Abstract
Ape1 is a molecule with dual functions in DNA repair and redox regulation of transcription factors. In Ape1-deficient mice, embryos do not survive beyond embryonic day 9, indicating that this molecule is required for normal embryo development. Currently, direct evidence of the role of Ape1 in regulating hematopoiesis is lacking. We used the embryonic stem (ES) cell differentiation system and an siRNA approach to knockdown Ape1 gene expression to test the role of Ape1 in hematopoiesis. Hemangioblast development from ES cells was reduced 2- to 3-fold when Ape1 gene expression was knocked down by Ape1-specific siRNA, as was primitive and definitive hematopoiesis. Impaired hematopoiesis was not associated with increased apoptosis in siRNA-treated cells. To begin to explore the mechanism whereby Ape1 regulates hematopoiesis, we found that inhibition of the redox activity of Ape1 with E3330, a specific Ape1 redox inhibitor, but not Ape1 DNA repair activity, which was blocked using the small molecule methoxyamine, affected cytokine-mediated hemangioblast development in vitro. In summary, these data indicate Ape1 is required in normal embryonic hematopoiesis and that the redox function, but not the repair endonuclease activity, of Ape1 is critical in normal embryonic hematopoietic development.
Introduction
Ape1/Ref-1 is a multifunctional protein involved in apurinic/apyrimidinic endonuclease DNA base excision repair activity, in proofreading exonuclease activity, and in modulating DNA binding activity of several transcription factors including NF-kB, Egr-1, p53, AP-1, CREB, HIF-α, and members of the Pax family.1 Ape1 acts on apurinic/apyrimidinic (AP) sites in DNA as a major member of the base excision repair (BER) pathway and is involved in oxidative DNA damage repair.2 Ape1 is ubiquitously expressed at high basal levels, and in previous studies to determine the function of Ape1 in vivo, Ape1 was disrupted using a gene targeting strategy.3 Embryos lacking expression of Ape1 die in utero between implantation and day 6.5, indicating that Ape1 is required for normal embryonic development. However, this approach does not allow for the determination of whether it is the redox or repair function of Ape1 that is crucial for embryonic development. Furthermore, this early lethality precludes analysis of the role of Ape1 in hematopoiesis.
Mouse embryonic stem (ES) cells are derived from the inner cell mass of the 3.5-day-old blastocyst.4,5 ES cells retain the differentiation ability of the inner cell mass after many passages in the presence of leukemia inhibitory factor (LIF).6 Removal of LIF results in ES cell differentiation into embryoid bodies (EBs).7 Using the ES/EB system, a precursor that responds to vascular endothelial growth factor (VEGF) and generates colonies consisting of undifferentiated blast cells was identified.8 These VEGF-responsive blast cell colonies were shown to contain endothelial, primitive erythroid, and various definitive hematopoietic precursors. These cells are referred to as blast colony-forming cells (BL-CFCs) and are thought to be cells that represent the hemangioblast precursor of blood and endothelial lineages.
Ape1 siRNA has previously been used to knock down target gene expression in several cell types.9–11 In this study, we used siRNA to knock down Ape1 gene expression in EB cells and discovered that reduction of Ape1 by siRNA results in a significant decrease in the frequency of hemangioblast formation and diminished formation of primitive and definitive hematopoietic colonies. Additionally, using an Ape1-specific redox inhibitor, E3330, we demonstrate that it is the redox function of Ape1 and not the repair activity that is involved in hematopoietic differentiation. These data suggest that Ape1 may play a critical role in embryonic hematopoiesis.
Materials and methods
Cells and reagents
The mouse CCE ES cells (passage no. 10) were kindly provided by Bill Carter in the mouse transgene and knockout core facility in our institute (Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis). Cytokines LIF, VEGF, SCF, FGF, and methylcellulose-based ES cell differentiation medium and collagenase were purchased from Stem Cell Technology (Vancouver, BC, Canada). Ape1 antibodies were purchased from Novus Biologicals (Littleton, CO). FITC-labeled anti–mouse Flk-1 mAb was purchased from BD Pharmingen (San Diego, CA). Oligofectamine 2000 was purchased from Invitrogen (Carlsbad, CA). All Ape1 dsRNA and scrambled Ape1 siRNAs were commercially obtained from Dharmacon (Lafayette, CO). The 21-base sequence was subjected to a BLAST12 search (NCBI) database of EST libraries to ensure that only one gene was targeted. Sense strand of RNA oligonucleotides corresponding to Ape1 (GTCTGGTAAGACTGGAATACC) was chemically synthesized using standard methods and high-performance liquid chromatography (HPLC) purification (Dharmacon). E3330, the redox-specific inhibitor of Ape1 molecule, was provided by Dr Rick Borch and Rod Nyland in the Department of Medical Chemistry, Purdue University (Lafayette, IN).
In vitro differentiation of ES cells
CCE ES cells were maintained on gelatinized tissue-culture dishes (100 mm; Costar, Cambridge, MA) in standard ES culture medium consisting of Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal calf serum (FCS; GIBCO, Grand Island, NY), 0.1 mM l-glutamine, 150 mM monothioglycerol (MTG), penicillin 100 U/mL, streptomycin 100 mg/mL, and LIF 1000 U/mL. The culture medium was changed every day, and cells were passaged every 2 or 3 days.13 To initiate differentiation into embryonic bodies (EBs), dissociated ES cells were added to IMDM medium (Invitrogen), 15% fetal bovine serum (FBS, HCC6900; Stem Cell Technology), 100 ng/mL stem cell factor (R&D System, Minneapolis, MN), and 450 μM monothioglycerol (Sigma, St Louis, MO) at a cell concentration of 5 to 10 × 103 cells/mL plated in 100-mm low-adhesion dishes.
Harvesting EBs
EBs were removed from liquid medium IMDM. Three-day-old EBs were collected, washed twice with PBSA (PBS with 1% albumin), and resuspended in 0.25% (wt/vol) trypsin mix in 15% FBS containing PBSA. The EBs were incubated at 37°C for 5 minutes and dissociated into a single-cell suspension by passing through a 20-gauge needle. Remaining small aggregates were removed by filtration through a 40-μm mesh (Falcon; Becton Dickinson, Lincoln Park, NJ). EB cells were spun and resuspended in 0.5% BSA (Path-O-Cyte 4; Miles, Kankakee, IL) in PBSA at concentration of 5 × 106 cells/mL.
Transfection of EB cells with oligos of dsRNA
Transient transfection of siRNAs was carried out as we previously reported.7,14 Total EB cells were diluted with fresh medium without antibiotics and transferred to 12-well plates at 1 × 105 cells/well (500 μL per well). Transient transfection of dsRNAs was carried out using Oligofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were untreated (control), or treated with scrambled Ape1 siRNA (25-50 nM) or Ape1 siRNA (25-50 nM).
Clonogenic cell assay
For hemangioblast assay, after siRNA transfection, day-3 EB cells were mixed with IMDM, 20% FCS, 10% BSA (Stem Cell Technology), 100 μg/mL bovine transferrin, 10 ng/mL bovine insulin, 0.1 mM MTG, 1% methylcellulose, and distributed to 35-mm bacterial-grade dishes at 104 cells/plate with VEGF, SCF, and DT4 conditional medium (CM). For primitive erythroid cell (Ery-P) assay, day-6 EB cells were cultivated with EPO at 5 IU/mL as previously described.15 For granulocyte-macrophage (CFU-GM) colony assay, day-10 EB cells were cultured with granulocyte-macrophage colony-stimulating factor (GM-CSF) at 10 ng/mL and macrophage colony-stimulating factor (M-CSF) at 5 ng/mL.15
Reverse-transcription–polymerase chain reaction (RT-PCR)
Total RNA was extracted from undifferentiated ES cells and EB cell populations using TRIZOL (Invitrogen), reverse transcription was performed using Superscript II reverse transcriptase (GIBCO, Carlsbad, CA), and PCR was performed using pyrobest DNA polymerase (Fisher Scientific, Pittsburgh, PA). Primer sequences for T, Flk1, and Scl genes have been reported.8 The primer sequence of β-actin gene has been reported.16
Western blot analysis
Whole cell extracts were prepared using nuclear protein extract buffer17 from both Ape1-siRNA– or scrambled siRNA–treated and untreated cells. The cell lysates were separated by SDS–polyacrylamide electrophoresis using a 10% (wt/vol) polyacrylamide resolving gel and transferred electrophoretically to a nitrocellulose membrane. The blots were blocked with 5% TBS/T buffer for 1 hour. Immunoblotting was performed using the Ape1 primary monoclonal antibody (Novus Biologicals) at a 1:400 dilution at room temperature, and the peroxidase-conjugated secondary Abs (Amersham Pharmacia, Arlington Heights, IL) overnight at 4°C. This antibody has been extensively used by us and others.11,18–22 All immunoblots were visualized by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech, Piscataway, NJ).
Inhibition of DNA repair domain of Ape1 with MX and redox domain with E3330
E3330 is a specific inhibitor of redox activity of Ape1 protein, while MX is a specific inhibitor of the DNA repair (AP endonuclease) function of Ape1 protein. MX does not specifically bind to Ape1, but binds to the abasic site that is recognized by Ape1 and prevents Ape1's ability to cleave the AP site.23–27
E3330 has been shown to be a very specific inhibitor of Ape1 redox function. Far-Western blots, and binding assays between radiolabeled E3330 and proteins renatured on membrane blots, demonstrated that 14C-labeled E3330 specifically bound to both recombinant Ape1 and purified Ape1 from cell nuclear extracts.28 The binding constant of Ape1 and E3330 was estimated by surface plasmon resonance analysis (SPR) resulting in a kinetic constant value of 1.6 nM, which suggests a specific interaction between Ape1 and E3330.28 Therefore, E3330 is a specific inhibitor of Ape1 redox function. E3330 does not affect the repair function of Ape1 (M.R.K. and M.-H.L., unpublished data, September 2005).
To verify the role of Ape1 in hemangioblast development via redox domain or DNA repair domain, various doses of E3330 (10-100 μM) or MX (1-10 mM) were added to standard hemangioblast culture. Hemangioblast colonies were scored after 4 days of culture.
Cell-cycle analysis
EB cells were treated with either scrambled siRNA or Ape1 siRNA. After 48 hours of culture, cells were washed with PBS, resuspended in 400 μL hypotonic buffer (0.15% Triton-X100 and 20 μg/mL RNase A) containing 50 μg/mL propidium iodide (PI), and analyzed using FACScan (BD Bioscience, Mountain View, CA).
Results
Ape1 is expressed in both undifferentiated and differentiated ES cells
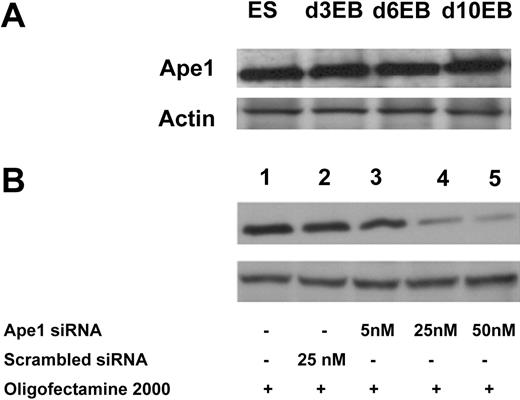
To examine whether Ape1 is expressed in ES cells and differentiated ES cells, we isolated total cell protein from ES cells, and day-3, -6, or -10 EB cells. Western blot analysis showed that Ape1 protein is detectable in both ES cells and EB cells (Figure 1A). Further experiments showed that Ape1 siRNA, but not scrambled control siRNA, is effective to knock down Ape1 gene expression in EB cells when probed by Western blot analysis after 48 hours of siRNA transfection (Figure 1B).
Ape1 siRNA is efficient to knock down target gene expression in EB cells. (A) Western blot analysis of Ape1 protein expression in both ES and EB cells at different ages (days 3, 6, and 10). (B) Silencing of endogenous Ape1 gene by Ape1 siRNA. Western blot analysis demonstrates that Ape1 siRNA efficiently knocks down Ape1 gene expression: lane 1 represents untreated cells; lane 2: cells treated with 25 nM scrambled siRNA; lane 3: cells treated with Ape1 siRNA at 5 nM; lane 4: cells treated with Ape1 siRNA at 25 nM; and lane 5: cells treated with Ape1 siRNA at 50 nM.
Ape1 siRNA is efficient to knock down target gene expression in EB cells. (A) Western blot analysis of Ape1 protein expression in both ES and EB cells at different ages (days 3, 6, and 10). (B) Silencing of endogenous Ape1 gene by Ape1 siRNA. Western blot analysis demonstrates that Ape1 siRNA efficiently knocks down Ape1 gene expression: lane 1 represents untreated cells; lane 2: cells treated with 25 nM scrambled siRNA; lane 3: cells treated with Ape1 siRNA at 5 nM; lane 4: cells treated with Ape1 siRNA at 25 nM; and lane 5: cells treated with Ape1 siRNA at 50 nM.
Normal hemangioblast development from ES cell development
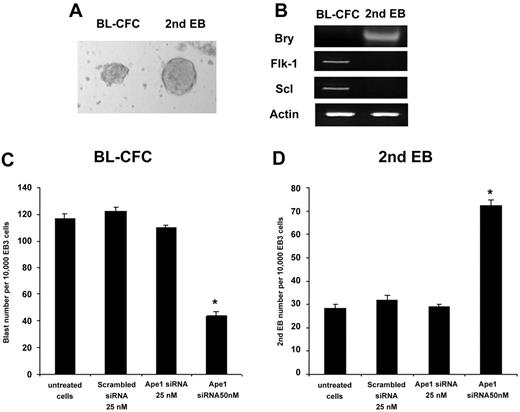
ES cells differentiate to EBs in the absence of LIF in culture. Day-3 EB cells replated into culture wells with VEGF and SCF results in the emergence of hemangioblast colonies. Some residual undifferentiated ES cells in EBs were able to differentiate into a cell mass called secondary EBs, which are round bodies with a smooth edge and are easy to distinguish from hemangioblast colonies (Figure 2A). We separately collected BL-CFCs or secondary EBs, and RT-PCR analysis revealed that they differ in their gene expression pattern (Figure 2B). Knockdown of Ape1 gene expression results in a decrease of BL-CFC formation and an increase in the secondary EB numbers in the culture (Figure 2C-D).
Knockdown of Ape1 expression results in a decreased frequency of BL-CFC formation. (A) The distinct morphology between BL-CFCs and secondary EBs is depicted (× 20). (B) The individual BL-CFC or secondary EB colonies were collected separately. mRNA was extracted and RT-PCR assay revealed the distinct phenotype between these 2 cell types; BL-CFCs express Flk-1 and scl genes, but not the brachyury gene. In contrast, secondary EB cells express the brachyury gene, but not Flk-1 and scl genes. (C) Hemangioblast progenitor assay indicated that knockdown of Ape1 gene expression with Ape1 siRNA, but not scrambled siRNA day-3 EB cells, resulted in a decrease in BL-CFC formation. *P < .05 comparing Ape1 siRNA to scrambled siRNA, treatment with transfection reagent (OFA2000), or no treatment. Data represent the mean ± SD for 3 experiments. (D) Knockdown of Ape1 gene expression of day-3 EB cells resulted in an increase in secondary EB formation in the culture. *P < .05 comparing Ape1 siRNA to scrambled siRNA, treatment with transfection reagent (OFA2000), or no treatment. Data represent the mean ± SD for 3 experiments. The image in panel A was obtained using a Leica MZ9.5 microscope equipped with a Planapo 6× objective and a 10×/2.1 B aperature (Leica, Allendale, NJ). Image was captured using a Leica DFC320 camera with Leica Application software version 2.4.0.
Knockdown of Ape1 expression results in a decreased frequency of BL-CFC formation. (A) The distinct morphology between BL-CFCs and secondary EBs is depicted (× 20). (B) The individual BL-CFC or secondary EB colonies were collected separately. mRNA was extracted and RT-PCR assay revealed the distinct phenotype between these 2 cell types; BL-CFCs express Flk-1 and scl genes, but not the brachyury gene. In contrast, secondary EB cells express the brachyury gene, but not Flk-1 and scl genes. (C) Hemangioblast progenitor assay indicated that knockdown of Ape1 gene expression with Ape1 siRNA, but not scrambled siRNA day-3 EB cells, resulted in a decrease in BL-CFC formation. *P < .05 comparing Ape1 siRNA to scrambled siRNA, treatment with transfection reagent (OFA2000), or no treatment. Data represent the mean ± SD for 3 experiments. (D) Knockdown of Ape1 gene expression of day-3 EB cells resulted in an increase in secondary EB formation in the culture. *P < .05 comparing Ape1 siRNA to scrambled siRNA, treatment with transfection reagent (OFA2000), or no treatment. Data represent the mean ± SD for 3 experiments. The image in panel A was obtained using a Leica MZ9.5 microscope equipped with a Planapo 6× objective and a 10×/2.1 B aperature (Leica, Allendale, NJ). Image was captured using a Leica DFC320 camera with Leica Application software version 2.4.0.
Ape1 is essential for both primitive and definitive hematopoiesis
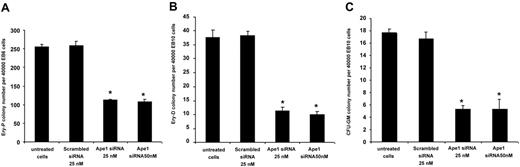
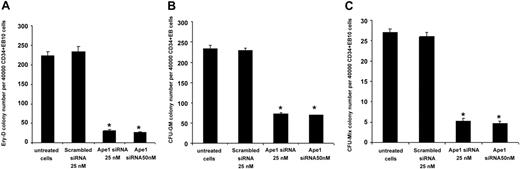
To test the role of Ape1 in primitive erythroid progenitor (Ery-P) cell development, day-6 EBs were dissociated and EB cells were replated in methylcellulose culture medium supplemented with plasma-derived serum and with erythropoietin. Knockdown of Ape1 expression in day-6 EB cells blunted their potential to form Ery-Ps (Figure 3A). To determine whether a reduction of Ape1 expression affected definitive hematopoiesis, Ape1 siRNA was introduced into dissociated day-10 EB cells followed by progenitor assays. Cells with reduced Ape1 expression generated significantly fewer definitive erythroid colonies (Ery-Ds) compared with untreated cells or cells transfected with scrambled siRNA (Figure 3B). Moreover, to examine myeloid differentiation, CFU-GMs were scored by replating control or Ape1 siRNA–treated day-10 EB cells in the presence of Epo, SCF, GM-CSF, and IL-3. Our results indicated that the frequencies of CFU-GM colony were decreased in the Ape1 siRNA–transfected group compared with the control scrambled siRNA–transfected group (Figure 3C) (P < .05). Consistent with the levels of Ape1 expression detected by Western blot analysis in cells transfected with 25 nM and 50 nM Ape1 siRNA (Figure 1B), no significant difference in hematopoietic colony frequency between cells treated with 25 nM or 50 nM Ape1 siRNA was observed (Figure 3A-C). As CD34+ day-10 EB cells contain hematopoietic progenitor cells,7,14 we then sorted CD34+ EB cells from day-10 EBs and treated cells with scrambled or Ape1 siRNA. Our data revealed that knockdown of Ape1 gene expression in these cells reduced their ability to form colonies of Ery-Ds, CFU-GMs, and CFU-Mix's (Figure 4A-C).
Reduction of Ape1 expression by siRNA abrogates primitive and definitive hematopoiesis. (A) Cells derived from day-6 CCE EBs were transfected with either Ape1 siRNA or scrambled siRNA, and plated for primitive progenitor assays (Ery-P). (B) Cells derived from day-10 EBs were transfected with 25 nM or 50 nM Ape1 siRNA1, or scrambled siRNA, and plated for either definitive erythroid progenitor assays (Ery-D) or (C) granulocyte-macrophage colony assays (CFU-GM). At both 25 nM and 50 nM Ape1 siRNA in all assays conducted, significantly fewer progenitors developed. (A-C) *P < .05 comparing cells transfected with Ape1 siRNA to that of scrambled siRNA for all progenitor assays conducted. Data represent the mean ± SD for 3 experiments.
Reduction of Ape1 expression by siRNA abrogates primitive and definitive hematopoiesis. (A) Cells derived from day-6 CCE EBs were transfected with either Ape1 siRNA or scrambled siRNA, and plated for primitive progenitor assays (Ery-P). (B) Cells derived from day-10 EBs were transfected with 25 nM or 50 nM Ape1 siRNA1, or scrambled siRNA, and plated for either definitive erythroid progenitor assays (Ery-D) or (C) granulocyte-macrophage colony assays (CFU-GM). At both 25 nM and 50 nM Ape1 siRNA in all assays conducted, significantly fewer progenitors developed. (A-C) *P < .05 comparing cells transfected with Ape1 siRNA to that of scrambled siRNA for all progenitor assays conducted. Data represent the mean ± SD for 3 experiments.
Reduction of Ape1 expression by siRNA in CD34+ day-10 EB cells abrogates definitive hematopoiesis. (A) CD34+ cells derived from day-10 CCE EBs were transfected with either Ape1 siRNA or scrambled siRNA, and plated for definitive erythroid progenitor (Ery-D) assays. (B) CD34+ EB cells derived from day-10 EBs were transfected with 25 nM or 50 nM Ape1 siRNA, or scrambled siRNA, and plated into granulocyte-macrophage colony (CFU-GM) assays, or (C) mixed lineage colony assays (CFU-Mix). CD34+ EB cells derived from day-10 EBs were transfected with 25 nM or 50 nM Ape1 siRNA, or scrambled siRNA, and plated into mixed progenitor colony (CFU-Mix) assay. Significantly fewer progenitors developed. (A-C) *P < .05 comparing cells transfected with Ape1 siRNA to that of scrambled siRNA for all progenitor assays conducted. Data represent the mean ± SD for 3 experiments.
Reduction of Ape1 expression by siRNA in CD34+ day-10 EB cells abrogates definitive hematopoiesis. (A) CD34+ cells derived from day-10 CCE EBs were transfected with either Ape1 siRNA or scrambled siRNA, and plated for definitive erythroid progenitor (Ery-D) assays. (B) CD34+ EB cells derived from day-10 EBs were transfected with 25 nM or 50 nM Ape1 siRNA, or scrambled siRNA, and plated into granulocyte-macrophage colony (CFU-GM) assays, or (C) mixed lineage colony assays (CFU-Mix). CD34+ EB cells derived from day-10 EBs were transfected with 25 nM or 50 nM Ape1 siRNA, or scrambled siRNA, and plated into mixed progenitor colony (CFU-Mix) assay. Significantly fewer progenitors developed. (A-C) *P < .05 comparing cells transfected with Ape1 siRNA to that of scrambled siRNA for all progenitor assays conducted. Data represent the mean ± SD for 3 experiments.
Knockdown of Ape1 gene expression in EB cells did not induce apoptosis in these cells
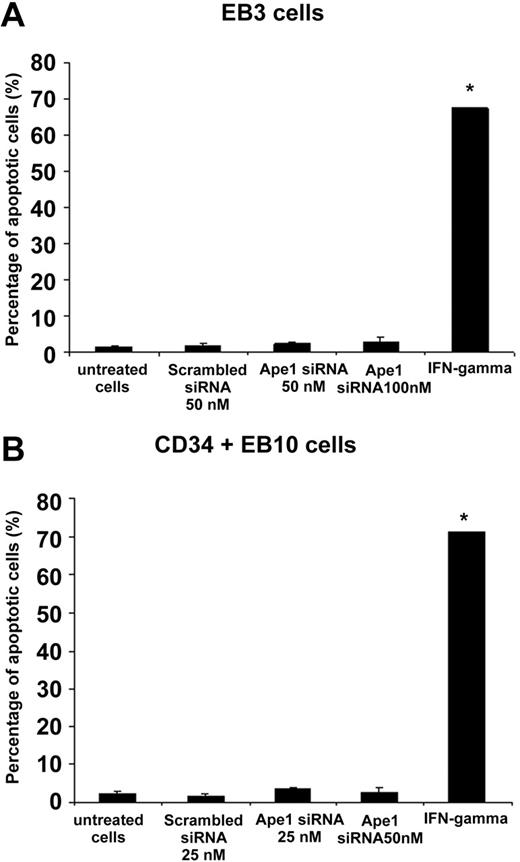
To determine whether knockdown of Ape1 gene expression in EB cells induced apoptosis, siRNA-transfected cells were harvested after 48 hours of transfection and stained with annexin V and PI. The cells were then analyzed by fluorescence-activated cell sorting (FACS). As shown in Figure 5, knockdown of Ape1 gene expression with siRNA did not trigger apoptosis in EB cells. In contrast, cells treated with IFN-γ displayed robust apoptosis (Figure 5).
Knockdown of Ape1 expression in day-3 EB cells or sorted CD34+ day-10 EB cells did not trigger apoptosis. (A) Day-3 or (B) CD34+ day-10 EB cells were transfected with control siRNA or Ape1 siRNA, and after 48 hours of transfection, cells were harvested and stained with annexin V and PI. Double annexin V– and PI–positive cells in staining were scored as apoptotic cells in FACS analysis. In a separate group, cells were treated with IFN-γ (500 IU) as positive control in apoptosis induction. (A-B) *P < .01. Bars represent mean ± SD of triplicate experiments.
Knockdown of Ape1 expression in day-3 EB cells or sorted CD34+ day-10 EB cells did not trigger apoptosis. (A) Day-3 or (B) CD34+ day-10 EB cells were transfected with control siRNA or Ape1 siRNA, and after 48 hours of transfection, cells were harvested and stained with annexin V and PI. Double annexin V– and PI–positive cells in staining were scored as apoptotic cells in FACS analysis. In a separate group, cells were treated with IFN-γ (500 IU) as positive control in apoptosis induction. (A-B) *P < .01. Bars represent mean ± SD of triplicate experiments.
Inhibition of Ape1 redox activity by E3330, but not DNA repair activity by methoxyamine, affects hemangioblast development in vitro
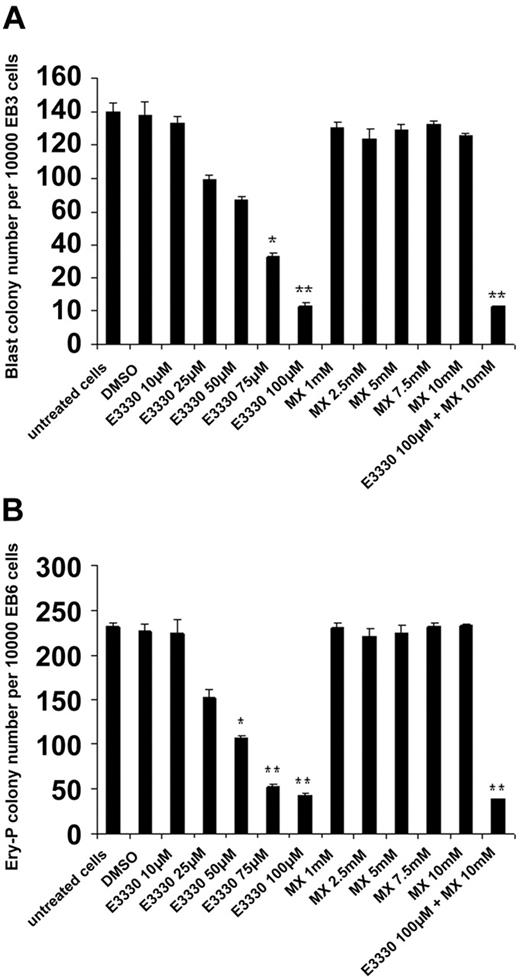
To address which functional domain in the Ape1 molecule may play the major role in regulation of hematopoiesis, we added the Ape1 redox inhibitor E3330 (0-100 μM) or the Ape1 abasic site excision inhibitor MX (0-10 mM) to add to the cultured EB cells. Inhibition of Ape1 redox activity by E3330, but not Ape1 DNA repair activity by MX, affected cytokine-mediated hemangioblast development in vitro (Figure 6). Furthermore, the inhibition of BL-CFC and Ery-P colony formation via E3330 was dose dependent.
Inhibition of redox activity of Ape1, but not base excision repair (BER) activity of Ape1, reduces BL-CFC and Ery-P formation. (A) In the BL-CFC assay, day-3 EB cells were treated with VEGF, SCF, and various doses of E3330 (0 to 100 nM) or MX (0 to 10 μM) as indicated in the culture. After 4 days of culture, the BL-CFC colonies were scored; the number is shown. Results shown are representative of 3 independent experiments, each performed in triplicate. Mean values significantly different from control cells (untreated or treated with DMSO) are indicated. *P < .05; **P < .01. (B) In Ery-P assay, day-6 EB cells were treated with Epo and various doses of E3330 (0 to 100 nM) or MX (0 to 10 μM). After 7 days of culture, Ery-P colonies were scored. Results shown are representative of 3 independent experiments, each performed in triplicate. Mean values significantly different from control cells (untreated or treated with DMSO) are indicated. *P < .05; **P < .01. Bars represent mean ± SD of triplicate experiments.
Inhibition of redox activity of Ape1, but not base excision repair (BER) activity of Ape1, reduces BL-CFC and Ery-P formation. (A) In the BL-CFC assay, day-3 EB cells were treated with VEGF, SCF, and various doses of E3330 (0 to 100 nM) or MX (0 to 10 μM) as indicated in the culture. After 4 days of culture, the BL-CFC colonies were scored; the number is shown. Results shown are representative of 3 independent experiments, each performed in triplicate. Mean values significantly different from control cells (untreated or treated with DMSO) are indicated. *P < .05; **P < .01. (B) In Ery-P assay, day-6 EB cells were treated with Epo and various doses of E3330 (0 to 100 nM) or MX (0 to 10 μM). After 7 days of culture, Ery-P colonies were scored. Results shown are representative of 3 independent experiments, each performed in triplicate. Mean values significantly different from control cells (untreated or treated with DMSO) are indicated. *P < .05; **P < .01. Bars represent mean ± SD of triplicate experiments.
Knockdown of Ape1 in EB cells induces G1 arrest in EB cells
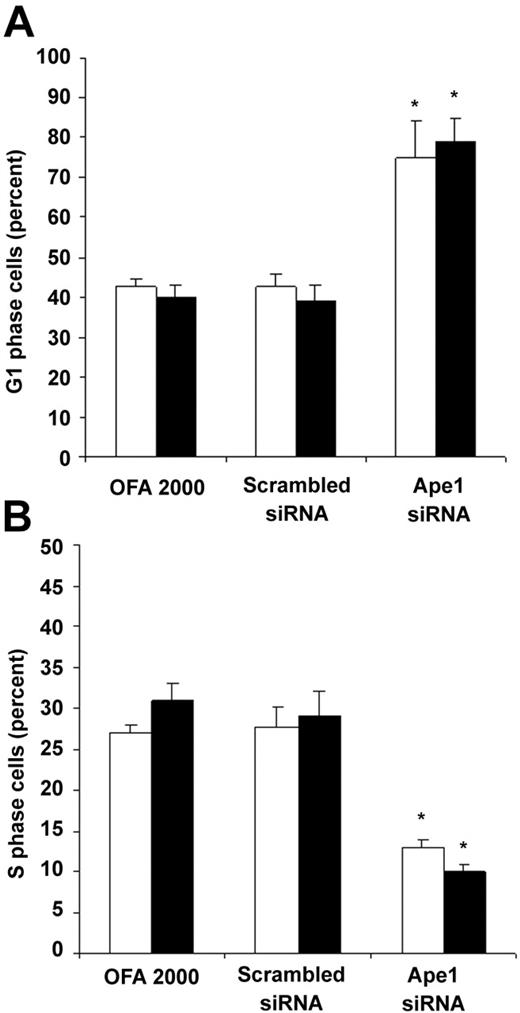
To examine whether the decrease in frequency of hematopoietic colonies was caused by low cell proliferation, we performed cell-cycle analysis of the cells treated with Ape1 siRNA. As shown in Figure 7, reduction of Ape1 gene expression with Ape1 siRNA, but not scrambled siRNA, significantly increased day-3 EB cells or Flk-1+ EB cells in the G1 phase and decreased the percent of cells in S phase. This indicates that knockdown of Ape1 gene expression in EB cells resulted in a decrease in the G1/S transition of the cell cycle in EB cells.
Ape1 regulates cell cycle G1-S transition in day-3 EB cells. Day-3 EB cells or Flk-1+ day-3 EB cells were treated with either transfection reagent (Oligofectamine 2000) alone, scrambled siRNA (25 nM), or Ape1 siRNA (25 nM) for 48 hours. Cells were collected, permeablized, and stained with PI prior to flow cytometry to determine the percentage of cells in G1 and S phase. Open bars represent day-3 EB cells and solid black bars represent Flk-1+ day-3 EB cells. (A) Percentage of G1 phase cells. (B) Percentage of S phase cells. (A-B) Bars represent means ± SD of triplicate experiments. *P < .05.
Ape1 regulates cell cycle G1-S transition in day-3 EB cells. Day-3 EB cells or Flk-1+ day-3 EB cells were treated with either transfection reagent (Oligofectamine 2000) alone, scrambled siRNA (25 nM), or Ape1 siRNA (25 nM) for 48 hours. Cells were collected, permeablized, and stained with PI prior to flow cytometry to determine the percentage of cells in G1 and S phase. Open bars represent day-3 EB cells and solid black bars represent Flk-1+ day-3 EB cells. (A) Percentage of G1 phase cells. (B) Percentage of S phase cells. (A-B) Bars represent means ± SD of triplicate experiments. *P < .05.
Discussion
We have demonstrated that the knockdown of Ape1 gene expression in mouse ES cells results in a decrease in the formation of hemangioblast, primitive, and definitive hematopoietic colony frequencies. Our study also reveals that knockdown of Ape1 gene expression in EB cells affected the G1/S transition of the cell cycle in these cells. Further studies showed that inhibition of only Ape1 redox activity, but not Ape1 DNA repair activity, diminished hematopoiesis.
ES cell differentiation is a valuable system to study molecular regulation in embryonic hematopoietic development. In the absence of LIF, ES cells differentiate into EBs after a 3-day differentiation.29 Day-3 EB cells include Flk-1+ BL-CFCs that form colonies in culture when VEGF is supplied.8 Flk1 and SCL are molecular determinants of the hemangioblast, and expression of both these molecules is required for colony formation.30 BL-CFCs represent the in vitro equivalent of the hemangioblast and, as such, one of the earliest stages of hematopoietic development described to date. Functional studies have demonstrated that some of these blast colonies contain primitive and definitive hematopoietic precursors as well as precursors that give rise to adherent endothelial-like cells. Hemangioblast development can be regulated by a number of factors. TPO alone supported BL-CFC formation and nearly doubled the number of BL-CFCs when added together with VEGF and SCF.31 The homeoprotein Hex is essential for hemangioblast differentiation into definitive embryonic hematopoietic progenitors.32 We recently described the positive regulatory role of the protein tyrosine phosphatase Shp-2 in hemangioblast development and implicated Shp-2 as an important mediator of FGF augmentation of BL-CFC activity in vitro.15 In the present study, we identified a novel role of Ape1 as a regulatory molecule in hemangioblast development. The reduction in BL-CFCs resulting from decreased Ape1 expression in the EB may result in diminished production of both primitive and definitive hematopoietic progenitors. It is interesting to contemplate that knockdown of Ape1 expression with siRNA may have affected emergence of the BL-CFC from mesoderm, whereas use of the Ape1 redox inhibitor molecule E3330 may have had a more direct effect on differentiation of the BL-CFC toward hematopoietic progenitors. To address whether a reduction in Ape1 expression directly influences progenitor differentiation, we isolated and knocked down Ape1 expression in this cell population that contains primary hematopoietic progenitor cells.
Changes in expression of Ape1 have been correlated with myeloid differentiation in HL60 leukemic cells induced by RA or DMSO.33 However, the regulatory role of Ape1 in myeloid differentiation of primary hematopoietic stem/progenitor cells has not been examined. Our study clearly demonstrated that Ape1 is essential for GM-CSF and other cytokines to stimulate myeloid hematopoiesis, as knockdown of Ape1 expression in CD34+ cells resulted in significant reduction of erythroid and myeloid progenitors in vitro. Therefore, we propose that Ape1 regulates normal erythromyelopoiesis.
In previous studies, PDGF-BB stimulated cell-cycle progression from G0/G1 to S in smooth muscle cells, with one of the postulated steps being redox regulation of AP-1 by Ape1 protein.34 Therefore, we wanted to see if Ape1 may play a role in growth factor–stimulated cell-cycle regulation. We report here that Ape1 regulates cell-cycle status in hematopoietic progenitors and that Ape1 also positively regulates G1/S transition in EB cells. The cell-cycle arrest of Ape1 siRNA–treated EB cells may be one mechanism to explain diminished hematopoietic CFC activity reported here. Future studies will need to prove how Ape1 may regulate cyclin-dependent kinases and cyclins that are critical for G/S transition.
In conclusion, this study presents direct evidence that Ape1 positively regulates embryonic hematopoiesis through its redox function. However, the target of Ape1's redox signaling has not been identified by this type of analysis, and additional studies will need to address which downstream target or targets are involved in this pathway. As this study was limited to investigation of the regulatory role of Ape1 in embryonic hematopoiesis using the ES cell system, further studies to identify a possible role for Ape1 in adult myelolymphoid development may prove insightful.
Authorship
Contribution: G.-M.Z. designed and performed research, analyzed data, and wrote the paper; M.-H.L. and A.R. performed research; and M.R.K. and M.C.Y. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mervin C. Yoder, Herman B. Well Center for Pediatrics Research, Indiana University School of Medicine, 1044 West Walnut St, Indianapolis IN 46202; e-mail: myoder@iupui.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants HL63069 (M.C.Y.) and CA094025, CA106298, ES03456, and P30 CA82709 (M.R.K.).
We thank Bill Carter in the transgene center for the CCE ES cells. We would like to thank Rod Nyland II and Rick Borch in Department of Medical Chemistry, Purdue University Cancer Center, for the E3330.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal