Abstract
Transcription factors are key regulators of hematopoietic stem cells (HSCs), yet the molecular mechanisms that control their expression are largely unknown. Previously, we demonstrated that expression of Scl/Tal1, a transcription factor required for the specification of HSCs, is controlled by Ets and GATA factors. Here we characterize the molecular mechanisms controlling expression of Lyl1, a paralog of Scl also required for HSC function. Two closely spaced promoters directed expression to hematopoietic progenitor, megakaryocytic, and endothelial cells in transgenic mice. Conserved binding sites required for promoter activity were bound in vivo by GATA-2 and the Ets factors Fli1, Elf1, Erg, and PU.1. However, despite coregulation of Scl and Lyl1 by the same Ets and GATA factors, Scl expression was initiated prior to Lyl1 in embryonic stem (ES) cell differentiation assays. Moreover, ectopic expression of Scl but not Lyl1 rescued hematopoietic differentiation in Scl−/− ES cells, thus providing a molecular explanation for the vastly different phenotypes of Scl−/− and Lyl1−/− mouse embryos. Furthermore, coregulation of Scl and Lyl1 later during development may explain the mild phenotype of Scl−/− adult HSCs.
Introduction
Appropriate control of cell type–specific gene expression underlies the development of all multicellular organisms. Developmental pathways by which pluripotent stem cells become committed to single lineages are thus reflected in dynamic changes of transcriptional programs. Hematopoiesis provides a prime example of such a process, in which transcription factors play central roles in cell fate specification and subsequent differentiation.1 Furthermore, aberrant expression of the same key transcription factors is often associated with the development of leukemia.2 Accurate transcriptional regulation of hematopoiesis is therefore critical even though the underlying molecular mechanisms remain, for the most part, poorly understood.
The Lyl1 gene3 represents the founding member of a family of basic helix-loop-helix (bHLH) transcription factors identified through their involvement in chromosome translocations in T-cell acute lymphoblastic leukemia (T-ALL). Translocation of the Lyl1 gene into the vicinity of regulatory elements of the T-cell receptor β chain gene resulted in the expression of full-length Lyl1 protein in T cells, which normally do not express Lyl1.4 Shortly after the discovery of Lyl1, several groups identified the highly similar bHLH transcription factor Scl (also known as Tal1) involved in chromosome translocations with the T-cell receptor δ chain gene (reviewed by Begley and Green5 ). Lyl1 and Scl exhibit more than 90% amino acid identity in their bHLH DNA binding domains, and both interact with the lim-only-domain leukemia oncogenes LMO1 and LMO2.6
Lyl1 and Scl display overlapping expression patterns across several hematopoietic lineages7 and are also both expressed in developing endothelial cells.8 Targeted deletion has shown that Scl is essential for the early specification of hematopoietic stem cells (HSCs) as well as vascular and neural development.9–13 Interestingly, analysis of conditional Scl−/− mice demonstrated that Scl is not required for self-renewal or long-term repopulation activity of adult HSCs but that short-term repopulating capacity of Scl-deleted HSCs is severely impaired.14–16 By contrast, Lyl1−/− mice are viable and have normal blood counts except for a reduced number of B cells.17 However, both fetal liver and adult bone marrow Lyl1−/− HSCs showed severe defects in competitive repopulation assays.17 Taken together, the loss of function experiments therefore suggest that Scl appears to be more important for the generation of HSCs, whereas Lyl1 may be more important to maintain stem cell properties. Alternatively, the knockout phenotypes may reflect the degree to which Lyl1 and Scl can compensate for each other as a result of overlapping yet distinct expression patterns or expression levels.
The key role of Scl during the specification of embryonic HSCs has led us in the past to focus our attention on how expression of Scl itself is regulated. A systematic dissection of Scl regulation resulted in the identification of 6 independent enhancers, each targeting expression to a specific subdomain of the normal Scl expression pattern.18–23 By contrast, the only study so far that has touched on the transcriptional regulation of Lyl1 is a comparative genomic study in which we reported that the Lyl1 promoter is conserved between eutherian and marsupial mammals and displays promoter activity in vitro in cell lines.8
In this paper, we report a comprehensive in vivo and in vitro analysis of the mouse Lyl1 promoter. We show that the Lyl1 promoter drives expression in transgenic mouse embryos in developing endothelial and blood cells and that, similar to Scl, Lyl1 is regulated by Ets and GATA family transcription factors in endothelial, hematopoietic progenitor, and megakaryocytic cells. Despite this overlap in upstream regulators, we find that Scl expression is initiated prior to Lyl1 during early hemangioblast specification. Moreover, forced expression of Lyl1 in embryonic stem (ES) cells did not rescue the hematopoietic defect of Scl−/− ES cells.
Materials and methods
Generation and analysis of transgenic mice
Transgenic mice were generated and maintained as described.21 For flow cytometry, E11.5 fetal livers were dissected and β-galactosidase activity was detected using the fluorescent substrate FDG as described.20 Phycoerythrin (PE)–conjugated monoclonal antibodies against surface markers Ter119, Mac1, and c-kit were purchased from BD Biosciences (San Jose, CA). Fluorescence-activated cell sorting (FACS) was performed using a FACSCalibur flow cytometer (BD Biosciences) and the results analyzed with FlowJo software (Tree Star, Ashland, OR). Whole-mount images were acquired using a Pixera Pro 150ES digital camera (Pixera, Los Gatos, CA) attached to a Nikon SM7800 microscope (Nikon, Kingston upon Thames, United Kingdom). Images of sections were acquired with the same camera attached to an Olympus BX51 microscope (Olympus, Southall, United Kingdom) using an Olympus UPlanApo 40×/0.85 numerical aperature (NA) or 100×/1.35 NA objective. ImagePro Express version 4.5 (Image Processing Solutions, North Reading, MA) was used for acquisition of both types of images. Digital images were processed using Adobe Photoshop version 6 (Adobe Systems, San Jose, CA).
Reporter constructs and transfection assays
A 464 bp fragment of the mouse Lyl1 promoter region was amplified by polymerase chain reaction (PCR) and cloned upstream of the luciferase gene in pGL2 (Promega, Madison, WI). The Lyl1-P1 and Lyl1-P2 subfragments were generated using a BssHII site within the Lyl1 promoter. Mutated promoter constructs were prepared by PCR as described.18 The sequences of the mutated constructs are shown in Figure S3 (available on the Blood website; see the Supplemental Figures link at the top of the online article). Transgenic reporter constructs were generated by inserting the Lyl1 promoter fragment either upstream of a lacZ cDNA or downstream of an SV40 minimal promoter/lacZ reporter cassette. Luciferase reporter assays were performed as described.24
In vivo DMS footprinting
Footprinting was performed as detailed previously.25 Briefly, cells or genomic DNA were incubated at room temperature in 0.2% DMS solution in PBS for 5 minutes. The reaction was stopped with multiple washes with ice-cold PBS. Following cell lysis and DNA extraction, DNA was cleaved with 0.1 M piperidine at 90°C for 10 minutes and analyzed by ligation-mediated (LM)–PCR. PCR products were labeled by primer extension using 32P-labeled nested primers and analyzed on 6% denaturing polyacrylamide gels. Linker and primer sequences are available on request. Experiments were performed on 3 separate occasions with material from 2 independent DMS-treated cell and genomic DNA preparations.
Chromatin immunoprecipitation and real-time PCR analysis
Chromatin immunoprecipitation (ChIP) assays were performed as described26 using the following antibodies: anti–Fli-1 (sc-356x), –Erg (sc-354x), –Elf-1 (sc-631x), –GATA2 (sc-9008x), and –PU.1 (sc-352x) from Santa Cruz Biotechnology (Santa Cruz, CA) and an anti–acetyl H3 antibody (06-599) from Upstate Biotechnology (Lake Placid, NY). Fold enrichments were calculated over those seen with the control IgG antibody and normalized for enrichment seen with a control region (α-fetoprotein promoter; Figure S4 shows results normalized to IgG control samples only). The following primers were used for quantitative PCR analysis of ChIP material: mLyl F: 5′AAGGGCGGGTCGGTCCAG 3′; mLyl R: 5′GGTCAGGTTTGTCAGTCCAGGTC 3′; mAFP F: 5′ TGTTTGCTCACTGAAGGTTACTAG 3′; mAFP R: 5′ AGTGCTGGAAGTGGGATGTTTC 3′.
Expression levels of endogenous Lyl1 mRNA were assessed in cell lines and RNA prepared from whole E11.5 mouse embryos. RNA was prepared using the Qiagen (Valencia, CA) RNeasy kit and analyzed by real-time PCR using Stratagene (La Jolla, CA) SYBR green reagents. Expression levels were normalized to β-actin using the following primers: mLyl F: 5′ AGATGAGGAAACGCCCTGTA 3′; mLyl R: 5′ AGCCACTGCAAGTAGCCTGT 3′; mActinb F: 5′TCCTGGCCTCACTGTCCAC 3′; mActinb R: 5′ GTCCGCCTAGAAGCACTTGC 3′. To directly compare expression levels of endogenous Scl and Lyl1 in Flk1+ cells isolated from differentiating ES cells, the amplification efficiencies for Scl and Lyl1 primer pairs were determined using serial dilutions of plasmids containing Scl and Lyl1 cDNAs, respectively. To account for the differences in primer efficiencies observed, standard curve equations were used to adjust threshold cycle values and calculate the amounts of transcripts in femtomoles per 50 ng total RNA.
ES cell transfection and differentiation
Flag-tagged Scl and Lyl1 expression constructs in pMSCV-IRES-puro were generated using standard cloning procedures (details available on request). ES cells were maintained and differentiated as previously described.27 Two independent Scl−/− ES lines kindly provided by professors Glenn Begley and Stuart Orkin were transfected with MSCV constructs using the GeneJuice (Novagen, Nottingham, United Kingdom) reagent following the manufacturer's recommendations and selected with puromycin at 0.5 μg/mL. Hematopoietic potential was assessed upon differentiation in methylcellulose replating assays as previously described.27
Results
The Lyl1 promoter region directs expression to endothelial and hematopoietic cells in transgenic mice
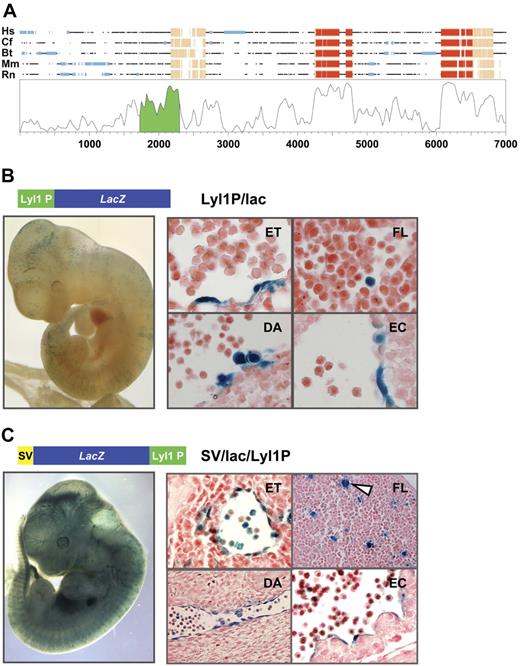
To identify Lyl1 candidate regulatory sequences, we aligned Lyl1 gene loci from human, dog, cow, mouse, and rat. This analysis demonstrated that the promoter proximal region of Lyl1 was highly conserved (Figure 1A). To assess potential in vivo gene regulatory activity of the conserved promoter proximal region, a 464 bp fragment (green region in Figure 1A; denoted as Lyl1P in Figure 1B-C) was subcloned both upstream of a promoterless lacZ reporter gene (Figure 1B) and downstream of a lacZ reporter cassette containing the SV40 minimal promoter (Figure 1C). Both constructs were used to generate transgenic mice that were analyzed by wholemount X-gal staining of midgestation E11.5 embryos. Two of 5 F0 transgenic embryos carrying the Lyl1P/lacZ transgene and 2 of 6 transgenic embryos carrying the SV/lac/Lyl1P construct showed staining. The SV/lac/Lyl1P construct was also used to generate 4 transgenic lines, 1 of which showed transgene expression. Based on visual inspection of wholemount stained embryos, transgene expression appeared to be localized to blood vessels in all embryos that expressed the lacZ reporter gene (Figure 1B-C). The overall staining patterns were similar regardless of whether the Lyl1 sequence was inserted upstream or downstream of the lacZ reporter gene, demonstrating that this region of the Lyl1 locus contains promoter activity as well as tissue-specific enhancer elements. Overall staining levels varied between lines (embryos shown in Figure 1B-C were at the lower and higher end of the spectrum, respectively), suggesting that the Lyl1 promoter construct did not confer position independence of transgene expression. Moreover, no staining was observed in adult hematopoietic cells, suggesting that sequences outside the Lyl1 core promoter are required to maintain expression in adult blood cells.
The Lyl1 promoter proximal region drives expression in hematopoietic and endothelial cells. (A) Synplot28 graphical representation of a Multi-LAGAN29 multiple sequence alignment of vertebrate Lyl1 loci where Hs is Homo sapiens, Cf is Canis familiaris, Bt is Bos taurus, Mm is Mus musculus, and Rn is Rattus norvegicus. Coding and noncoding exons are highlighted in red and pink, respectively. Repetitive sequences are indicated by light blue shading. The base pair numbering along the horizontal axis includes gaps introduced by the alignment program. The segment of the homology profile shaded in green corresponds to the Lyl1 proximal promoter fragment used to generate transgenic mice. (B) The Lyl1 promoter region is active in transgenic mice. Shown is a representative E11.5 transgenic embryo expressing lacZ under control of the Lyl1 proximal promoter region (wholemount view, left; histologic sections, right). Endothelial staining discernible from wholemount analysis was confirmed by analysis of a blood vessel (ET = endothelium). Staining was also observed in round hematopoietic cells in the fetal liver (FL), clusters of round cells attached to the ventral wall of the dorsal aorta (DA), and the endocardium (EC). (C) The Lyl1 proximal promoter region functions as an enhancer in transgenic mice. Shown is a representative E11.5 transgenic embryo where the Lyl1 proximal promoter region drives lacZ expression from the SV40 minimal promoter (wholemount view, left; histologic sections, right). The staining pattern was similar to that observed in Figure 1B. The arrowhead indicates a β-galactosidase–positive cell with megakaryocyte morphology.
The Lyl1 promoter proximal region drives expression in hematopoietic and endothelial cells. (A) Synplot28 graphical representation of a Multi-LAGAN29 multiple sequence alignment of vertebrate Lyl1 loci where Hs is Homo sapiens, Cf is Canis familiaris, Bt is Bos taurus, Mm is Mus musculus, and Rn is Rattus norvegicus. Coding and noncoding exons are highlighted in red and pink, respectively. Repetitive sequences are indicated by light blue shading. The base pair numbering along the horizontal axis includes gaps introduced by the alignment program. The segment of the homology profile shaded in green corresponds to the Lyl1 proximal promoter fragment used to generate transgenic mice. (B) The Lyl1 promoter region is active in transgenic mice. Shown is a representative E11.5 transgenic embryo expressing lacZ under control of the Lyl1 proximal promoter region (wholemount view, left; histologic sections, right). Endothelial staining discernible from wholemount analysis was confirmed by analysis of a blood vessel (ET = endothelium). Staining was also observed in round hematopoietic cells in the fetal liver (FL), clusters of round cells attached to the ventral wall of the dorsal aorta (DA), and the endocardium (EC). (C) The Lyl1 proximal promoter region functions as an enhancer in transgenic mice. Shown is a representative E11.5 transgenic embryo where the Lyl1 proximal promoter region drives lacZ expression from the SV40 minimal promoter (wholemount view, left; histologic sections, right). The staining pattern was similar to that observed in Figure 1B. The arrowhead indicates a β-galactosidase–positive cell with megakaryocyte morphology.
To further define the specificity of transgene expression, Lyl1P/lac and SV/lac/Lyl1P transgenic embryos were sectioned for histologic analysis. For both constructs, transgene expression was observed in endothelial and hematopoietic cells (Figure 1B-C). Endothelial activity was found in large and small vessels and included endocardial staining. Hematopoietic activity was restricted to a minority of fetal liver cells, including large cells with megakaryocyte morphology (Figure S2 presents a high-magnification view). Interestingly, we detected reporter gene activity in small clusters of round cells attached to the ventral floor of the dorsal aorta (Figure 1B). Such clusters are thought to represent the site from which the first HSCs emerge during development,30,31 suggesting that the Lyl1 promoter region may be active in hematopoietic stem and/or progenitor cells. Taken together, these results demonstrated that the Lyl1 promoter proximal region is active in vivo in endothelial cells as well as a subset of hematopoietic cells. Moreover, a similar pattern of staining was observed in Lyl1 lacZ knock-in embryos, demonstrating that the Lyl1 promoter region recapitulates Lyl1 expression during embryonic development (Figure S1).
The Lyl1 promoter region contains 2 independent core promoters
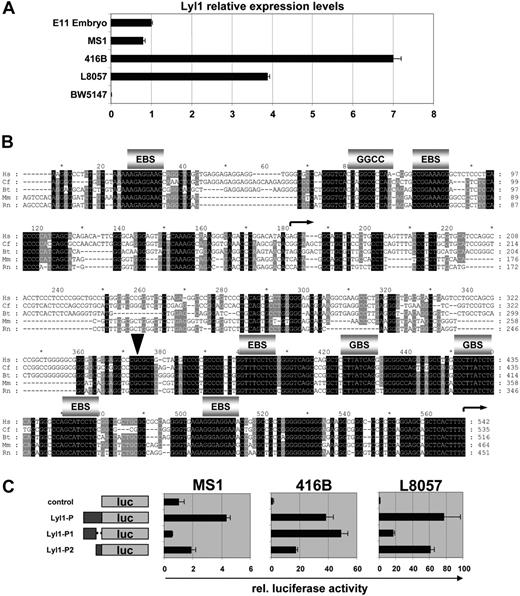
The transgenic analysis described in the previous section had identified the Lyl1 promoter sequence as a key regulatory region controlling Lyl1 expression in endothelium, megakaryocytes, and hematopoietic progenitors. To dissect the underlying molecular mechanisms, cellular models needed to be established. To this end, we performed quantitative reverse transcriptase (RT)–PCR analysis to monitor Lyl1 expression levels in MS1, 416B, and L8057 cell lines representing endothelial, hematopoietic progenitor, and megakaryocytic cells, respectively. RNA from whole E11.5 mouse embryos and the T-cell line BW5147 served as positive and negative controls, respectively. MS1, 416B, and L8057 cells were found to express significant levels of Lyl1 mRNA, whereas expression in BW5147 was negligible (Figure 2A). These results therefore suggested that MS1, 416B, and L8057 represented suitable cellular model systems with which to perform a molecular dissection of the Lyl1 promoter proximal region.
The Lyl1 promoter proximal region contains 2 evolutionarily conserved core promoters. (A) Lyl1 is expressed in endothelial (MS1), hematopoietic progenitor (416B), and megakaryocytic L8057 cell lines but not in a T-cell line (BW5147). Total cDNAs were subjected to real-time PCR analysis. The level of Lyl1 in RNA prepared from whole E11.5 mouse embryos, normalized by the level of actin, was assigned a value of 1. The relative abundance of Lyl1, normalized by the level of actin, is depicted by black bars ± SD. (B) Nucleotide sequence alignment of the Lyl1 proximal promoter region (species as described in Figure 1). Conserved Ets binding sites (EBS), GATA binding sites (GBS), and the GGCC motif as well as the 2 transcriptional start sites (curved arrows) are indicated. The arrowhead designates the cloning site used to separate the 2 core promoters Lyl1P1 and Lyl1P2. (C) The Lyl1 proximal promoter contains 2 core promoters. Shown on the left are the reporter constructs in which either the entire Lyl1P proximal promoter region or the 2 core promoters Lyl1P1 and Lyl1P2 were inserted upstream of the luciferase gene in the promoterless pGL2B vector. Following from left to right are the results of transient transfection assays in MS1, 416B, and L8057 showing luciferase activities corrected for transfection efficiency with the pEF-BOS LacZ plasmid. The luciferase activities are presented as fold increase over the activity of the control (pGL2B) vector, which was assigned a value of 1. Each bar represents the mean relative luciferase activity from at least 2 experiments performed in triplicate ± SD.
The Lyl1 promoter proximal region contains 2 evolutionarily conserved core promoters. (A) Lyl1 is expressed in endothelial (MS1), hematopoietic progenitor (416B), and megakaryocytic L8057 cell lines but not in a T-cell line (BW5147). Total cDNAs were subjected to real-time PCR analysis. The level of Lyl1 in RNA prepared from whole E11.5 mouse embryos, normalized by the level of actin, was assigned a value of 1. The relative abundance of Lyl1, normalized by the level of actin, is depicted by black bars ± SD. (B) Nucleotide sequence alignment of the Lyl1 proximal promoter region (species as described in Figure 1). Conserved Ets binding sites (EBS), GATA binding sites (GBS), and the GGCC motif as well as the 2 transcriptional start sites (curved arrows) are indicated. The arrowhead designates the cloning site used to separate the 2 core promoters Lyl1P1 and Lyl1P2. (C) The Lyl1 proximal promoter contains 2 core promoters. Shown on the left are the reporter constructs in which either the entire Lyl1P proximal promoter region or the 2 core promoters Lyl1P1 and Lyl1P2 were inserted upstream of the luciferase gene in the promoterless pGL2B vector. Following from left to right are the results of transient transfection assays in MS1, 416B, and L8057 showing luciferase activities corrected for transfection efficiency with the pEF-BOS LacZ plasmid. The luciferase activities are presented as fold increase over the activity of the control (pGL2B) vector, which was assigned a value of 1. Each bar represents the mean relative luciferase activity from at least 2 experiments performed in triplicate ± SD.
A close inspection of the sequence alignment of the 464 bp Lyl1P transgenic fragment revealed several blocks of high sequence conservation in both the 5′ and 3′ portions of this fragment with relatively less conservation in the central core (Figure 2B). The DataBase of Transcriptional Start Sites (DBTSS32 ) contains 2 distinct clusters of start sites for the murine Lyl1 gene separated by approximately 300 bp (Figure 2B, arrows), suggesting that the Lyl1P fragment may contain 2 distinct minimal promoter elements. To directly test this possibility, luciferase reporter constructs containing the entire 464 bp as well as the 5′ 273 bp and 3′ 191 bp of the Lyl1P fragment were generated and transfected into MS1, 416B, and L8057 cells (Figure 2C). These experiments showed that the whole Lyl1P fragment had significant promoter activity in all 3 cell lines. Moreover, the 5′ subfragment Lyl1P1 was active in 416B and L8057, and the 3′ Lyl1P2 fragment was active in all 3 cell lines. These experiments therefore demonstrated that the Lyl1 promoter region contains 2 independent core promoters.
Evolutionarily conserved sequence motifs of the Lyl1 promoter region are occupied in vivo in hematopoietic cells
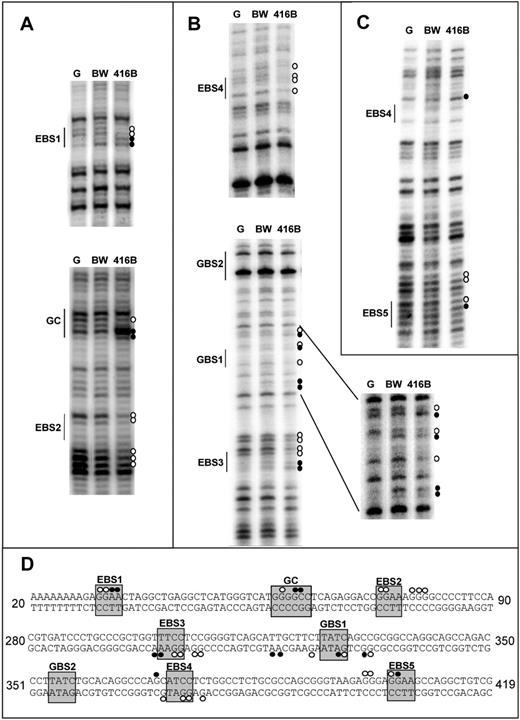
The comparative sequence analysis discussed in the previous section revealed several blocks of sequence conservation within the Lyl1 P1 and P2 promoter regions, including 2 conserved Ets family binding sites in promoter P1 as well as 3 conserved Ets and 2 conserved GATA binding sites in promoter P2 (Figure 2 B). To assess whether these or any of the other conserved sequence blocks were targets for protein binding in vivo, the P1 and P2 promoter regions were studied using in vivo DMS footprinting, which highlights purine (mostly G(N7)) contacts between proteins and DNA, with both protections and enhancements of reactivity indicating protein-DNA binding at single nucleotide resolution. Figure 3 shows representative experiments comparing the Lyl1-expressing hematopoietic progenitor cell line 416B with the nonexpressing T-cell line BW5147 and the control naked genomic DNA. The data are summarized in Figure 3D and represent at least 3 independent experiments from 2 independent cell/genomic DNA preparations.
In vivo DMS footprinting of the Lyl1 promoter proximal region reveals extensive protections and enhancements of DMS reactivity on both DNA strands in hematopoietic progenitor (416B) but not T-cell (BW) cell lines. (A) In vivo DMS footprinting of Lyl1-P1 core promoter (sense strand). Protections (○) and enhancements (•) were observed over both Ets binding sites (EBS1 and EBS2) as well as the conserved GGCC motif in 416B compared with naked DNA (G reaction; G) and the nonexpressing T-cell line BW. (B) In vivo DMS footprinting of Lyl1-P2 core promoter (antisense strand). Protections (○) and/or enhancements (•) were observed over the 3 conserved Ets binding sites (EBS3, EBS4, and EBS5) as well as 1 of the conserved GATA sites (GBS1) in 416B compared with naked DNA (G reaction; G) and the nonexpressing T-cell line BW. (C) In vivo DMS footprinting of Lyl1-P2 core promoter (sense strand). Protections (○) and/or enhancements (•) indicate the positions of conserved Ets binding sites EBS4 and EBS5. (D) The DNA sequence of the mouse Lyl1 promoter proximal region with protections and enhancements is indicated as white and black circles, respectively. The numbers indicate the nucleotide positions relative to the alignment shown in Figure 2B.
In vivo DMS footprinting of the Lyl1 promoter proximal region reveals extensive protections and enhancements of DMS reactivity on both DNA strands in hematopoietic progenitor (416B) but not T-cell (BW) cell lines. (A) In vivo DMS footprinting of Lyl1-P1 core promoter (sense strand). Protections (○) and enhancements (•) were observed over both Ets binding sites (EBS1 and EBS2) as well as the conserved GGCC motif in 416B compared with naked DNA (G reaction; G) and the nonexpressing T-cell line BW. (B) In vivo DMS footprinting of Lyl1-P2 core promoter (antisense strand). Protections (○) and/or enhancements (•) were observed over the 3 conserved Ets binding sites (EBS3, EBS4, and EBS5) as well as 1 of the conserved GATA sites (GBS1) in 416B compared with naked DNA (G reaction; G) and the nonexpressing T-cell line BW. (C) In vivo DMS footprinting of Lyl1-P2 core promoter (sense strand). Protections (○) and/or enhancements (•) indicate the positions of conserved Ets binding sites EBS4 and EBS5. (D) The DNA sequence of the mouse Lyl1 promoter proximal region with protections and enhancements is indicated as white and black circles, respectively. The numbers indicate the nucleotide positions relative to the alignment shown in Figure 2B.
Figure 3A shows the sense strand of the P1 promoter, with both conserved ETS sites (EBS1 and EBS2) and an intervening conserved GGCC region occupied in the Lyl1-expressing 416B cells but not the nonexpressing BW5147 cells. Figure 3B-C shows the antisense and sense strands of the P2 promoter, respectively. Again, with 416B cells, there is evidence of occupancy of each of the conserved ETS binding sites (EBS3, EBS4, EBS5), with an additional footprint observed at one of the conserved GATA sites (GBS1). No evidence of protein binding was observed at the second conserved GATA site (GBS2) in any of the repeat experiments.
The GGCC motif identified by in vivo footprinting did not conform to any consensus binding sites present in the JASPAR database of manually curated eukaryotic transcription factor binding consensus sequences,33 suggesting that an as yet uncharacterized transcription factor might be involved in Lyl1 regulation. Taken together, the footprinting experiments demonstrated that all the conserved ETS binding sites and one of the conserved GATA sites are occupied in vivo, thus identifying the Ets and GATA transcription factor families as likely regulators of Lyl1.
Activity of Lyl1 promoters P1 and P2 depends on Ets and GATA motifs bound in vivo by Fli1, Elf1, Erg, PU.1, and GATA2
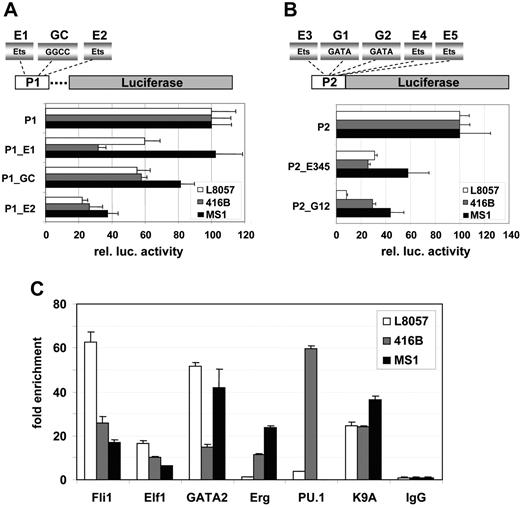
To assess the potential significance of the Ets, GGCC, and GATA motifs identified by comparative sequence analysis and in vivo footprinting, we mutated these motifs in the context of the Lyl1-P1 or Lyl1-P2 reporters. Promoter activity of wild-type and mutant Lyl1-P1 constructs was quantified using transient transfection assays in L8057, 416B and MS1 (Figure 4A). Mutation of the first Ets site and the GGCC motif (P1_E1 and P1_GC constructs) resulted in 40% to 70% reduction in luciferase activity in L8057 and 416B cells but no significant reduction in MS1 cells. Mutation of the second Ets site (P1_E2 construct) produced a 60% to 80% reduction of activity in all 3 cell lines (Figure 4A). These results demonstrate that activity of the Lyl1 P1 promoter depended on intact GGCC and Ets motifs.
The Lyl1 core promoters depend on conserved Ets, GATA, and GGCC motifs, and the Lyl1 promoter proximal region is bound in vivo by Ets and GATA factors. (A) The 2 Ets binding sites and the GGCC motif are important for activity of the Lyl1P1 promoter. Shown are the results from reporter assays of a series of Lyl1P1 promoter mutation constructs in which either the 2 Ets binding sites (P1_e1 and P1_E2) or the GGCC motif (P1_GC) were mutated. Constructs were transfected into L8057, 416B, and MS1 cells as described in Figure 2. (B) Ets and GATA binding sites are necessary for activity of the Lyl1P2 promoter. Shown are the results from reporter assays of Lyl1P2 promoter constructs in which either all 3 Ets (P2_E345) or the 2 GATA binding sites (P2_G12) were mutated. Constructs were transfected into L8057, 416B, and MS1 cells as described in Figure 2. (C) Fli-1, Elf-1, GATA2, Erg, and PU.1 bind the Lyl1 proximal promoter region. Chromatin immunoprecipitation assays were performed in L8057, 416B, and MS1 cell lines with anti-Fli1, -Elf2, -GATA2, -Erg, -PU.1, -GATA2, –acetylated lysine K9 of histone H3, and control IgG antibodies. The DNA content of the immunoprecipitates was analyzed by real-time PCR. The level of enrichment with each antibody was normalized to the levels obtained with the control IgG and plotted as fold increase over the level of enrichment at a control region (see “Materials and methods”). MS1 cells do not express PU.1. Error bars indicate SD.
The Lyl1 core promoters depend on conserved Ets, GATA, and GGCC motifs, and the Lyl1 promoter proximal region is bound in vivo by Ets and GATA factors. (A) The 2 Ets binding sites and the GGCC motif are important for activity of the Lyl1P1 promoter. Shown are the results from reporter assays of a series of Lyl1P1 promoter mutation constructs in which either the 2 Ets binding sites (P1_e1 and P1_E2) or the GGCC motif (P1_GC) were mutated. Constructs were transfected into L8057, 416B, and MS1 cells as described in Figure 2. (B) Ets and GATA binding sites are necessary for activity of the Lyl1P2 promoter. Shown are the results from reporter assays of Lyl1P2 promoter constructs in which either all 3 Ets (P2_E345) or the 2 GATA binding sites (P2_G12) were mutated. Constructs were transfected into L8057, 416B, and MS1 cells as described in Figure 2. (C) Fli-1, Elf-1, GATA2, Erg, and PU.1 bind the Lyl1 proximal promoter region. Chromatin immunoprecipitation assays were performed in L8057, 416B, and MS1 cell lines with anti-Fli1, -Elf2, -GATA2, -Erg, -PU.1, -GATA2, –acetylated lysine K9 of histone H3, and control IgG antibodies. The DNA content of the immunoprecipitates was analyzed by real-time PCR. The level of enrichment with each antibody was normalized to the levels obtained with the control IgG and plotted as fold increase over the level of enrichment at a control region (see “Materials and methods”). MS1 cells do not express PU.1. Error bars indicate SD.
To circumvent possible redundancy of multiple sites within the Lyl1 P2 promoter, constructs with mutations in all 3 Ets motifs or the 2 GATA motifs were assayed in L8057, 416B, and MS1 (Figure 4B). Mutation of all 3 Ets sites (P2_E123 construct) resulted in a 70% reduction in luciferase activity in L8057 and 416B cells and a 40% reduction in MS1 cells. Mutation of the 2 GATA sites (P2_G12 construct) produced a 90% reduction of activity in L8057 cells, 70% reduction in 416B, and 60% reduction in MS1 (Figure 4 B). These assays therefore identified a critical role for Ets and GATA motifs in regulating activity of the Lyl1 P2 promoter in all 3 cell types.
We had shown previously that GATA-2 and the Ets family transcription factors Fli-1, Erg, and Elf-1 regulate expression of Scl in endothelial and blood progenitor cells.18,22,34 To investigate whether these factors as well as Pu.1 are involved in regulating Lyl1 expression, we performed chromatin immunoprecipitation (ChIP) assays in L8057, 416B, and MS1 cells. Rabbit IgG and an antibody against acetylated lysine K9 of histone H3 served as negative and positive controls, respectively. Immunoprecipitated chromatin samples were analyzed by quantitative real-time PCR. The levels of enrichment were normalized to that obtained with the control rabbit IgG and plotted as a fold increase over that measured at a control region (see “Materials and methods”). These experiments demonstrated that Fli1, Elf1, and GATA-2 were bound to the Lyl1 promoter in all 3 cell lines. Erg was bound in 416B and MS1, and PU.1 strongly bound in 416B and to a much lesser extent in L8057 (Figure 4C). None of the Ets and GATA factors analyzed here were bound to the Lyl1 promoter in NIH3T3 cells, which do not express Lyl1. Taken together, our results suggest that the molecular mechanisms controlling Lyl1 expression in progenitor, megakaryocytic, and endothelial cells are highly related (ie, GATA and Ets dependent), yet the precise combinations of Ets factors bound to the Lyl1 promoter vary between different lineages.
The Lyl1 promoter proximal region is active in early hematopoietic progenitors
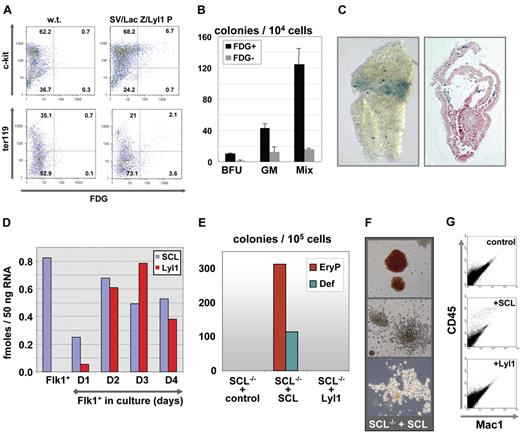
The mutation and ChIP studies suggested that Lyl1 and Scl operate within the same transcriptional hierarchy, which would be consistent with a model in which these genes may compensate for each other in hematopoietic stem/progenitor cells. To directly investigate whether the Lyl1 promoter targeted hematopoietic progenitor cells, we characterized the surface marker phenotype of lacZ-expressing fetal liver cells from SV/lac/Lyl1P transgenic mice. FACS analysis demonstrated that approximately 5% of E11.5 fetal liver cells expressed the lacZ reporter gene (Figure 5A). Most lacZ-expressing cells coexpressed the hematopoietic progenitor marker c-kit. Less than half of lacZ-expressing cells coexpressed the Ter119 marker for differentiated erythroid cells. These results are therefore consistent with the hypothesis that cells targeted in vivo by the Lyl1 promoter proximal fragment include hematopoietic progenitor cells.
The Lyl1 promoter region and endogenous Lyl1 are expressed early during hematopoietic specification, yet Lyl1 cannot compensate for loss of Scl in hematopoietic differentiation assays using Scl−/−ES cells. (A) Flow cytometric analysis of E11.5 fetal liver demonstrates that enhancer-positive cells are largely c-kit+ and Ter119− consistent with a hematopoietic progenitor phenotype. FDG indicates fluorescein di-β-D-galactopyranoside fluorescent β-galactosidase/lacZ substrate. (B) E11.5 fetal liver cells expressing the Lyl1P transgene are enriched for hematopoietic progenitors; lacZ-positive and lacZ-negative cells were sorted by FACS and assessed for hematopoietic colony-forming activity. BFU indicates erythroid burst-forming units; GM, granulocyte-macrophage colonies; Mix, multipotent colonies. Error bars indicate SD. (C) Expression of the Lyl1P transgene in E7.5 mouse embryos; X-gal staining of a representative E7.5 transgenic embryo. Staining in the extraembryonic mesoderm seen by wholemount analysis (left panel) was confirmed on histologic sections (right panel). (D) Analysis of Scl and Lyl1 expression by real-time PCR at the onset of hematopoietic development. Flk1+ cells were isolated from day 3.3 embryoid bodies generated from E14.1 ES cells and grown in hemangioblast conditions for up to 4 days. RNA and cDNA were prepared for Flk1+ cells and day 1 to day 4 cultures and analyzed by real-time PCR. The results are represented as femtomole template per 50 ng total RNA (see “Materials and methods”) and are representative of 3 independent experiments. (E) Scl−/− ES cells were transfected with MSCV constructs expressing Scl or Lyl1 cDNA or MSCV control vector. Day 5 embryoid bodies derived from transfected ES cells were harvested, and single-cell suspensions were replated in methylcellulose-containing cytokines to induce hematopoietic colony formation. EryP indicates primitive erythroid colonies; Def, definitive hematopoietic colonies including macrophage, macrophage-erythrocyte, mix, and granulocyte-macrophage colonies. (F) Representative pictures of Scl-rescued colonies showing primitive colonies (top panel) and definitive colonies (middle and bottom panels). (G) Colonies from methylcellulose cultures were harvested, stained for CD45 and Mac1 expression, and analyzed by flow cytometry.
The Lyl1 promoter region and endogenous Lyl1 are expressed early during hematopoietic specification, yet Lyl1 cannot compensate for loss of Scl in hematopoietic differentiation assays using Scl−/−ES cells. (A) Flow cytometric analysis of E11.5 fetal liver demonstrates that enhancer-positive cells are largely c-kit+ and Ter119− consistent with a hematopoietic progenitor phenotype. FDG indicates fluorescein di-β-D-galactopyranoside fluorescent β-galactosidase/lacZ substrate. (B) E11.5 fetal liver cells expressing the Lyl1P transgene are enriched for hematopoietic progenitors; lacZ-positive and lacZ-negative cells were sorted by FACS and assessed for hematopoietic colony-forming activity. BFU indicates erythroid burst-forming units; GM, granulocyte-macrophage colonies; Mix, multipotent colonies. Error bars indicate SD. (C) Expression of the Lyl1P transgene in E7.5 mouse embryos; X-gal staining of a representative E7.5 transgenic embryo. Staining in the extraembryonic mesoderm seen by wholemount analysis (left panel) was confirmed on histologic sections (right panel). (D) Analysis of Scl and Lyl1 expression by real-time PCR at the onset of hematopoietic development. Flk1+ cells were isolated from day 3.3 embryoid bodies generated from E14.1 ES cells and grown in hemangioblast conditions for up to 4 days. RNA and cDNA were prepared for Flk1+ cells and day 1 to day 4 cultures and analyzed by real-time PCR. The results are represented as femtomole template per 50 ng total RNA (see “Materials and methods”) and are representative of 3 independent experiments. (E) Scl−/− ES cells were transfected with MSCV constructs expressing Scl or Lyl1 cDNA or MSCV control vector. Day 5 embryoid bodies derived from transfected ES cells were harvested, and single-cell suspensions were replated in methylcellulose-containing cytokines to induce hematopoietic colony formation. EryP indicates primitive erythroid colonies; Def, definitive hematopoietic colonies including macrophage, macrophage-erythrocyte, mix, and granulocyte-macrophage colonies. (F) Representative pictures of Scl-rescued colonies showing primitive colonies (top panel) and definitive colonies (middle and bottom panels). (G) Colonies from methylcellulose cultures were harvested, stained for CD45 and Mac1 expression, and analyzed by flow cytometry.
To directly assess the biologic function of hematopoietic cells targeted by the Lyl1P promoter proximal fragment, we performed colony assays on lacZ-expressing cells isolated by FACS using the fluorescent β-galactosidase substrate FDG. The frequency of erythroid, myeloid, and mixed colonies was found to be 3- to 10-fold higher in FDG-positive relative to FDG-negative fractions (Figure 5B). These results were consistent with our immunophenotype analysis and directly demonstrated that the Lyl1P fragment directed lacZ expression to a significant proportion of fetal liver erythroid and myeloid progenitors. In addition, histologic analysis of E7.5 transgenic embryos demonstrated that the Lyl1 promoter proximal region was already active at this early stage of development where lacZ expression was observed in mesodermal cells within the extraembryonic region, which will form the yolk sac blood islands (Figure 5C).
Lyl1 cannot compensate for the early hematopoietic defect in Scl−/− ES cells
The results described in the prevous section are consistent with possible compensation between Lyl1 and Scl at the fetal liver stage but raise the question of why Lyl1 does not compensate for loss of Scl during the earliest stages of hematopoietic specification. These earliest stages are best defined in ES cell differentiation systems.35 The earliest identified hematopoietic precursors, the hemangioblasts, are present in embryoid bodies between 2.5 and 4 days of differentiation and express Flk1. Hemangioblast precursors can be induced to develop further into endothelium and more mature hematopoietic precursors in response to VEGF. To assess Scl and Lyl1 expression during these earliest stages of hematopoietic specification, RNA was isolated from Flk1+ cells sorted from day 2.5 embryoid bodies and also from the same Flk1+ cells cultured for 1, 2, 3, or 4 days in the presence of VEGF in methylcellulose. The levels of Lyl1 and Scl transcripts were measured using quantitative RT-PCR (Figure 5D). This analysis demonstrated that both Lyl1 and Scl were induced early during hematopoietic specification. However, in sharp contrast to Scl, Lyl1 expression was never detected in the Flk1+ hemangioblast population and initiated 1 day later than Scl when hemangioblasts were induced to differentiate (Figure 5D). These results therefore suggested that the delayed expression of Lyl1 may be responsible for its inability to compensate for loss of Scl in Scl−/− ES cell differentiation assays. However, they did not exclude the possibility that functional differences between Lyl1 and Scl proteins may also contribute to the failure of Lyl1 to compensate for loss of Scl.
We therefore tested directly whether Lyl1 protein could rescue the hematopoietic defect in Scl−/− ES cells. To this end we transduced Scl−/− ES cells with retroviral constructs expressing Scl and Lyl1, respectively, and subjected transduced cells to hematopoietic differentiation assays. As reported before,9,36 transduction of Scl into Scl−/− ES cells resulted in robust hematopoietic rescue with the appearance of both primitive and definitive hematopoietic cells (Figure 5E-G). By contrast, no hematopoietic colony-forming cells could be detected when Scl−/− cells were transduced with Lyl1-expressing retrovirus even though expression levels of Lyl1 and Scl in transduced Scl−/− cells were comparable (Figure S5). The same results were obtained using 2 independently derived Scl−/− ES cell lines and MSCV expression vectors encoding tagged as well as nontagged Scl and Lyl1 proteins (data not shown). These results therefore demonstrate that Lyl1 expression during early hematopoietic specification is later in onset and lower in magnitude than Scl, consistent with an early window of hematopoietic development specifically controlled by Scl. Moreover, the rescue experiments identified important functional differences between Scl and Lyl1 proteins.
Discussion
Ectopic expression of Lyl1 in T cells is associated with the development of T-cell leukemia,3,37 while Lyl1−/− adult HSCs show severe functional defects.17 Appropriate transcriptional regulation of Lyl1 therefore is critical for both normal blood stem cell function as well as preventing the development of hematologic malignancies. In this paper, we provide the first detailed analysis of the transcriptional regulation of the mouse Lyl1 gene during mouse embryogenesis. We also provide new insights into the earliest stages of blood specification by demonstrating why Lyl1 is unable to compensate for loss of Scl during ES cell in vitro differentiation.
Using extensive transgenic and molecular analyses, we have characterized the mechanisms responsible for normal expression of Lyl1 in endothelial, hematopoietic progenitor, and megakaryocytic cells. A fragment of fewer than 500 bp encompassing 2 closely spaced promoters with functionally important Ets and GATA sites was active in all 3 cell types. A close developmental relationship between endothelium and blood stem/progenitor cells has long been recognized.38 Several other regulatory elements identified by us (in the Scl, Fli1, and Hex gene loci18,39 ) and others (ie, in the Gata-2 gene locus40 ) are also active in blood progenitor/stem cells and endothelium. Somewhat less expected is our observation that a single element active in hematopoietic progenitor/endothelial cells was also active in megakaryocytes. However, other parallels between endothelium, hematopoietic stem/progenitor cells, and megakaryocytes have been identified previously. For example, human endothelial cells and megakaryocyte precursors express the hematopoietic stem/progenitor surface markers CD34 and c-kit.41,42 Moreover, within the myeloid compartment GATA and Ets factors cross-antagonize each other during erythroid and granulocyte/monocyte differentiation, respectively,43,44 whereas megakaryocytes, just like HSCs, are known to depend on simultaneous GATA and Ets activity.45
Our data demonstrate that within endothelial and hematopoietic progenitor cells, Lyl1 and Scl operate in parallel, both being controlled by GATA-2 and several Ets factors. Interestingly, the recent analysis of conditional Scl-null mice demonstrated that Scl is largely dispensable for HSC function.14 Moreover, even though Scl initially seemed absolutely required for red blood cell differentiation,16 it has recently been reported that, over time, mice are able to compensate for loss of Scl.46 The authors of the latter study speculated that Lyl1 might be compensating for loss of Scl. A comparative discussion of Scl−/− and Lyl1−/− HSCs also concluded that Scl and Lyl1 are functionally related in adult HSCs yet play distinct roles during B-lymphoid and myeloid development.17 Our new data, which suggest that Lyl1 and Scl operate in parallel within a GATA/Ets transcriptional hierarchy, would be consistent with compensatory mechanisms, at least at the progenitor stages. A genetic interaction between Lyl1 and Scl is also implied by our recent observation that, at least on a mixed genetic background, Lyl1−/−/Scl+/− mice die shortly after birth (S.K. and B.G., unpublished observations, January 2005).
Using an ES cell differentiation assay, which is able to capture the earliest stages of mesoderm specification toward the hematopoietic lineage, we were able to demonstrate that Scl expression is initiated prior to expression of Lyl1. It is therefore possible that the initiation of Lyl1 expression represents the time point when Ets/GATA-responsive endothelial/hematopoietic regulatory elements are activated. Accordingly, initiation of Scl expression in ES cell differentiation assays was unaffected when the Ets/GATA responsive +19 enhancer was deleted from the Scl gene locus.22 This finding suggested that there may be as yet unidentified regulatory pathways responsible for initiating Scl expression during the specification of hematopoietic cells from developing mesoderm. Further characterization of these pathways is likely to reveal key aspects of the earliest stages of blood cell development.
The results presented in the current paper also lay the foundation for future studies aiming to identify specific perturbations of transcriptional pathways in patients with Lyl1-positive T-ALL. Expression analyses of large cohorts of T-ALL patients by quantitative RT-PCR and microarray expression profiling demonstrated that T lymphoblasts in 22% to 35% of T-ALL patients express high levels of Lyl1 without any overt chromosomal abnormalities involving the Lyl1 gene locus.47,48 Importantly, Lyl1 expression was linked with bad response to treatment and poor prognosis.48 Identification of the mechanisms underlying ectopic expression in T-ALL T cells therefore has potential therapeutic implications. Moreover, elevated levels of Lyl1 expression have recently also been implicated in the drug resistance and disrupted differentiation of acute myeloblastic leukemia (AML),49 suggesting that transcriptional control of Lyl1 may represent a potential drug target in T-ALL as well as AML.
Despite the wealth of published data on the role of Lyl1 as a powerful leukemia oncogene, comparatively little has been published on the function of the normal Lyl1 protein. Lyl1 and Scl share more than 90% sequence identity within the bHLH DNA binding domain. Not unsurprisingly, therefore, proteins such as LMO2 and E2A, which have been shown to interact with Scl through the bHLH domain, also bind to Lyl1.6 Moreover, the hematopoietic phenotype in Scl−/− ES cells could be rescued by expressing the Scl bHLH domain alone as well as a hybrid protein containing the Scl N- and C-terminal domains flanking the Lyl1 bHLH domain.36 Our observation that full-length Lyl1 was unable to rescue the same phenotype was therefore surprising. Importantly, our results indicate that sequences outside the bHLH domain (which are not conserved between Scl and Lyl1) confer distinct functions on the 2 proteins. The notion of Lyl1- or Scl-specific protein interactions is consistent with a previous report that demonstrated that NF-κB1 p105 interacts with Lyl1 but not Scl.50 Given that the Scl bHLH region alone could rescue hematopoiesis,36 it is possible that N- or C-terminal domains of Lyl1 destabilize one or more crucial protein-protein interactions. Identification of potentially Scl-specific protein complexes involved in early hematopoietic specification will be important to understand how Scl specifies HSC formation. Moreover, putative Scl-specific complexes would be consistent with the very different phenotypes of Scl and Lyl1 during the early stages of HSC specification.
Authorship
Contribution: W.Y.I.C., G.A.F., G.L., J.E.P., J.-R.L., S.K., K.K., S.P., I.J.D., V.K., L.G., and B.G. performed the research and analyzed the data; and W.Y.I.C., A.R.G., F.S., V.K., and B.G. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Berthold Göttgens, Department of Haematology, Cambridge Institute for Medical Research, Cambridge University, Hills Road, Cambridge, CB2 2XY, United Kingdom; e-mail: bg200@cam.ac.uk.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by the Leukemia Research Fund (LRF), Wellcome Trust, Biotechnology and Biological Sciences Research Fund (BBSRC), and Cancer Research UK (CRUK). J.-R.L holds a fellowship from the Canadian Institutes of Health Research, and J.E.P. is a CJ Martin/RG Menzies fellow of the NHMRC of Australia. We are grateful to Glenn Begley and Stuart Orkin for the 2 Scl−/− ES cell lines and to Catherine Porcher for the pMSCV-Scl-puro construct.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal