Abstract
The last 2 years have seen much excitement in the field of genetics with the identification of a formerly unappreciated level of “structural variation” within the normal human genome. Genetic structural variants include deletions, duplications, and inversions in addition to the recently discovered, copy number variants. Single nucleotide polymorphisms are the most extensively evaluated variant within the genome to date. Combining our knowledge from these studies with our rapidly accumulating understanding of structural variants, it is apparent that the extent of genetic dissimilarity between any 2 individuals is considerable and much greater than that which was previously recognized. Clearly, this more diverse view of the genome has significant implications for allogeneic hematopoietic stem cell transplantation, not least in the generation of transplant antigens but also in terms of individual susceptibility to transplant-related toxicities. With advances in DNA sequencing technology we now have the capacity to perform genome-wide analysis in a high throughput fashion, permitting a detailed genetic analysis of patient and donor prior to transplantation. Understanding the significance of this additional genetic information and applying it in a clinically meaningful way will be one of the challenges faced by transplant clinicians in the future.
Introduction
Structural variation in the human genome
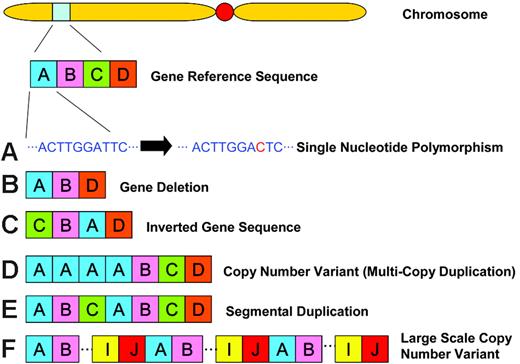
Recently, much focus has been directed at documenting the extent of normal human genomic variation, particularly in the form of single nucleotide polymorphisms (SNPs; Table 1). In October 2005, phase 1 of the International Hapmap Project was published.2 This project catalogued more than one million SNPs, genome-wide, into a publicly accessible database (www.hapmap.org). Approximately 11 500 of the SNPs recorded in phase 1 are nonsynonymous coding SNPs. Other examples of genomic variation include gene deletions, inverted gene sequences, multiple copy gene duplications, segmental duplications and large-scale copy number variants (CNVs). Each of these structural variants is shown schematically in Figure 1.
Genomic variation definitions. (A) A single nucleotide polymorphism (SNP) occurs as a result of a single base substitution at an individual site in the DNA sequence along the chromosome. Nonsynonymous coding SNPs occur in the coding region of a gene where the alternate SNP allele results in the coding of a different amino acid. (B) A deletion is the loss/absence of DNA sequence. (C) An inversion is a rearrangement causing a segment of DNA to be present in reverse orientation. (D) A copy number variant (CNV) is a segment of DNA that is 1 kb or larger and is present at a variable copy number in comparison with a reference genome. CNVs can be either deletion variants where there is loss of copy number relative to the reference sequence or multicopy duplications where there is gain of copy number relative to the reference sample. (E) A segmental duplication is a segment of DNA at least 1 kb in size that occurs in 2 or more copies per haploid genome, with the different copies sharing at least 90% sequence identity. (F) A large-scale copy number variant is a CNV that involves a segment of DNA more than 50 kb in size.
Genomic variation definitions. (A) A single nucleotide polymorphism (SNP) occurs as a result of a single base substitution at an individual site in the DNA sequence along the chromosome. Nonsynonymous coding SNPs occur in the coding region of a gene where the alternate SNP allele results in the coding of a different amino acid. (B) A deletion is the loss/absence of DNA sequence. (C) An inversion is a rearrangement causing a segment of DNA to be present in reverse orientation. (D) A copy number variant (CNV) is a segment of DNA that is 1 kb or larger and is present at a variable copy number in comparison with a reference genome. CNVs can be either deletion variants where there is loss of copy number relative to the reference sequence or multicopy duplications where there is gain of copy number relative to the reference sample. (E) A segmental duplication is a segment of DNA at least 1 kb in size that occurs in 2 or more copies per haploid genome, with the different copies sharing at least 90% sequence identity. (F) A large-scale copy number variant is a CNV that involves a segment of DNA more than 50 kb in size.
The extent of human genomic variation
| Variable . | Value . | Source . |
|---|---|---|
| Whole genome | ||
| No. BPs in the human genome | 3 × 109 | Kruglyak and Nickerson1 |
| No. SNPs in the human genome | 1 × 107 | Kruglyak and Nickerson1 |
| No. validated HapMap SNPs | 4 × 106 | HapMap P2 |
| No. validated HapMap exonic SNPs | 7 × 104 | HapMap P2 |
| No. validated HapMap NS coding SNPs | 3 × 104 | HapMap P2 |
| Estimated no. BPs affected by CNVs | > 3 × 108 | Redon et al101 |
| Individual genome | ||
| SNP frequency in the genome | 1/300 BP | Kruglyak and Nickerson1 |
| Estimated no. CNVs per individual genome | > 100 | Feuk et al8 |
| Variable . | Value . | Source . |
|---|---|---|
| Whole genome | ||
| No. BPs in the human genome | 3 × 109 | Kruglyak and Nickerson1 |
| No. SNPs in the human genome | 1 × 107 | Kruglyak and Nickerson1 |
| No. validated HapMap SNPs | 4 × 106 | HapMap P2 |
| No. validated HapMap exonic SNPs | 7 × 104 | HapMap P2 |
| No. validated HapMap NS coding SNPs | 3 × 104 | HapMap P2 |
| Estimated no. BPs affected by CNVs | > 3 × 108 | Redon et al101 |
| Individual genome | ||
| SNP frequency in the genome | 1/300 BP | Kruglyak and Nickerson1 |
| Estimated no. CNVs per individual genome | > 100 | Feuk et al8 |
BP indicates base pair; SNP, single nucleotide polymorphism; NS, nonsynonymous; CNV, copy number variant; P2, phase 2.
In the year prior to the Hapmap publication, 2 landmark studies reported that large-scale CNVs are distributed widely throughout the human genome and that a high proportion of these encompass known genes.3,4 These studies were groundbreaking because they challenged the previously held view that the complete DNA sequences of any 2 individuals are 99.9% identical, the 0.1% difference being largely attributed to SNPs. Subsequently, several studies on CNVs followed,5-7 culminating in a large international study of the Hapmap population by Redon et al101 that identified more than 1400 CNVs spanning approximately 12% of the human genome. Even with this comprehensive evaluation, it is likely that this present index of CNVs is far from complete and new variants are continually being discovered.8 In 2006, 3 studies specifically characterized deletion variants in the human genome.9-11 Combined, these investigations described approximately 1000 deletions of varying size, with the majority being less than 10 000 bp. McCarroll et al identified 10 genes that appeared to be consistently deleted in a homozygous fashion, in regions overlapping coding exons of the gene.9 Over half of the CNVs documented by Redon et al101 also overlap known expressed genes.
The CNV studies indicate that genes subject to structural variation appear to be significantly enriched in immune response, chemosensation, and drug detoxification functions.4,7,10,12 Accordingly, it has been suggested that these genes may have roles in the adaptability and fitness of an organism in response to external pressures and that structural variation represents a process of adaptive evolution.8 Indeed, selection on gene copy number has been reported for CCL3L1, an immune response gene, where relatively low copy number is associated with increased susceptibility to HIV/AIDS.13 More recently, low FCGR3B copy number has been associated with increased susceptibility to immunologically mediated glomerulonephritis in patients with lupus.14
The implications of the studies mentioned for our understanding of human genetic diversity and for disease susceptibility are considerable. In addition, these studies are significant for the field of allogeneic hematopoietic stem cell transplantation (HSCT). Nonsynonymous coding SNPs and deletion polymorphisms hold importance as ways of generating potentially immunogenic transplantation antigens. SNPs in non-HLA genes can influence immune responses. Genes subject to CNV include MHC, KIR, and the genes encoding Fc and immunoglobulin receptors. Genes involved in drug detoxification, which are prone to structural variation and are of potential relevance in HSCT, include those involved in the metabolism of busulfan (GST family), cyclophosphamide (cytochrome P450, GST family), and calcineurin inhibitors (cytochrome P450, UGT2B family). It is highly probable, therefore, that normal structural genetic variation has an impact on individual patient outcomes during HSCT.
Minor histocompatibility antigens
Minor histocompatibility antigens (mHAs) were originally described by Snell in 194815 as non-MHC genetic loci sufficient to initiate allogeneic tissue rejection in congenic mouse strains (ie animals bred to be identical at the MHC locus but disparate at other areas of the genome). In 1978, Korngold et al demonstrated that lethal graft-versus-host disease (GVHD) could occur across minor histocompatibility barriers in mice and that GVHD appeared to be mediated by T cells.16 In a seminal paper in 1990, Wallny et al demonstrated that mHAs were peptides derived from normal cellular proteins presented by self-MHC and that they were recognized by cytotoxic T lymphocytes (CTLs) in a minor histoincompatible mouse strain.17 In humans also, mHAs have been defined by allospecific CTLs obtained from individuals primed in vivo through organ or bone marrow grafting and blood transfusions18 (Table 2 provides a current list of known human mHAs). The molecular structure of a human mHA was first characterized in 1995.19,20 Although mHAs have been identified as peptide epitopes presented by MHC molecules and recognized by donor T cells, recent studies have shown that antibodies are also capable of recognizing human mHA.21 These antibodies are capable of distinguishing between recipient and donor-derived peptide sequences in soluble form21 and the presence of antibodies has been associated with chronic GVHD rather than acute GVHD.22 Because mHAs are intracellular proteins, the role of mHA antibodies as mediators of GVHD is unknown and further studies are needed to define the mechanisms by which mHA antibodies may contribute to tissue injury.
Human mHAs
| Mechanism underlying immunogenicity and name . | Gene/chromosome . | Tissue distribution . |
|---|---|---|
| Nonsynonymous coding SNP(s) | ||
| HA-1 | KIAA0223/19p13.3 | Hematopoietic |
| HA-2 | MYO1G/7p12p13 | Hematopoietic |
| HA-3 | LBC/15q24q25 | Ubiquitous |
| HA-8 | KIAA0020/9p24.2 | Ubiquitous |
| HB-1 | 5q32 | B-lymphoid cells |
| BCL2A1 | BCL2A1/15q24.3 | Hematopoietic |
| Polymorphism with chromosome X | ||
| SMCY | Yq11.1 | Ubiquitous |
| DFFRY | Yq11 | Ubiquitous |
| UTY | Yq11.1 | Ubiquitous |
| DBY | Yq11 | Ubiquitous |
| RPS4Y | Yp11.3 | Ubiquitous |
| Homozygous gene deletion | ||
| UGT2B17 | UGT2B17/4q13 | Ubiquitous |
| Deletion SNP causing a frameshift | ||
| LRH-1 | P2X5/17p13.2 | Hematopoietic |
| NS cSNP causing a stop codon | ||
| PANE1 | PANE1/22q13 | B-lymphoid cells |
| NS cSNP in an alternatively translated peptide | ||
| ECGF-1 | ECGF-1/22q13 | Hematopoietic |
| NS cSNP resulting in alternate splicing | ||
| SP110 | SP11O/2q37 | Hematopoietic |
| Unconventional ORF | ||
| TMSB4Y | Yq11 | Ubiquitous |
| Mechanism underlying immunogenicity and name . | Gene/chromosome . | Tissue distribution . |
|---|---|---|
| Nonsynonymous coding SNP(s) | ||
| HA-1 | KIAA0223/19p13.3 | Hematopoietic |
| HA-2 | MYO1G/7p12p13 | Hematopoietic |
| HA-3 | LBC/15q24q25 | Ubiquitous |
| HA-8 | KIAA0020/9p24.2 | Ubiquitous |
| HB-1 | 5q32 | B-lymphoid cells |
| BCL2A1 | BCL2A1/15q24.3 | Hematopoietic |
| Polymorphism with chromosome X | ||
| SMCY | Yq11.1 | Ubiquitous |
| DFFRY | Yq11 | Ubiquitous |
| UTY | Yq11.1 | Ubiquitous |
| DBY | Yq11 | Ubiquitous |
| RPS4Y | Yp11.3 | Ubiquitous |
| Homozygous gene deletion | ||
| UGT2B17 | UGT2B17/4q13 | Ubiquitous |
| Deletion SNP causing a frameshift | ||
| LRH-1 | P2X5/17p13.2 | Hematopoietic |
| NS cSNP causing a stop codon | ||
| PANE1 | PANE1/22q13 | B-lymphoid cells |
| NS cSNP in an alternatively translated peptide | ||
| ECGF-1 | ECGF-1/22q13 | Hematopoietic |
| NS cSNP resulting in alternate splicing | ||
| SP110 | SP11O/2q37 | Hematopoietic |
| Unconventional ORF | ||
| TMSB4Y | Yq11 | Ubiquitous |
NS indicates nonsynonymous; cSNP, coding single nucleotide polymorphism; ORF, open reading frame.
All mHAs arise as a consequence of normal genetic variation but the mechanisms by which they occur are varied.23 Most autosomal mHAs identified thus far are generated as a result of nonsynonymous coding SNPs, which lead to differences in the amino acid sequences of homologous proteins between donor and recipient cells. Single amino acid substitutions can result in altered proteosomal cleavage, peptide transport, HLA binding or T-cell receptor (TCR) contact that ultimately triggers donor T cells to recognize these peptides as foreign. The recently identified mHA SP100 occurs as a result of alternate splicing in the proteosome.24 Autosomal mHAs may also arise when the recipient is confronted with a peptide for which there is no allelic homologue in the donor. In the single instance where this has been described in humans, immunogenicity arose as a consequence of a homozygous gene deletion in the donor, which resulted in expression of the protein in recipient cells but not in donor cells.25 Several mHAs are encoded by genes on the Y chromosome, which show significant polymorphism with homologous regions of the X chromosome. This becomes immunologically relevant in the setting of sex-mismatched transplants when male patients receive stem cell grafts from female donors.26 It has recently been demonstrated that human mHAs can also arise from cryptic polypeptides derived from noncoding regions of the genome, such as untranscribed regions (UTRs) or introns.27
The absolute risk of acute GVHD in HLA-identical sibling transplants is approximately 40%, with the risk of chronic GVHD exceeding 50%.28 This provides strong evidence that mHAs significantly influence HSCT outcome. It has been estimated that several hundred mHAs exist in mice29 and the number in humans is likely to be at least this large. Experiments in mice indicate that a hierarchy of immunogenicity exists among mHAs, with a limited number of “immunodominant” antigens responsible for an oligoclonal T-cell response in any MHC-identical donor-recipient pair.30 Clinical studies of mHA disparity have largely been restricted to evaluations of single mHAs (most commonly HA-1) and have been limited by small patient numbers due to the fact that mHAs are HLA restricted and the frequency of mHA disparity between recipient and donor is often low (10%-15% for HA-1). A study of 148 HLA-identical sibling donor-recipient pairs showed a significant association between mismatch of HA-1 and the development of GVHD after bone marrow transplantation.31 However, this finding has not been consistently seen, with subsequent studies observing both similar32-34 and conflicting results.35-37 HA-1–specific CTLs have been demonstrated in the peripheral blood of HSCT recipients as early as 14 days after transplantation with significantly increased levels during acute GVHD.38 A study of 3 patients treated with donor lymphocyte infusion (DLI) for hematologic malignancy after allogeneic HSCT showed that a high percentage of the leukemia-reactive T cells in the recipients were directed against HA-1 or HA-2.39 Both of these studies suggest that HA-1 functions as an immunodominant mHA in the HLA-matched allogeneic setting. Male patients receiving a hematopoietic stem cell (HSC) transplant from female donors have a higher risk of GVHD and a lower incidence of leukemia relapse, implying immunodominance for at least some mHAs encoded on the Y chromosome.26 The importance of mHAs on the Y chromosome was further underscored recently with the demonstration that transplants from male donors to female recipients for aplastic anemia are associated with decreased survival as a result of increased rejection.40
Knowledge of the molecular structure of mHAs brings with it the possibility of genotyping for these antigens, in addition to typing for HLA, prior to transplantation. In fact, the development of a liquid bead array for multiplexed genotyping of the mHAs, HA-1, HA-2, HA-3, HA-8, and HB-1, was recently described.41 With the increased frequency of matched unrelated transplantations, mHAs are likely to become increasingly relevant in selecting an optimal HSC donor because a choice among multiple potential donors often occurs in the matched unrelated setting. Clearly, however, before typing for mHAs becomes part of the routine transplantation evaluation, we need to identify additional human mHAs and ultimately establish a core set of “immunodominant” mHAs with maximal clinical relevance. In the meantime, preferentially choosing male donors for male recipients is appropriate.
Homozygous gene deletions are also a potential source of mHAs. UGT2B17, identified as homozygously deleted in 30% of individuals of European ancestry,9 has been recognized as an mHA in the setting of HSCT.25 A Japanese study of 435 HSC recipients and their HLA-matched unrelated donors found that UGT2B17 was homozygously deleted in 85% of the donors and 82% of recipients.42 However, there was no significant association between UGT2B17 mismatch in the GVHD direction and incidence of acute or chronic GVHD, relapse, or survival. The authors suggested that this may be a result of absent processing or presentation of UGT2B17 peptides on the cell surface or homology between UGT2B17 and other UGT2B family members resulting in overlapping substrate specificity. The same group reported similar findings when they investigated 2 additional genes, GSTM1 and GSTT1, in 373 HLA-matched unrelated HSCT pairs.43 GSTM1 and GSTT1 were also identified as homozygous null at appreciable frequency in the Hapmap population.9 GSTT1 has been identified as a potential alloantigen mediating de novo immune hepatitis in liver transplant recipients who are homozygous null for the gene, but receive a GSTT1+ graft.44 Six of the 10 exonic gene deletions described by McCarroll et al were determined to be homozygous null in 30% or greater of the European/North American Hapmap population. Redon et al101 recently documented 33 homozygous deletions genome-wide, 8 of which involved expressed genes.
As the number of known clinically important mHAs expands, their relevance for clinical practice will increase. One promising application of mHAs is in the field of cellular immunotherapy. The mHAs whose tissue distribution is limited to hematopoietic cells provide candidate target antigens for adoptive immunotherapy of leukemia, lymphoma, and myeloma.45 Fontaine et al demonstrated in mice that CTLs primed against a single MHC class I-restricted immunodominant mHA could eradicate leukemia cells without causing GVHD.46 In humans, Dickinson et al determined that CTLs specific for mHAs whose expression was limited to hematopoietic cells induced no or low GVH reactions.47 In the clinical setting, CTLs recognizing the mHAs, HA-1 and HA-2, on the recipient's hematopoietic cells coincide with the development of complete disease remission and restoration of 100% donor chimerism after DLI.48 Recently, an in vivo translational mouse model was used to show that CTLs specific for a single mHA, HA-1, could eradicate leukemia, thus providing direct evidence of the in vivo antileukemic effects of HA-1–specific CTLs.49 The generation of mHA-specific CTLs from the stem cell donor ex vivo as a potential therapy for relapsed leukemia has been explored.50 It may also be possible to generate mHA-specific CTLs ex vivo from umbilical cord blood.51 Another potential therapeutic strategy is to use mHA peptides to vaccinate either patients after HSCT or donors prior to transplantation,52 in an attempt to augment the donor graft-versus-leukemia (GVL) response. The immunotherapeutic potential of tissue-restricted mHAs underlies the importance of identifying additional members of this group. In fact, in the last 6 months, 3 such mHAs, each with immunotherapeutic potential in chronic myelogenous leukemia (CML),53 chronic lymphocytic leukemia (CLL),54 and myeloma,55 respectively, have been discovered.
Immune response polymorphisms
Although we know from syngeneic transplants that antigen disparity between host and donor is essential for the development of GVHD, the pathophysiology of the disease is complex. It is now generally accepted that the development of acute GVHD can be summarized into a 3-phase process.56 In phase 1 the conditioning regimen causes host tissue damage resulting in activation of inflammatory cytokines. In this inflammatory milieu there is increased expression of MHC and adhesion molecules leading to enhanced recognition of alloantigens by donor T cells, resulting in their activation and proliferation. In the final phase, T cells induce further target tissue damage through cell-mediated cytotoxicity. In this model, inflammatory cytokines play a key role. It is now well established that many of the genes encoding cytokines are polymorphic and polymorphisms in these genes and in genes associated with innate immune response are among those most extensively studied in HSCT. It has been suggested that polymorphisms in these immune effector genes may be as important as the degree of HLA mismatch in determining GVHD severity and that typing for such allelic polymorphisms may, in the future, be an important consideration when selecting an HSCT donor.57
Cytokine polymorphisms
A wide range of cytokine gene polymorphisms have been studied in the context of HSCT, including polymorphisms in IL10, TNF, IFNG, IL6, IL1, and TGFB1 (reviewed by Dickinson et al58 ). Typically, studies have focused on SNPs in regulatory regions of these genes because these regions may influence the amount of cytokine produced at a steady state or after stimulation. Most of these studies have been performed at single institutions in patients who received stem cells from HLA-matched siblings after myeloablative conditioning. The majority have attempted to correlate SNP genotype with risk of acute GVHD.
The largest study (almost 1000 patient-donor pairs) was performed by Lin et al who found that the presence of an SNP in the promoter region of the recipient's IL10 gene was significantly associated with a decreased risk of severe acute GVHD and death after HSCT.59 A haplotype analysis showed that the IL-10 SNP in question was a specific marker for a particular IL-10 promoter haplotype. The authors hypothesized that the presence of this haplotype in the recipient caused increased IL-10 production and reduced GVHD-induced inflammation, although IL-10 levels were not measured in the study. This group has recently evaluated a nonsynonymous coding SNP in the IL10RB gene in the same cohort of patient-donor pairs.60 On this occasion they found that the presence of this SNP in the donor was associated with a reduced incidence of severe acute GVHD, suggesting that genetic regulation of the IL-10 pathway plays a role in the development of acute GVHD. The functional significance of the IL-10RB SNP is currently unknown.
These 2 studies are among the best conducted in this area in that they evaluated a large sample population, they studied both donors and recipients, they investigated a homogenous population (HLA-identical siblings, T cell–replete transplants, identical GVH prophylaxis regimens), and at least in the first study, an attempt was made to validate results in an independent cohort (low frequency of the SNP of interest ultimately precluded this). Yet, they also serve as a striking illustration of the problems encountered within this area of research.
Perhaps the greatest weakness of these studies is the absence of a clear understanding of the biologic significance of the findings. In their original study Lin et al showed that the presence of the IL-10-592A/A genotype was a specific marker for a particular haplotype, which they proposed was associated with high IL-10 production. However, earlier studies had suggested increased IL-10 production with the alternative IL-10-592C/C genotype in autologous GVHD.61 As mentioned previously IL-10 levels were not measured in the initial Lin study. Had they been checked and found to be high (as predicted), this finding would have been at odds with 2 earlier (albeit much smaller studies) showing an association between high IL-10 levels after HSCT and more severe acute GVHD62 and increased mortality.63
Recently, high recipient IL-10 mRNA levels in peripheral blood mononuclear cells on day +14 after nonmyeloablative HSCT were found to be associated with a reduced incidence of acute GVHD.64 This finding is in keeping with an earlier study showing that high endogenous IL-10 production prior to myeloablative HSCT is protective against acute GVHD.65 The cytokine-releasing profile of donor cells in response to recipient alloantigen stimulation has been shown to correlate with the development of acute GVHD, with a high frequency of donor cells producing IL-10 associated with the absence of acute GVHD after myeloablative HSCT.66 In contrast to the studies noted previously,62,63 these studies suggest that higher levels of IL-10 in the peritransplantation period confer protection against the subsequent development of GVHD.
At the time of the Lin study it was suggested that IL-10 genotyping be included as part of the standard evaluation of a patient being considered for HSCT from an HLA-identical sibling donor.67 Clearly, given the biologic complexity of IL-10, we need a much better understanding of the functional significance of immune polymorphisms in this gene before this recommendation can be incorporated into clinical practice. As has been suggested by others, we also need a more extensive evaluation of as many genetic polymorphisms as possible, in larger numbers of patients, so that the most informative variants, predictive of outcome are identified.68
Polymorphisms in innate immunity genes
Although highly conserved across a wide range of species, innate immunity genes demonstrate substantial interindividual variability predominantly in the form of SNPs.69 The nucleotide oligomerization domain 2 gene (NOD2) encodes a protein involved in the immune response to intracellular bacterial lipopolysaccharides (LPS). In 2001, NOD2 was identified as the first “susceptibility” gene for inflammatory bowel disease (IBD).70,71 Clinical evidence indicates that the innate immune system contributes to the development of acute GVHD through its response to the translocation of bacterial toxins across the gastrointestinal mucosa.72 Indeed, LPS antagonism has been shown to reduce GVHD severity in a mouse model.73 In this context, polymorphisms in NOD2 have been evaluated in the setting of HSCT. Holler at al recently demonstrated a significant association between the presence of SNPs in NOD2 and the development of severe acute GVHD, higher transplant-related mortality (TRM), and reduced survival after transplantation.74
A number of genetic polymorphisms in innate immunity genes have been evaluated for an association with risk of infection after HSCT. For example, mannose-binding lectin (MBL2) is a pathogen recognition molecule that activates complement independently of specific antibody. As a result of MBL2 polymorphism, MBL levels vary up to 1000-fold in an unselected population, with coding mutation homozygotes having negligible functional MBL activity.75 Mullighan et al found that MBL2-coding polymorphisms associated with low MBL levels were linked to a high risk of major infections after HSCT, and that MBL2 promoter polymorphisms associated with high blood levels of MBL were protective.76 Rocha et al observed that donor myeloperoxidase (MPO) genotype influenced the incidence of severe bacterial infection after HSCT.77 MPO is a lysosomal protein responsible for the microbicidal activity of polymorphonuclear leukocytes (PMNs). In addition, they noted that recipients with a coding polymorphism in FcγRIIa (CD32) had an increased incidence of first severe infection of any type. FcγRIIa is a cell surface receptor found on PMNs, dendritic cells, and endothelial cells. Interestingly, FcγRIIa is also expressed on the surface of acute myeloid leukemia (AML) cells, leading to its evaluation as a potential mHA. A retrospective study of 73 AML patients and their donors found a clear association between FcγRIIa mismatch and acute GVHD with no significant effect on relapse or overall survival.78 Finally, polymorphisms in the bactericidal/permeability-increasing (BPI) gene were recently noted to be significantly associated with the risk of developing bronchiolitis obliterans after HSCT.79 BPI belongs to a family of lipid transport proteins that have a critical role in the innate immune response to bacterial infection.
Killer cell immunoglobulin-like receptor genes
Natural killer (NK) cells are innate immune lymphocytes that recognize self-MHC class I through a unique class of receptors, NK cell receptors (NKRs). The killer cell immunoglobulin-like receptor (KIR) family of NKRs comprises 2 functionally distinct sets: inhibitory and activating. NK cell responses occur as a result of competing signals mediated through these inhibitory and activating receptors. In general, it appears that inhibitory KIRs have a greater affinity for the corresponding HLA class I alleles than do the activating KIRs.80 Variations in gene content, gene copy number, and allelic polymorphism within individual KIR genes result in significant diversity in KIR haplotypes between individuals81 (Figure 2). The KIR gene locus comprises 15 genes in total. Some of these genes are present on all haplotypes, whereas the presence/absence of others varies with the haplotype. Individual genes present on a particular haplotype may differ in copy number and further diversity exists as a result of polymorphisms within distinct KIR genes. The KIR (chromosome 19) and HLA genes (chromosome 6) segregate independently of each other in a conventional mendelian inheritance pattern. KIR genotype is the dominant factor determining the repertoire of KIR expression on NK cells, that is, the influence of HLA genotype on NK cell KIR expression is subordinate to that of KIR.82 The impact of HLA is to change the frequency of KIR-expressing cells in the NK repertoire. In this way, HLA class I genotype imposes selection during normal development such that NK cells expressing inhibitory KIRs specific for autologous HLA class I become relatively increased in number.
KIR variability. (A) The KIR gene locus on chromosome 19 consists of 15 genes and 2 pseudogenes (shown in blue). Three conserved “framework” genes are located one at either end and one in the middle of the locus (red). These are present on all haplotypes and are inhibitory. The regions between the framework genes (between the brackets) contain a variable number of activating (green) and inhibitory (pink) KIR genes. Based on gene content KIR haplotypes have been divided into 2 primary sets: A and B. (B) Most group A haplotypes have the gene organization shown here. They contain the least genes and only a single activating KIR, 2DS4. (C) Group B haplotypes display a much greater variety of subtypes than group A. Two examples of at least 20 currently reported B haplotypes are shown here. An individual's KIR genotype is the sum of 2 haplotypes, one from each parent. (D) Multiple copies of the same KIR gene have been identified in single individuals as shown here for 2DL4. (E) Further diversification of KIR haplotypes occurs as a result of allelic polymorphism within individual genes. This is a prominent feature of group A haplotypes and 3 possible separate haplotypes arising as a result of polymorphisms within the 2DL3, 2DL1, 3DL1, and 3DL2 genes are shown here. More than 20 group A haplotypes emerging as a result of allelic polymorphism have been described. (Figure reproduced by kind permission of Dr Steven Marsh, Anthony Nolan Research Institute, London, from the IPD-KIR database [www.ebi.ac.uk/ipd/kir/introduction.html]).
KIR variability. (A) The KIR gene locus on chromosome 19 consists of 15 genes and 2 pseudogenes (shown in blue). Three conserved “framework” genes are located one at either end and one in the middle of the locus (red). These are present on all haplotypes and are inhibitory. The regions between the framework genes (between the brackets) contain a variable number of activating (green) and inhibitory (pink) KIR genes. Based on gene content KIR haplotypes have been divided into 2 primary sets: A and B. (B) Most group A haplotypes have the gene organization shown here. They contain the least genes and only a single activating KIR, 2DS4. (C) Group B haplotypes display a much greater variety of subtypes than group A. Two examples of at least 20 currently reported B haplotypes are shown here. An individual's KIR genotype is the sum of 2 haplotypes, one from each parent. (D) Multiple copies of the same KIR gene have been identified in single individuals as shown here for 2DL4. (E) Further diversification of KIR haplotypes occurs as a result of allelic polymorphism within individual genes. This is a prominent feature of group A haplotypes and 3 possible separate haplotypes arising as a result of polymorphisms within the 2DL3, 2DL1, 3DL1, and 3DL2 genes are shown here. More than 20 group A haplotypes emerging as a result of allelic polymorphism have been described. (Figure reproduced by kind permission of Dr Steven Marsh, Anthony Nolan Research Institute, London, from the IPD-KIR database [www.ebi.ac.uk/ipd/kir/introduction.html]).
In the setting of allogeneic HSCT, the importance of KIR inhibitory receptors was first demonstrated in HLA haplotype-mismatched transplant recipients. In a subset of these patients, donor NK cell clones could not engage their class I inhibitory ligand as it was absent in the recipient, as a result of donor-recipient mismatch at the HLA class I group, resulting in the development of NK cell alloreactivity against host cells.83,84 Because NK cell alloreactivity appears to target primarily lymphohematopoietic cells, a remarkable GVL effect was observed (at least in patients with AML) in these early studies, and it occurred in the absence of GVHD. In a follow-up study by the same group donor-versus-recipient NK cell alloreactivity did not effect the rate of relapse in patients with AML.85 The original Perugia studies defined KIR ligand incompatibility in the GVL direction as absence in recipients of HLA class I epitopes present in the donor, that is, the “ligand-ligand” model. More recently, a pediatric haploidentical transplantation study demonstrated improved accuracy in predicting relapse by evaluating the KIR repertoire of the donor in addition to HLA genotyping the recipient, that is, “receptor-ligand” model.86 In this model, potential NK alloreactivity is present if the recipient does not have HLA class I alleles for any of the donor's inhibitory KIR. The potency of the antileukemic effect increased as the number of receptor-ligand mismatch pairs rose suggesting that a donor with more mismatched pairs may be a better donor. Leung et al also demonstrated that KIR expression on NK cells derived from donor CD34+ cells always adopted a donor-specific pattern within 3 months of HSCT, despite the fact that transplantation had occurred into an HLA-disparate recipient. This suggests that a mismatched donor could be selected on the basis of a single evaluation of donor KIR repertoire prior to transplantation.
Because the genes that encode HLA and KIR segregate independently, the likelihood of KIR disparity approaches 100% in allogeneic HSCT between unrelated individuals and exceeds 75% between family members regardless of HLA identity.82 This suggests that NK alloreactivity can occur in HLA-identical sibling transplants if the recipient lacks an HLA ligand for the donor's inhibitory KIR. For NK alloreactivity to occur in this setting, an NK clone with autoreactive potential present in the donor (lacking self-HLA class I ligand) would have to expand in the recipient after HSCT. NK cells with autoreactive potential have been observed at low frequency in the blood of healthy donors.81 Recently, these NK cells have been demonstrated to be capable of exerting cytotoxic activity against autologous CD34+ HSCs, although it has been speculated that in healthy individuals these cells remain quiescent and thus are not overtly autoreactive.87 Hsu et al demonstrated in a study of 178 T cell-depleted (TCD) HLA-identical sibling transplants that more than 60% of the pairs exhibited missing KIR ligand in the recipient, that this resulted in an increased overall survival in the patients who received transplants for AML or myelodysplastic syndrome (MDS), and that the increased overall survival occurred as a result of a decrease in disease relapse.88 The potential clinical significance of activating KIRs in autoimmune disease is just beginning to receive attention, where it has been shown that the presence of an activating KIR in the absence of the ligand for its inhibitory counterpart is associated with an increased susceptibility to psoriatic arthritis.89 This prompted Verheyden et al to evaluate the role of activating KIRs in 65 HLA-identical sibling transplant pairs (34 TCD).90 They found that the presence of 2 activating KIRs, 2DS1 and 2DS2, in the donor was significantly associated with a decreased leukemic relapse rate, but this did not appear to be mediated through an HLA ligand interaction.
Overall, these studies suggest that genotyping for KIR to predict NK alloreactivity and optimize donor selection may be useful in the mismatched, unrelated, and sibling transplant settings. Further investigation in this area is needed, however, because the results to date have not been uniformly consistent.
Pharmacogenetic polymorphisms
Our increasing knowledge of genetic variability has seen a rapid expansion in the field of pharmacogenomics. The folate metabolism pathway, and more specifically, a key regulatory enzyme in this pathway, methylenetetrahydrofolate reductase (MTHFR), have received the most attention in the HSCT arena. Two nonsynonymous SNPs in the MTHFR gene result in reduced enzymatic activity. The C677T polymorphism results in an alanine-to-valine substitution at codon 222, with the variant TT genotype producing an enzyme with approximately 30% of the activity of the wild-type CC genotype, with heterozygotes (CT) possessing about 60% of normal enzymatic activity.91 The A1298C polymorphism causes alanine to be replaced by glutamic acid at codon 429, leading to reduced MTHFR activity, but to a lesser extent than that seen with C677T.92 Methotrexate (MTX) exerts its cytotoxic effect by inhibiting dihydrofolate reductase (DHFR) and other related enzymes of folate metabolism, including MTHFR.
The C677T polymorphism was first evaluated in 220 patients with CML who received a myeloablative HSCT and MTX for GVHD prophylaxis.93 After controlling for other risk factors, recipients with lower MTHFR activity (TT genotype) were found to have experienced higher MTX toxicity, as evidenced by increased oral mucositis and a trend toward delayed platelet recovery. Recently, Robien et al performed expanded genotyping of additional genes in the folate pathway and again found the MTHFR 677TT genotype to be associated with higher rates of mucositis, as was a deletion polymorphism in the thymidylate synthase (TS) gene.94
Polymorphisms of the glutathione S-transferase (GST) family of genes have also been investigated in the setting of HSCT. The major route of detoxification of busulfan is through conjugation with glutathione (GSH) which is catalyzed by the GST enzymes. A recent study of pediatric patients undergoing myeloablative HSCT for β-thalassemia found a significantly increased incidence of hepatic veno-occlusive disease (VOD) in patients with the GSTM1 null genotype compared with those with the GSTM1+ genotype.95 In this study, patients with the GSTM1 null genotype showed a significantly higher clearance and a lower steady-state concentration of busulfan after the first dose compared with those with a positive genotype. The authors proposed that this faster rate of busulfan metabolism depleted the GSH pool, allowing the busulfan metabolites generated to damage the sinusoidal endothelial cells and hepatocytes of the liver, culminating in the development of hepatic VOD.
Conclusions
As we delve deeper into the human genome, it is clear that the extent of normal variation therein is considerable. Genetic heterogeneity has an impact on HSCT by generating host-donor antigen disparity and by providing functional polymorphisms that may influence individual patient responses. Identifying additional clinically relevant transplant antigens is a priority because of the therapeutic possibilities and the potential for improved donor selection. Genotyping for KIR as a means to predict NK cell alloreactivity is also becoming increasingly relevant.
Investigation of functional genetic polymorphisms is an area of expanding interest within the field. Other genes which have been evaluated in this regard and which were not included in this review include VDR,58 ACE,96 CPSI,96 and HFE.96 The existing studies in this domain have a number of limitations, not least being that the biologic significance of the genetic polymorphisms in these genes remains largely unknown. Almost all of these analyses have assessed a limited number of arbitrarily chosen SNPs in a single or limited number of genes. In this setting, the influence of SNPs in neighboring genes is unclear. Evaluation of candidate gene pathways using a linkage disequilibrium approach targeting SNPs in a particular haplotype is a more rational and recently applied method of investigation in this area.79 Similarly unclear are the importance of SNPs determined to be of significance in individual studies, relative to that of SNPs identified in other studies. In fact, many studies have demonstrated conflicting results. Also, many polymorphic genes known to play an important role in the immune response have never been evaluated in the context of HSCT (eg, CTLA4,97 FOXP398 ). The recent appreciation of the extent of undiscovered structural variation within the genome adds a further layer of complexity to the situation.
We now possess the ability to perform large scale SNP genotyping using high throughput technologies such as microarrays,99 pyrosequencing,100 and liquid bead arrays.41 In the future, evaluation before transplantation is likely to comprise a more detailed genetic analysis of patient and donor with the ultimate goal of individualized patient treatment based on predictive risk scoring. Before this goal can be realized we need a more comprehensive examination of multiple genomic variants, undertaken in a prospective fashion, with validation in independent HSCT populations. Understanding the biologic and clinical implications of the findings will be the challenge we confront in the postgenomic era.
Authorship
Contribution: A.M. designed the format of the article and wrote the draft; and J.R. critically reviewed and edited the text, tables, and figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jerome Ritz, MD, Dana-Farber Cancer Institute, 44 Binney St, Mayer 530, Boston, MA 02115; e-mail:jerome_ritz@dfci.harvard.edu.
Acknowledgments
This work was supported by the Ted and Ilene Pasquarello Research Fund.

![Figure 2. KIR variability. (A) The KIR gene locus on chromosome 19 consists of 15 genes and 2 pseudogenes (shown in blue). Three conserved “framework” genes are located one at either end and one in the middle of the locus (red). These are present on all haplotypes and are inhibitory. The regions between the framework genes (between the brackets) contain a variable number of activating (green) and inhibitory (pink) KIR genes. Based on gene content KIR haplotypes have been divided into 2 primary sets: A and B. (B) Most group A haplotypes have the gene organization shown here. They contain the least genes and only a single activating KIR, 2DS4. (C) Group B haplotypes display a much greater variety of subtypes than group A. Two examples of at least 20 currently reported B haplotypes are shown here. An individual's KIR genotype is the sum of 2 haplotypes, one from each parent. (D) Multiple copies of the same KIR gene have been identified in single individuals as shown here for 2DL4. (E) Further diversification of KIR haplotypes occurs as a result of allelic polymorphism within individual genes. This is a prominent feature of group A haplotypes and 3 possible separate haplotypes arising as a result of polymorphisms within the 2DL3, 2DL1, 3DL1, and 3DL2 genes are shown here. More than 20 group A haplotypes emerging as a result of allelic polymorphism have been described. (Figure reproduced by kind permission of Dr Steven Marsh, Anthony Nolan Research Institute, London, from the IPD-KIR database [www.ebi.ac.uk/ipd/kir/introduction.html]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/4/10.1182_blood-2006-06-030858/4/m_zh80040708240002.jpeg?Expires=1769126794&Signature=kJD8Sf-8edWz5syluTTYOqK~X0lAnXxtMRnexBryfJ3AhbNYxzOLhSMdOBruSntEtL6ATH8yd1IpJEM-WYJCdiU1cB~~WTCjkA3LTba6WWk58HQ2eGXOvXlECUSd5hccpBpvAYwWzRImIZ3iqjq3UF2Xj2~5RcmQT5q2wg4PBgMPmvgCF2SLN-z3FfhvXiOR0aNdbrSS5KioVIeazHO1xTPOGFD2MZ0Ak4erYkebdTrcQ2UNx2EePIsrpJEOj3E7w9uBfgLHLT5uTvYKKXQQ04olh6Y~ZH8LjsxGLEs5h2GEkMLL9H~ilZD-gUz3EO2xnMmuRxpyzX6GH3JmBDdj2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal