive-repopulation experiments. Small numbers of cells with distinguishable phenotypes are transplanted into a recipient animal in which hematopoiesis has been ablated. After the HSCs engraft and restore blood cell production, the percentage of marrow progenitor cells or blood granulocytes of each phenotype is tracked over time, because this reflects the phenotype of contributing (differentiating) HSC clones.1,2 Using such data, the frequency of HSCs in mice and cats was estimated, as were their average replication and differentiation rates.3–5 Importantly, the estimated frequency of HSCs in mice derived with stochastic analyses was similar to frequencies estimated with independent methods.6–9 In addition, the results from transplantation studies overlapped the results from studies of unperturbed hematopoiesis, including 5-bromodeoxyuridine (BrdU)–labeling experiments.6,9,10
Given the significant differences in estimates of the frequency of HSCs (number of HSCs/number of nucleated marrow cells; 4-8/105 vs. 6/107) and the average HSC-replication rate (once/2.5 weeks vs once/8-10 weeks) between mice and cats, we hypothesized that underlying biologic principles might explain these discrepancies. For example, our data and additional observations in rats11 suggest that the total number of HSCs per animal (frequency of HSCs × total number of nucleated cells in marrow) is similar in all mammals.12 This indicates that the total number of stem cells in each animal is evolutionarily conserved, yet the differentiation and proliferative capacities of individual HSCs are not. We wondered if the total number of HSC self-renewal divisions per animal lifetime were similarly conserved.
To test this hypothesis, we studied hematopoiesis in baboons. Adult baboons (Papio cynocephalus and Papio anubis) weigh 11 to 26 kg and have life spans of 30 to 45 years in captivity (http://pin.primate.wisc.edu/factsheets).13,14 Similar analyses were also performed in rhesus macaques (Macaca mulatta; weight, 5-8 kg; life span, 25 years). These nonhuman primates can be thought of as intermediates between cats (including safari cats, on which previous studies were performed; safari cats weigh 3.5-8.5 kg and have life spans of 12-18 years) and humans (weight of 50-100 kg and life spans of 70-100 years).15–17
Limiting-dilution competitive-repopulation experiments are not feasible in primates. BrdU-labeling studies, although a powerful tool in mice,6,9,10 are imprecise because of their inability to accurately identify primate HSCs with flow-cytometry markers.18 Therefore, surrogate assays are needed to study HSC behavior in vivo; also, because the experimental observations are indirect, it is important that all results are confirmed with an independent experimental approach. In this study we used 2 experimental approaches (retroviral gene-marking experiments and studies of telomere lengths in granulocytes) to estimate the replication rate of HSCs in baboons and macaques.
Retroviral vector gene-marking experiments are similar to competitive-repopulation experiments in that the phenotype of granulocytes is followed over time and reflects the phenotype of HSCs (or short-term repopulating cells [STRCs]) contributing to hematopoiesis.19,20 Retroviral vector gene-marking data were collected from 11 baboons and 7 macaques. The percentage of transduced granulocytes 2 to 8 weeks after transplantation was used to estimate the percentage of transduced STRCs in each animal, and the percentage of transduced granulocytes when the marking frequencies plateaued was used to estimate the percentage of transduced HSCs. Telomeres (defined as the ends of eukaryotic chromosomes consisting as tandem repeats of [TTAGGG]) decrease with somatic cell division and, hence, with age.21–23 Therefore, observations of the change in the mean telomere lengths in granulocytes over time are surrogate markers for the replication rate of HSCs under assumptions that the HSC telomere shortening per replication is roughly constant and that cumulative telomere shortening during differentiation from HSCs to granulocytes is constant.24 Telomere-length data were collected from 132 baboons25 and 14 macaques.
Our basic approach was to simulate each specific experimental design by using an arbitrary HSC-replication rate (λ). We next asked if the data points that were observed in the specific baboon or macaque experiment could be a random draw from 1000 simulated outcomes. If yes, then λ was considered a possible result. We then repeated this process and tested additional values for λ until we derived which λ values were feasible (range) and which λ value was optimal.
These analyses showed that baboon HSCs replicate more slowly than both cat and mouse HSCs and suggest that the replication rate of macaque HSCs is similarly slow. Our results further provide evidence that the mean number of times that an HSC will replicate during a mammal's life span is reasonably constant across species. In addition, if one extrapolates the relationship between HSC-replication rate and animal longevity to man, one obtains an estimate for the average replication rate of human HSCs that is very similar to that (once per 45 weeks) estimated previously by the direct analysis of granulocyte telomere-length shortening.26
Materials and methods
Gene-marking data
Gene-marking data were collected from 11 baboons and 7 macaques. HSCs were transduced with retrovirus vectors that contained a marker gene. The percentage of cells expressing the transgene was measured longitudinally. Because the goal of these experiments was to optimize methods for HSC gene transfer, experimental conditions were not uniform across animals; specifically, different cultures and vectors had different transduction success rates (initially and long-term), and follow-up time varied between experiments. Measurements taken less than 1 week after transplantation were excluded. Analyses (see below) accounted for these differences in experimental design.
Experimental details have been described previously for baboons J00116, M00081, and T00024,27 F01007, F01064, and M01044,28 F99074,29 and F99310 and M99267.30 For animals M00165 and M00228, CD34+ cells were enriched from baboon marrow leukocytes by using immunoglobulin M monoclonal antibody 12.8 and an immunomagnetic column and prestimulated for 48 hours in medium containing 10% fetal bovine serum, stem cell factor, granulocyte colony-stimulating factor, interleukin 3 (IL-3), IL-6 (no IL-6 was present for M00228 for prestimulation or virus transduction), Flt3-L, and Tpo at 100 ng/mL each. After prestimulation, cells were transferred to flasks coated with CH-296 and preloaded twice with virus-containing medium. Cells next were exposed for 4 hours to virus supplemented with the same serum and cytokines and then collected and resuspended in fresh medium with cytokines. The following day, another 4-hour exposure to virus was performed before the cells were reinfused into the irradiated baboon (1020 cGy total-body irradiation). The range for total numbers of CD34 cells transplanted across all baboons was 3.8 × 107 to 24 × 107.
HSC mobilization, apheresis, retroviral transduction, and transplantation in the young rhesus macaques used in this study has been described previously (RQ3636, RQ3590, RQ2851, and RC909,31 RC708 and RQ2287,32 and RQ223733 ). To determine in vivo gene marking after transplantation, real-time polymerase chain reaction (PCR) was performed as described previously31 by using genomic DNA extracted from peripheral blood mononuclear cells and granulocytes collected at different time points after transplantation.
Telomere-length data
All telomere lengths were computed by using equivalent methods, specifically fluorescence in situ hybridization and flow cytometry.34
Telomere-length decrease per T-cell population doubling was computed by using T cells, grown in culture separately, from 5 baboons.35 Telomere length and cell doublings were measured longitudinally (2-9 measurements per animal, excluding measurements before culture establishment at time 0), and slopes describing their relationship were computed per baboon. The number of base pairs that T-cell telomere lengths decrease per population doubling was estimated as the weighted average of these slopes, weighting by the inverse of slope standard errors. Granulocyte telomere lengths were measured for 132 baboons aged between 0.92 and 24 years.25 Granulocytes were used because they are readily obtained and purified to homogeneity and are short-lived (circulation life spans, < 5-10 hours).
T-cell culturing and computational techniques for 5 rhesus macaques (2-9 measurements per animal) were identical to those of baboons.36 Total white blood cell telomere lengths were measured for 14 rhesus macaques aged between 2.8 and 15.3 years. Results using total white blood cells instead of granulocytes should be similar but perhaps less precise because of cellular heterogeneity.
Stochastic methods
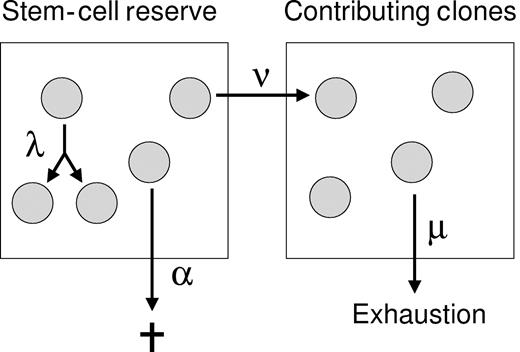
The stochastic model of HSC behavior used here has been described previously4 and is described by Figure 1. The HSC-replication rate (λ) and other parameters that define hematopoiesis (ν, α, and μ; Figure 1) were estimated through simulations. We simulated gene-marking and telomere-length data in populations of virtual primates, inputting plausible parameter values. HSC parameters were then estimated as those input values that yielded simulated results that were similar to observed outcomes, according to the criteria specified below.
Stochastic model of HSC behavior.26 HSCs in the stem-cell reserve have 3 potential fates: replication (1 HSC becomes 2 HSCs), differentiation (1 HSC becomes 1 STRC), or apoptosis (1 HSC becomes 0). These occur at average rates of λ, ν, or α, respectively. The maximum number of HSCs in the stem-cell reserve is K. If an HSC differentiates, then it enters the second compartment and contributes to hematopoiesis (becomes an STRC). The average length of time from differentiation until the clone ends its contribution is μ.
Stochastic model of HSC behavior.26 HSCs in the stem-cell reserve have 3 potential fates: replication (1 HSC becomes 2 HSCs), differentiation (1 HSC becomes 1 STRC), or apoptosis (1 HSC becomes 0). These occur at average rates of λ, ν, or α, respectively. The maximum number of HSCs in the stem-cell reserve is K. If an HSC differentiates, then it enters the second compartment and contributes to hematopoiesis (becomes an STRC). The average length of time from differentiation until the clone ends its contribution is μ.
Gene-marking simulations
Simulation procedures for baboons and macaques were identical and are described for baboons only. We simulated gene-labeling studies in populations of 1000 virtual baboons, inputting different values for the parameters λ, ν, α, μ, K (the maximum number of HSCs in the stem-cell reserve), and R0 (the number of transplanted HSCs) and following the percentage labeled over time. Input values are shown in Table 1 and discussed at the end of this section. At time 0, R0 HSCs start in the stem-cell reserve. The times until replication, differentiation, and apoptosis are selected randomly for each HSC from independent exponential distributions with average rates for λ, ν, and α, respectively. If a replication event occurs, times until replication, differentiation, and apoptosis again are selected randomly for each of the 2 resulting HSCs. C0 STRCs are in the contributing-clones compartment at time 0; because hematopoiesis in the donor animal is in steady state, C0 = R0ν/(λ − ν − α + μ).5 The STRC contributes to hematopoiesis (produces granulocytes) until a time randomly selected from an exponential distribution with an average rate of 1/μ. The key assumption generating exponential distributions is the Markov assumption that the time until an HSC's next replication, differentiation, or apoptosis event does not depend on any previous replication event or time.
Simulation plan for gene-marking experiments
| A. Test compatibility of mouse and cat parameter values assuming R0 = 100, 300, and 500 |
| λ = 1 replication per 2.5 wk; ν = 1 differentiation per 3.4 wk; α = 1 apoptosis per 20 wk; and μ = 6.9 wk for mouse5 |
| λ = 1 replication per 8.3 wk; ν = 1 differentiation per 12.5 wk; α = 1 apoptosis per 50 wk; and μ = 6.7 wk for cat4 |
| B. Obtain a range for λ, assuming μ = 6.7 wk, R0 = 300, ν = 0.71λ, and α = 0.14λ |
| C. Obtain a range for λ with same assumptions as in B, but R0 = 100 |
| D. Obtain a range for λ with same assumptions as in B, but R0 = 500 |
| E. Obtain a range for μ at best λ, with same assumptions as in B |
| F. Obtain a range for (α, ν) at best λ, with μ = 6.7 wk, R0 = 300, and ν + α = 0.85λ |
| G. Obtain a range for ν + α at best λ with μ = 6.7 wk, R0 = 300, and α = 0.2 ν |
| A. Test compatibility of mouse and cat parameter values assuming R0 = 100, 300, and 500 |
| λ = 1 replication per 2.5 wk; ν = 1 differentiation per 3.4 wk; α = 1 apoptosis per 20 wk; and μ = 6.9 wk for mouse5 |
| λ = 1 replication per 8.3 wk; ν = 1 differentiation per 12.5 wk; α = 1 apoptosis per 50 wk; and μ = 6.7 wk for cat4 |
| B. Obtain a range for λ, assuming μ = 6.7 wk, R0 = 300, ν = 0.71λ, and α = 0.14λ |
| C. Obtain a range for λ with same assumptions as in B, but R0 = 100 |
| D. Obtain a range for λ with same assumptions as in B, but R0 = 500 |
| E. Obtain a range for μ at best λ, with same assumptions as in B |
| F. Obtain a range for (α, ν) at best λ, with μ = 6.7 wk, R0 = 300, and ν + α = 0.85λ |
| G. Obtain a range for ν + α at best λ with μ = 6.7 wk, R0 = 300, and α = 0.2 ν |
In our simulations we accounted for experiment-to-experiment variability. The gene-marking data for each animal were described by the initial percentage of transduced cells (pi; first recorded posttransplantation marking percentage), the long-term percentage of transduced cells (ps; mean of all measurements after the time at which the marking percentage stabilizes), and the amount of follow-up time (T). (If the change point is not significant [described in the next paragraph], then ps is the average of all measurements. If there is drift [described in the next paragraph], then ps equals the percentage labeled at the change-point estimate.) These 3 characteristics are intrinsic to a specific experiment in an individual animal. Because the goal is to simulate gene-labeling studies in populations of virtual baboons and incorporate all experimental variability, for each simulated baboon we randomly selected 1 of 11 sets (pi, ps, T), each set corresponding to estimates of (pi, ps, T) from 1 of the 11 observed baboons, and generated virtual data for that baboon under experimental conditions defined by the randomly selected (pi, ps, T). In each simulation the number of marked HSCs at time 0 is drawn from Bin(ps, R0) (binomial distribution with R0 realizations with the probability of being marked equal to ps), and the number of marked STRCs at time 0 is drawn from Bin(pi, C0). The percentage of marked granulocytes is set equal to the percentage of marked STRCs. Therefore, our simulated studies control for the variability in experimental design of the observed data.
To estimate HSC parameters, before simulating we defined formal criteria for comparing simulated percentage-marked trajectories and observed data. The criteria are based on the following descriptive properties:
Time until the percentage labeled stabilizes: This is the maximum-likelihood estimate for the change point in a 2-segmented linear regression model given that it is a better fit than a linear model (P < .10)37 ; otherwise, there is no estimated stabilization time. The change point was restricted to be after the third measurement.
Presence or absence of drift after stabilization: Drift was defined as a poststabilization slope significantly different from 0 (P < .01) and estimated as at least 2% per 100 days. (A conservative P value was used to prevent overdiagnosis of drift, and negative drift was not considered because it is difficult to distinguish from stabilization.) Of 9 baboons, 2 (22%) met the criteria for drift (baboons J00116 and F99310; baboons M00165 and F01064 had no estimated stabilization time).
Amount of residual variation after stabilization: This amount was defined as the variance about a smoothed fit to the data38 after the change-point estimate or, if the change-point model was not chosen, then over the entire data range. It was computed after applying the standard variance-stabilizing transformation for binomial data, arcsine of square root, to make variance estimates more comparable across baboons.39
Each of these quantities was estimated for each baboon in the gene-marking datasets. In all simulations, for each of the 1000 virtual animals these 3 quantities were also computed.
Three criteria based on the 3 descriptive quantities determined if observed data could be a random draw from simulated data. Assuming the true proportion of baboons with drift was the proportion observed in our simulation, then if the probability of observing drift in 2 of 9 randomly selected baboons was more than .05, then simulated and observed data were compatible with respect to criterion 2. Kolmogorov-Smirnov goodness-of-fit tests were used for criteria 1 and 3, with P values less than .05 defining incompatibility (for example, criterion 3 compared the distribution of residual variation calculated in the 11 observed baboons with the distribution of residual variation computed in 1000 virtual baboons). If simulated and observed data were compatible using all 3 criteria, then the corresponding HSC parameters were ruled compatible with the data. Such an approach accounts for the variability in our baboon estimates.
Table 1 shows our simulation plan. In general, we fixed all parameters except the one to be estimated (typically λ); robustness was examined by varying a single parameter (or function of parameters) fixing all other parameters. Initial simulations were performed with R0 = 300. This estimate derives from inverse PCR analyses of individual clones of macaque marrow-derived granulocyte/monocyte colony-forming cells (CFU-GM) colonies.40 Thirty-two unique insertion sites were detected, and 8% of the cells were marked, implying that approximately 32/0.08 = 400 were transplanted; thus, estimates of 300 to 500 HSCs seemed most reasonable for macaque and, by extension, baboon simulations. This value is consistent with estimates from baboon and macaque linear amplification-mediated polymerase chain reaction (LAM-PCR) studies, although these estimates are less precise and require additional extrapolation, because only 25% of identified bands were sequenced to confirm uniqueness.41 In addition, R0 = 300 is consistent with estimates derived by assuming that the frequency of HSCs in nonhuman primates is bounded by values computed in cat and estimated for humans12 and the numbers of CD34+ cells transduced and transplanted in the gene-marking studies. We also performed simulations assuming R0 = 100 and 500.
In both cats and mice, μ has been estimated as approximately 6.7 weeks.4,5 This value was assumed in these simulations, with robustness to different values of μ tested in step E (Table 1). The best estimate for λ was chosen in step B as the value with the largest minimum P value for all 3 criteria. The choices for relationships between ν, α, and λ were based on approximate relationships observed in the best estimates of cat and mouse parameters.4,5 Robustness of estimates to these choices is examined in steps F and G.
Telomere simulations
We simulated granulocyte telomere lengths in populations of virtual baboons and macaques in a manner identical to that reported previously.26 Briefly, at time 0 (birth), each baboon had K HSCs of equal telomere length. With each replication, the resulting 2 HSC telomeres were shortened d bp. Granulocyte telomere lengths were taken as equal to HSC telomere lengths minus a constant (which does not need to be estimated). The recorded telomere length for each virtual baboon was obtained by computing the average telomere length of all HSCs at each virtual baboon's measurement age (48, 212, 652, 896, and 1248 weeks; results were similar for different ages). The relationship between the amount telomeres shorten in vivo per HSC replication, d, and in vitro telomere shortening per T-cell population doubling was assumed to be the same in baboons as estimated previously in cats, 18.4%.26 Hence, HSC telomeres in vivo were assumed to decrease per replication approximately d = 0.184 × (telomere-length decrease per T-cell population doubling).
The HSC-replication rate was estimated as the input value that resulted in the slope of simulated telomere length by age data equal to the slope of observed telomere length by age data. Simulations were performed by assuming ν = 0.71λ, α = 0.14λ, and K = 11 000 HSCs (on the basis of the cat and mouse parameter relationships discussed earlier). We assumed for all simulations that if there were a replication and K HSCs, then one of the 2 newly created HSCs dies. Estimates are similar if a random HSC of the K + 1 HSC dies.26 Results are robust to different assumptions for the relationship between ν, α, and λ, as well as different values for K.26 A range for λ was estimated by using the 95% confidence interval (CI) for the amount that T-cell telomere lengths decrease per population doubling and the 95% CI for the slope of the relationship between granulocyte telomere length and age.
All animals were housed under conditions approved by the American Association for Accreditation of Laboratory Animal Care, and protocols for each study were approved by the proper Animal Care and Use Committee/Institutional Review Board (National Heart, Lung, & Blood Institute; University of Washington; Southwest Regional Primate Research Center).
Results
Baboon gene-marking simulations
Gene-marking data were simulated for populations of 1000 virtual baboons using the stochastic model of Figure 1. For each simulation we input values for the average rates at which HSCs replicate (λ), differentiate (ν), and apoptose (α), the average time a differentiated cell contributes to hematopoiesis (μ), the number of transplanted HSCs (R0), and the percentage of HSCs and progenitors labeled (transduced) at transplantation. We then longitudinally followed the percentage of labeled granulocytes in our simulated baboons.
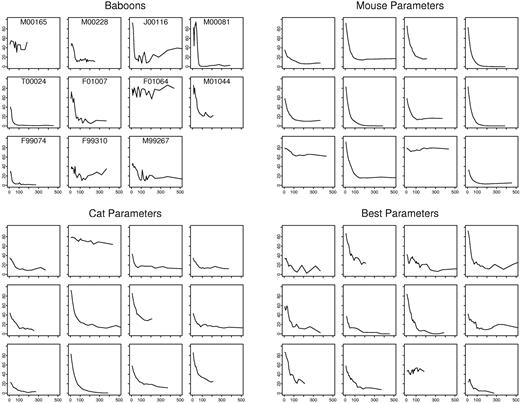
Figure 2 shows the percentage of labeled granulocytes over time for the baboons (ie, the observed data from retroviral gene-transfer experiments) and the first 12 (of 1000) simulations using HSC parameters for mouse and for cat. We determined if simulated data were compatible to observed data using 3 criteria: the time until the percentage of marked granulocytes stabilized (criterion 1), the presence or absence of a persisting drift (criterion 2), and the extent of variation over time (criterion 3).
Observed and simulated gene-marking data. Shown is the percentage of marked granulocytes as a function of time (from 0 to 500 days) in 11 baboons, in 12 virtual baboons simulated by using mouse parameters (λ = 1 replication per 2.5 weeks; ν = 1 differentiation per 3.4 weeks; α = 1 apoptosis per 20 weeks; μ = 6.9 weeks5 ; R0 = 300), in 12 virtual baboons simulated by using cat parameters (λ = 1 replication per 8.3 weeks; ν = 1 differentiation per 12.5 weeks; α = 1 apoptosis per 50 weeks; μ = 6.7 weeks4 ; R0 = 300), and in 12 virtual baboons simulated by using the best estimates of baboon parameter values (λ = 1 replication per 36 weeks; ν = 1 differentiation per 51 weeks; α = 1 apoptosis per 257 weeks; μ = 6.7 weeks; R0 = 300). For simulated data, the trajectories shown are the first 12 of 1000 virtual baboons.
Observed and simulated gene-marking data. Shown is the percentage of marked granulocytes as a function of time (from 0 to 500 days) in 11 baboons, in 12 virtual baboons simulated by using mouse parameters (λ = 1 replication per 2.5 weeks; ν = 1 differentiation per 3.4 weeks; α = 1 apoptosis per 20 weeks; μ = 6.9 weeks5 ; R0 = 300), in 12 virtual baboons simulated by using cat parameters (λ = 1 replication per 8.3 weeks; ν = 1 differentiation per 12.5 weeks; α = 1 apoptosis per 50 weeks; μ = 6.7 weeks4 ; R0 = 300), and in 12 virtual baboons simulated by using the best estimates of baboon parameter values (λ = 1 replication per 36 weeks; ν = 1 differentiation per 51 weeks; α = 1 apoptosis per 257 weeks; μ = 6.7 weeks; R0 = 300). For simulated data, the trajectories shown are the first 12 of 1000 virtual baboons.
The compatibility between observed and simulated data is shown in Table 2. Simulations that were based on mice and cat HSC parameter values did not result in simulated data compatible with the observed data. For example, the variation of the percentage-marked granulocytes over time in the simulations generated by using mice and cat HSC parameters is much smaller than the variation seen in the actual baboon experiments. For mice parameters, criterion 3 was rejected for R0 (the assumed number of transplanted HSCs) between 100 and 500 and criterion 2 was rejected for R0 = 500. For cat parameters, criteria 2 and 3 were rejected for R0 = 300 and 500. The sole exception was using cat parameters with R0 = 100 (criteria 3; P = .07).
Compatibility of simulated and observed baboon gene-marking data
| . | Probability values . | Compatible . | ||
|---|---|---|---|---|
| Criterion 1 . | Criterion 2 . | Criterion 3 . | ||
| A. Other mammalian HSC parameters | ||||
| Mouse | ||||
| R0 = 100 (HSC) | .49 | .25 | < .0001 | No |
| R0 = 300 | .31 | .06 | < .0001 | No |
| R0 = 500 | .28 | .02 | < .0001 | No |
| Cat | ||||
| R0 = 100 | .41 | .25 | .07 | Yes |
| R0 = 300 | .43 | .047 | < .0001 | No |
| R0 = 500 | .30 | .02 | < .0001 | No |
| B. Estimating replication rate, R0 = 300 | ||||
| λ = 1 replication/20 wk | .39 | .15 | .04 | No |
| λ = 1 replication/21 wk | .42 | .10 | .054 | Yes |
| λ = 1 replication/36 wk | .41 | .22 | .31 | Yes |
| λ = 1 replication/70 wk | .32 | .30 | .07 | Yes |
| λ = 1 replication/72 wk | .48 | .31 | .04 | No |
| C. Replication rate, R0 = 100 | ||||
| λ = 1 replication/6 wk | .43 | .23 | .03 | No |
| λ = 1 replication/7 wk | .5 | .24 | .054 | Yes |
| λ = 1 replication/25 wk | .29 | .30 | .057 | Yes |
| λ = 1 replication/29 wk | .24 | .31 | .04 | No |
| D. Replication rate, R0 = 500 | ||||
| λ = 1 replication/32 wk | .40 | .08 | .048 | No |
| λ = 1 replication/33 wk | .39 | .13 | .050 | Yes |
| λ = 1 replication/120 wk | .32 | .30 | .068 | Yes |
| λ = 1 replication/125 wk | .34 | .30 | .047 | No |
| E. Robustness to changes to μ | ||||
| μ = 3.0 wk | .07 | .31 | < .0001 | No |
| μ = 4.5 wk | .75 | .29 | .03 | No |
| μ = 5.0 wk | .60 | .29 | .050 | Yes |
| μ = 8.5 wk | .07 | .15 | .10 | Yes |
| μ = 9.0 wk | .046 | .16 | .07 | No |
| F. Robustness to changes to ν | ||||
| ν = 0.40 γ | .47 | .30 | .049 | No |
| ν = 0.45 γ | .41 | .30 | .08 | Yes |
| ν = 1.00 γ | .45 | .17 | .23 | Yes |
| G. Robustness to changes to γ | ||||
| γ = 0.35 λ | .61 | .29 | .04 | No |
| γ = 0.40 λ | .60 | .28 | .06 | Yes |
| γ = 1.00 λ | .38 | .23 | .46 | Yes |
| . | Probability values . | Compatible . | ||
|---|---|---|---|---|
| Criterion 1 . | Criterion 2 . | Criterion 3 . | ||
| A. Other mammalian HSC parameters | ||||
| Mouse | ||||
| R0 = 100 (HSC) | .49 | .25 | < .0001 | No |
| R0 = 300 | .31 | .06 | < .0001 | No |
| R0 = 500 | .28 | .02 | < .0001 | No |
| Cat | ||||
| R0 = 100 | .41 | .25 | .07 | Yes |
| R0 = 300 | .43 | .047 | < .0001 | No |
| R0 = 500 | .30 | .02 | < .0001 | No |
| B. Estimating replication rate, R0 = 300 | ||||
| λ = 1 replication/20 wk | .39 | .15 | .04 | No |
| λ = 1 replication/21 wk | .42 | .10 | .054 | Yes |
| λ = 1 replication/36 wk | .41 | .22 | .31 | Yes |
| λ = 1 replication/70 wk | .32 | .30 | .07 | Yes |
| λ = 1 replication/72 wk | .48 | .31 | .04 | No |
| C. Replication rate, R0 = 100 | ||||
| λ = 1 replication/6 wk | .43 | .23 | .03 | No |
| λ = 1 replication/7 wk | .5 | .24 | .054 | Yes |
| λ = 1 replication/25 wk | .29 | .30 | .057 | Yes |
| λ = 1 replication/29 wk | .24 | .31 | .04 | No |
| D. Replication rate, R0 = 500 | ||||
| λ = 1 replication/32 wk | .40 | .08 | .048 | No |
| λ = 1 replication/33 wk | .39 | .13 | .050 | Yes |
| λ = 1 replication/120 wk | .32 | .30 | .068 | Yes |
| λ = 1 replication/125 wk | .34 | .30 | .047 | No |
| E. Robustness to changes to μ | ||||
| μ = 3.0 wk | .07 | .31 | < .0001 | No |
| μ = 4.5 wk | .75 | .29 | .03 | No |
| μ = 5.0 wk | .60 | .29 | .050 | Yes |
| μ = 8.5 wk | .07 | .15 | .10 | Yes |
| μ = 9.0 wk | .046 | .16 | .07 | No |
| F. Robustness to changes to ν | ||||
| ν = 0.40 γ | .47 | .30 | .049 | No |
| ν = 0.45 γ | .41 | .30 | .08 | Yes |
| ν = 1.00 γ | .45 | .17 | .23 | Yes |
| G. Robustness to changes to γ | ||||
| γ = 0.35 λ | .61 | .29 | .04 | No |
| γ = 0.40 λ | .60 | .28 | .06 | Yes |
| γ = 1.00 λ | .38 | .23 | .46 | Yes |
Criteria 1, 2, and 3 compared the compatibility of marking percentages on the basis of the time until stabilization, the presence of drift, and the amount of variation, respectively.
We next determined the most optimal parameter estimates. With R0 = 300, estimates for λ between 1 replication per 21 weeks and 1 replication per 70 weeks met all 3 criteria. The best estimate was 1 replication per 36 weeks. The first 12 (of 1000) simulations with this estimate and ν = 1 differentiation per 51 weeks, α = 1 apoptosis event per 257 weeks, μ = 6.7 weeks, and R0 = 300 are also shown in Figure 2.
We then analyzed the dependencies and robustness of this result. Results were dependent on R0: for R0 = 100, the range for λ was 1 replication per 7 to 25 weeks; for R0 = 500, the range was 1 replication per 33 to 120 weeks. In contrast, results were robust to the differentiation and apoptosis rates. If one kept the exit rate (γ = ν + α) constant and varied ν and α (ie, varying the proportion of the exit rate due to differentiation), simulations using the other best parameter estimates yielded data compatible with the observed data between ν = 0.45γ and ν = γ. Results were also robust to keeping the proportion of the exit rate due to differentiation constant (ie, set ν = 0.835γ) and varying the value of the exit rate: simulated data were consistent with observed data for all exit rates between γ = 0.4λ and γ = λ. Thus, these results do not depend on (and provide little information about) the rate at which HSCs exit the stem-cell compartment and the manner in which they exit (differentiation or death). Results were similarly robust to the choice of K, the maximum number of HSCs (data not shown).
Besides providing an estimate of the average replication rate of baboon HSCs (λ), these simulations also provide information about μ (the time STRCs contribute to hematopoiesis), because the time until stabilization (criterion 1) is driven by the choice of μ. At our best estimate for λ, criterion 1 was only rejected for μ outside the range of 3 to 8.5 weeks. This range of μ is fairly constant regardless of the other parameters: using cat parameters, criterion 1 was rejected for μ only outside 4 to 10 weeks; using the best estimates for λ when R0 = 100 and 500, criterion 1 was rejected for μ only outside 4 to 10 and 3.3 to 9 weeks, respectively. Therefore, μ = 6.7 weeks is justified by this analysis, as well as previous data (“Materials and methods, Gene-marking simulations”).
As seen in Table 2, estimation of λ is particularly driven by criterion 3 (the amount of residual variation), because data simulated with less-frequent replication rates tend to be more variable. To ensure that the excess variation in the baboon data were a result of stochastic contributions of HSCs to hematopoiesis and not measurement error, we computed the variation of 6 samples from baboon M00165 between days 472 and 483 after transplantation. Because these measurements were taken long after transplantation and over a short time interval, any differences in the percentage of marked granulocytes should be a result of measurement error. Variation was small; measurements ranged from 44.5% to 47.3%. We subtracted the variance of these percentages from the observed residual variation in each baboon and reevaluated criterion 3 using this smaller residual variation estimate. The resulting estimated replication rate (assuming R0 = 300) was essentially the same, with a best estimate of once per 35 weeks (compared with 36 weeks) and an identical range (once per 21 to 70 weeks).
Baboon telomere-length simulations
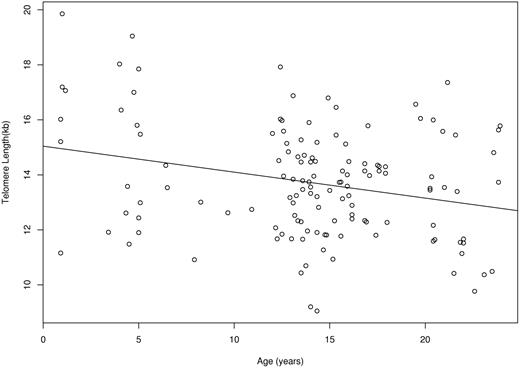
Baboon T cells were estimated to decrease 121 bp per population doubling (95% CI, 92-150 bp). As discussed in “Materials and methods,” in vivo HSC telomere loss per replication was estimated in a previous study26 as 18.4% of in vitro T-cell telomere loss per population doubling. Hence, d (the HSC telomere-length decrease per replication) was set equal to 22.2 bp. On the basis of cross-sectional observations on 132 baboons, granulocyte telomere lengths were estimated to shorten 94.4 bp/y (95% CI, 35-154 bp/y) (Figure 3).
Granulocyte telomere lengths for 132 baboons as a function of age. The relationship between telomere length and age is shown with the best-fitting line to the data, slope = −94.4 bp/year.
Granulocyte telomere lengths for 132 baboons as a function of age. The relationship between telomere length and age is shown with the best-fitting line to the data, slope = −94.4 bp/year.
Granulocyte telomere lengths were simulated in populations of virtual baboons assuming d = 22.2 bp/replication. Simulated data generated with λ = 1 replication per 23 weeks yielded a slope in simulated data equal to that in observed baboon data (94.4 bp/y). Hence, the best estimate for the HSC-replication rate using telomere lengths is once per 23 weeks (range, 1 per 11-75 weeks). As demonstrated elsewhere,26 this estimate was robust to different choices of K and γ = ν + α but dependent on the choice of d.
Studies in rhesus macaques
Gene-marking and telomere-length data were also collected in rhesus macaques. The stochastic methods described in this article were applied to analyze both sources of data (data and analyses not shown). Simulations using mouse HSC parameters were inconsistent with the observed data. However, the estimated range for the replication rate was very wide (1 per 2.5-200 weeks), because only 3 of 7 macaques had a high-enough transduction efficiency (eg, > 3%) to be able to observe if the percentage of marked granulocytes fluctuated. Telomere-length analyses yielded a best estimate of 1 HSC replication per 31 weeks. However, because the granulocyte telomere lengths derived from 14 (not 132) animals and because our estimate for the amount macaque T cells decrease per population doubling was wide (103 bp per population doubling; 95% CI, 3.2-202 bp), the range for the HSC-replication rate was too broad (between 0 and 3.6 replications per week) to consider this a reliable estimate. Therefore, results are compatible with the baboon studies, but more data (both gene-marking and telomere-length determinations) need to be collected to yield accurate estimates of the HSC-replication rate in rhesus macaques.
Discussion
We have used stochastic methods and 2 independent experimental approaches to determine the average replication rate of baboon HSCs. Our best estimates are once per 36 weeks (range, 21-70 weeks) and once per 23 weeks (range, 11-75 weeks), which are based on gene-marking and telomere-shortening analyses, respectively. Both analyses demonstrate that the baboon HSC-replication rate is less frequent than that of cats and mice. Similar but less-precise estimates were obtained for the replication rate of HSCs in rhesus macaques.
In “Results” we discussed robustness and dependencies of these estimates. Although gene-marking simulations depend on the number of transplanted HSCs, the assumption that R0 = 300 is well supported (see “Materials and methods”), and the general conclusion that HSCs replicate less frequently in baboons than mice or cats holds for all R0 in the prespecified range of 100 to 500. We also assumed that the behavior of marked HSCs is similar to the behavior of nonmarked HSCs. Similarly, telomere analyses rely on the assumption that the average telomere base pairs lost per HSC replication, d, is 18.4% of the amount that T-cell telomeres shorten per population doubling. This relationship is unknown but was extrapolated from cats, assumed a priori, and used in a previous analysis.26 We did not consider potential complexities resulting from developmental changes in HSC and telomerase activity.42–44 If telomerase activity substantially varies during an animal's lifetime, the telomere results could be affected, because we assume that the amount that telomeres decrease per replication remains roughly constant.
Because we must use surrogate assays to study HSC behavior in nonhuman primates, the strength of our conclusions comes from the combined message and trends seen using 2 independent sources of experimental data. With both types of data, specific, reasonable assumptions were made before performing the analyses (ie, R0 = 300 and d = 0.184 × telomere-length decrease per T-cell population doubling), and both simulation results were consistent with each other.
Our baboon results correspond with previous BrdU experiments, which showed that in steady-state hematopoiesis, baboon CD34+ cells (and, by extension, HSCs) require prolonged periods to cycle.18 Specifically, if the HSC-replication rate in baboons is once per 23 to 36 weeks (our estimate), then the percentage of HSCs that would cycle in 1 year is 90% to 76.5%. For comparison, during a year-long period of BrdU labeling in 2 baboons, Mahmud et al18 found that 56% and 84% of CD34+ cells underwent self-replication.
As expected, the HSC-replication rate in nonhuman primates seems to be more frequent than that of humans, estimated by using telomere lengths and identical methods to be once per 45 weeks (range, 1 per 19-83 weeks).26 The concordance of the estimated baboon HSC-replication rate using granulocyte telomere lengths and an independent technique (gene marking) strengthens our confidence that the HSC-replication rate in humans is, indeed, on the order of once per year.21
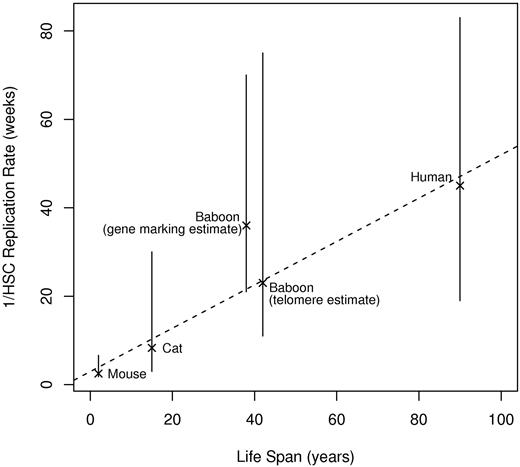
It is interesting to compute the total number of replications (self-renewal divisions) that an HSC remaining in the stem-cell compartment will make over a lifetime for each species (derived through simulations26 ). Assuming HSC-replication rates of once per 2.5, 8.3, 36, 23, and 45 weeks and life spans of 2, 15, 40, 40, and 90 years for mice, cats, baboons, baboons, and humans, the estimated number of lifetime replications for an HSC is 78, 170, 107, 167, and 192, respectively. (Two numbers are included for baboons, corresponding to the 2 different estimates of the HSC-replication rate.) These numbers are remarkably consistent. Figure 4 shows a plot of animal life span versus the inverse of the HSC-replication rate (which equals the average length of time between a single HSC's replications); the relationship appears linear. Therefore, our studies suggest that self-renewal capacity may be a defined, evolutionarily conserved property of mammalian HSCs. It seems that stem cells in vivo have a mitotic clock that is not species specific.
Inverse of HSC-replication rate (1/λ) and range as a function of life span. The dotted line is the best-fitting line to the point estimates. Life spans are approximate; actual values are unknown.17,18
In 1961, Hayflick and Moorhead reported that normal diploid human cells (in contrast to cancer cell lines) have finite life spans in in vitro culture.45 The relative importance of telomere shortening, oxidative stress, and genomic stability in defining the proliferative capacity of cells is unclear and controversial.46,47 Our results indirectly support the concept that HSCs (similar to other somatic cells) undergo a finite number of replications before cellular senescence.48
Because mice, cats, and baboons can be ordered not only by longevity but also by size, we cannot rule out the possibility that the biologic mechanism that drives slower HSC-replication rates in primates is size. Similar to that shown in Figure 4, we constructed a plot of λ−1 versus animal weight. The linear relationship of λ−1 with weight was good (r = 0.94, Pearson's correlation) but not as strong as seen with life span (r = 0.98) (figure not shown).
Thus, these results imply that to maintain hematopoiesis in these larger/longer-living animals, either (1) their HSCs must give rise to many more mature blood cells and/or (2) there must be much less progenitor/mature blood cell death. One or 2 more divisions of the earliest multilineage progenitor cells in larger animals could account for differences in the number of blood cells,49 and apoptosis is a feature of progenitor cell differentiation.50
The baboon gene-labeling studies also allowed estimation of μ, the amount of time that differentiated HSCs (ie, STRCs) contribute to hematopoiesis. This approximate range (3-10 weeks) is consistent with estimates in mice5,51 and cats4 and with data that tracked the contribution of retrovirally transduced primitive progenitors to hematopoiesis after autologous transplantation in rhesus macaques.40 Therefore, μ seems to be conserved in mammals.
In summary, various experimental approaches and stochastic simulations have revealed general hematopoiesis and stem cell principles. Together with previous experiments, these results provide evidence that the length of time that differentiated HSCs contribute to hematopoiesis is fairly constant across species, as is the number of times that an HSC replicates (self-renews) during an animal's lifetime. These general principles may provide insights into human hematopoiesis and influence strategies for gene therapy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants RO1 HL046598, HL082933, HL53750, HL54881, and AI29524.
National Institutes of Health
Authorship
Contribution: H.-P.K., P.M.L., G.A., C.E.D., A.L., and R.S. collected the data; B.E.S. performed data analysis; B.E.S., P.G., and J.L.A. designed the research; and B.E.S. and J.L.A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Janis L. Abkowitz, University of Washington, Box 357710, Seattle, WA 98195-7710; e-mail: janabk@u.washington.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal