Abstract
Transduction with recombinant adeno-associated virus (AAV) vectors is limited by the need to convert its single-stranded (ss) genome to transcriptionally active double-stranded (ds) forms. For AAV-mediated hemophilia B (HB) gene therapy, we have overcome this obstacle by constructing a liver-restricted mini–human factor IX (hFIX) expression cassette that can be packaged as complementary dimers within individual AAV particles. Molecular analysis of murine liver transduced with these self-complementary (sc) vectors demonstrated rapid formation of active ds-linear genomes that persisted stably as concatamers or monomeric circles. This unique property resulted in a 20-fold improvement in hFIX expression in mice over comparable ssAAV vectors. Administration of only 1 × 1010 scAAV particles led to expression of hFIX at supraphysiologic levels (8I U/mL) and correction of the bleeding diathesis in FIX knock-out mice. Of importance, therapeutic levels of hFIX (3%-30% of normal) were achieved in nonhuman primates using a significantly lower dose of scAAV than required with ssAAV. Furthermore, AAV5-pseudotyped scAAV vectors mediated successful transduction in macaques with pre-existing immunity to AAV8. Hence, this novel vector represents an important advance for hemophilia B gene therapy.
Introduction
The liver is an important target for gene therapy of a variety of genetic disorders, one of which is hemophilia B (HB), a life-threatening bleeding disorder that arises from mutations in the blood coagulation factor IX (FIX) gene. By maintaining plasma FIX levels above 1% of normal (> 0.05 μg/mL), the incidence of spontaneous hemorrhage is dramatically reduced and so the therapeutic end point for HB gene therapy is modest.1 Currently, adeno-associated virus (AAV) vectors are the most promising for HB gene therapy and have been the focus of 2 recent clinical trials.2 Efficient transduction with AAV is, however, limited by the need to convert its single-stranded (ss) genome into transcriptionally active double-stranded (ds) forms in target cells because of its dependence on host-cell–mediated DNA synthesis of the leading strand3 or annealing of complementary genomes derived from separate virions.4 Coinfection with adenovirus5 or priming the target tissues with genotoxic agents6,7 can enhance ds transgene formation, but the clinical use of these approaches is limited by potential toxicity. Rapid uncoating of the viral genome, as recently described with AAV8 vectors, allows efficient annealing of the ssAAV provirus to form double-stranded genomes. This unique biologic property is responsible for the 10- to 100-fold higher transduction of the liver with rAAV8 when compared with AAV2 vectors in murine models.8-10 Even so, almost 1013 AAV8 vector particles are required to achieve 100% hepatocyte transduction in mice, a level that is required for successful gene therapy of some metabolic disorders of the liver.11 This high vector dose is problematic because it may (1) cause hepatocellular toxicity in humans as observed in the recent phase 1/2 study,12 (2) result in a broader tissue distribution of vector,13 (3) elicit stronger anticapsid immunity due to the large viral load, and (4) put a tremendous burden on vector production. Needed, therefore, are novel strategies that can enhance liver transduction efficiency beyond that achieved with rAAV8 vectors, thereby allowing the use of lower vector doses to achieve therapeutic end points.
The ability to package double-stranded proviral DNA into individual AAV particles offers such an opportunity, as this would overcome a major limitation of this vector system. A number of groups have shown, using small reporter genes such as alkaline phosphatase or green fluorescent protein, that replicating provirus that is half the size of the wild-type genome is naturally packaged as 2 complementary strands within a single AAV particle. This enhances the efficiency with which transcriptionally active ds molecules are formed and has been shown to increase the transduction efficiency of hepatocytes, muscle, and retina by 10- to 100-fold.14-17 scAAV vectors, however, remain largely untested in large outbred animal models with respect to efficacy and toxicity. In addition, their smaller packaging capacity (∼ 2.5 kb) has limited their use for clinical gene therapy despite recent endeavors to improve the efficiency with which the AAV genome is packaged as dimers.14,15
Using hemophilia B gene therapy as a model, we have overcome this obstacle by constructing a novel mini-hFIX expression cassette on a modified AAV2 vector backbone that can be efficiently packaged as a tail-to-tail dimer in scAAV vectors. These vectors when critically evaluated in a murine model of HB and in large outbred nonhuman primates demonstrated significantly higher potency than their single-stranded counterparts. Indeed, expression of hFIX at therapeutic levels was observed in nonhuman primates using vector doses previously regarded as subtherapeutic in animal models as well as humans.12 Hence scAAV vectors provide a promising approach for gene therapy of HB and other disorders affecting the liver.
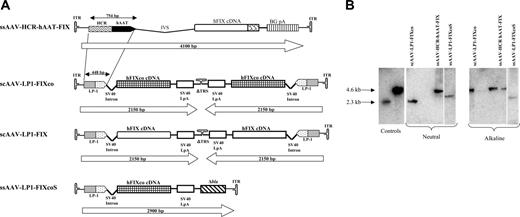
Construction and characterization of scAAV vectors. (A) Structure of rAAV hFIX vectors. Each vector is shown schematically as it is packaged in the virion, with scAAV vectors shown as dimers. ssAAV-HCR-hAAT-FIX consists of the human apolipoprotein E/C-I gene locus control region (HCR) and the human α1 antitrypsin promoter (hAAT), a chicken β actin/rabbit β globin composite intron (IVS), 1.6-kb human FIX cDNA (hFIX), and a bovine growth hormone polyadenylation signal (BGpA) flanked by the AAV internal terminal repeats (ITRs shown as hairpin loop). Self-complementary scAAV-LP1-hFIXco vector containing the LP1 promoter consisting of core liver-specific elements from HCR (base pairs 134 to 442 of GenBank record HSU32510) and hAAT (base pairs 1747 to 2001 of GenBank record K02212), modified SV40 small t antigen intron (base pairs 4644 to 4552 of GenBank record J02400), codon-optimized hFIX (hFIXco), and SV40 late polyA (SV40 LpA; base pairs 2600 to 2733 of GenBank record J02400), and a deleted 3′ trs (Δtrs). scAAV-LP1-hFIX contains the wild-type human FIX cDNA from which the 3′ untranslated region has been deleted (ΔhFIX) instead of hFIXco. ssAAV-LP1-hFIXcoS has intact 5′ and 3′ ITR and in addition contains 760 bp noncoding stuffer sequence from the β-lactamase (Δbla). (B) Characterization of ss and scAAV2/8 viral particles. Viral particles were electrophoresed on a 1% neutral or alkaline agarose gel, Southern blotted, and hybridized with a vector-specific probe. The controls consisted of approximately 4.6- and 2.3-kbp fragments that were derived from ssAAV HCR-hAAT FIX and scAAV LP1-hFIXco plasmids cut with a double cutter, which cuts outside the ITR regions. The scAAV genome migrated with the 4.6-kb marker on the alkaline agarose gel and 2.3-kb marker on neutral gel, while the migration pattern of ssAAV vectors of differing size on these 2 gels did not change.
Construction and characterization of scAAV vectors. (A) Structure of rAAV hFIX vectors. Each vector is shown schematically as it is packaged in the virion, with scAAV vectors shown as dimers. ssAAV-HCR-hAAT-FIX consists of the human apolipoprotein E/C-I gene locus control region (HCR) and the human α1 antitrypsin promoter (hAAT), a chicken β actin/rabbit β globin composite intron (IVS), 1.6-kb human FIX cDNA (hFIX), and a bovine growth hormone polyadenylation signal (BGpA) flanked by the AAV internal terminal repeats (ITRs shown as hairpin loop). Self-complementary scAAV-LP1-hFIXco vector containing the LP1 promoter consisting of core liver-specific elements from HCR (base pairs 134 to 442 of GenBank record HSU32510) and hAAT (base pairs 1747 to 2001 of GenBank record K02212), modified SV40 small t antigen intron (base pairs 4644 to 4552 of GenBank record J02400), codon-optimized hFIX (hFIXco), and SV40 late polyA (SV40 LpA; base pairs 2600 to 2733 of GenBank record J02400), and a deleted 3′ trs (Δtrs). scAAV-LP1-hFIX contains the wild-type human FIX cDNA from which the 3′ untranslated region has been deleted (ΔhFIX) instead of hFIXco. ssAAV-LP1-hFIXcoS has intact 5′ and 3′ ITR and in addition contains 760 bp noncoding stuffer sequence from the β-lactamase (Δbla). (B) Characterization of ss and scAAV2/8 viral particles. Viral particles were electrophoresed on a 1% neutral or alkaline agarose gel, Southern blotted, and hybridized with a vector-specific probe. The controls consisted of approximately 4.6- and 2.3-kbp fragments that were derived from ssAAV HCR-hAAT FIX and scAAV LP1-hFIXco plasmids cut with a double cutter, which cuts outside the ITR regions. The scAAV genome migrated with the 4.6-kb marker on the alkaline agarose gel and 2.3-kb marker on neutral gel, while the migration pattern of ssAAV vectors of differing size on these 2 gels did not change.
Materials and methods
AAV-hFIX vector production and purification
An scAAV backbone plasmid was constructed by ligating MscI-BsaI and BsaI-Tsp45I fragments from AAV2-HCR-hAAT-FIX10 to the simian virus 40 late polyA (SV40 LpA). The resulting plasmid contained the modified AAV2 backbone with an intact 5′ terminal resolution site (trs) and a deleted 3′ trs analogous to that described previously.14,15 The LP1 enhancer/ promoter was constructed using standard polymerase chain reaction (PCR) methods with amplification of consecutive segments of the human apolipoprotein hepatic control region (HCR) the human alpha-1-antitrypsin (hAAT) gene promoter including the 5′ untranslated region and cloned upstream of a modified SV40 small t antigen intron (SV40 intron) modified at positions 4582 (g to c), 4580 (g to c), 4578 (a to c), and 4561 (a to t) into the modified AAV-2 backbone (Figure 1). The wild-type hFIX cDNA without the 3′ untranslated region (UTR) regions was PCR amplified from AAV-HCR-hAAT-hFIX10 and inserted downstream of the modified SV40 intron to make scAAV-LP1-hFIX. A codon-optimized hFIX was generated using codons most frequently found in highly expressed eukaryotic genes,18 synthesized as oligonucleotides, and subsequently assembled by ligation, PCR amplified, and sequenced prior to cloning into the AAV-LP1 backbone to create scAAV-LP1-hFIXco. The full sequence of scAAV-LP1-hFIXco has been outlined in Figure S1, available on the Blood website; click on the Supplemental Figure link at the top of the online article. ssAAV-LP1-hFIXcoS contained a reconstructed normal 3′trs and 760 bp of stuffer noncoding sequence from the β-lactamase gene inserted downstream of the SV40-LpA. ss and scAAV vectors were made by the adenovirus-free transient transfection method described before.19 AAV5-pseudotyped vector particles were generated using a chimeric AAV2 Rep-5Cap packaging plasmid called pLT-RCO3, which is based on XX220 and pAAV5-221 and similar in configuration to that described before.22 Additionally, AAV8-pseudotyped vectors were made using the packaging plasmid pAAV8-2.8 AAV2/5 and 2/8 vectors were purified by the previously described ion exchange chromatography method.19 Vector genome (vg) titers were determined by our previously described quantitative slot-blot method using supercoiled plasmid DNA as standards.13 Of importance, each scAAV particle was calculated as containing 2 copies of ss viral genomes. The purified vector stocks were consistently free of contamination with wt-AAV and cellular and adenoviral proteins as judged by our previously described methods.13,19 To determine the molecular configuration of scAAV, vector stock was incubated with an equal volume of sample loading buffer (60 mM NaOH, 2 mm EDTA, 20% Ficoll, and 0.06% Bromocresol green) and separated on a 1% agarose gel containing 30 mM NaOH and 1 mM EDTA, transferred to nitrocellulose by Southern blotting, and hybridized with an α32P-labeled 424-bp BstApI fragment from the scAAV-LP1-FIXco at 42°C. The intensity of the hybridization was determined using the STORM phosphorimager (Amersham Biosciences, Amersham, United Kingdom).
Animal studies
All procedures were performed in accordance with institutional guidelines under protocols approved by the Institutional Biosafety Committee and the Institutional Animal Care and Use Committee at St Jude Children's Research Hospital and the University of Tennessee, Memphis. All animal work carried out in the United Kingdom was performed under the authority of the UK Home Office Project and Personal Licenses regulations and was compliant with the guidelines of the University College and Imperial College ethics review committee. The HB mouse strain, based on 129/sv mice with disruption of the FIX gene, was obtained from Inder Verma (Salk Institute, La Jolla, CA).23 Tail-vein administration of rAAV vector particles was performed in 7- to 10-week-old male mice as described before.13 A two-thirds partial hepatectomy was performed at 16 weeks after tail-vein administration of 1 × 1010 scAAV2/8-LP1-hFIXco as previously described.24 Four weeks later, at a time when the liver mass was fully reconstituted, the mice were killed to harvest the liver. Captive bred male Macaca mulatta aged between 2.5 to 6.6 years and weighing between 1.6 and 7.6 kg were purchased from Covance Research Products (Alice, TX) and housed in the dedicated primate facility at the University of Tennessee Health Science Center. Vector particles in phosphate-buffered saline (PBS) were infused into the mesenteric circulation of male macaques, as described before.25 The CBC, serum chemistry, and coagulation profile were performed by ANTECH Diagnostics (Southaven, MS). Serum IL-6 levels were assessed using an IL-6 immunoassay kit (R&D Systems, Oxford, United Kingdom) as per the manufacturer's instruction. To eradicate anti-hIX antibody in M1-sc treatment with rituximab (250 mg/m2, 2 doses at 3 weekly intervals) and daily oral cyclosporine (30-100 mg/kg per day, adjusted to maintain through therapeutic levels at 100-500 ng/mL) was started approximately 23 weeks after gene transfer and maintained for a period of 4 weeks.
Determination of transduction efficiency and vector biodistribution
Human FIX antigen levels in murine and rhesus samples were determined by enzyme-linked immunosorbent assay (ELISA) according to the previously described methods.25 The probability of statistical difference between experimental groups was determined by one-way ANOVA and paired student t test using GraphPad Prizm version 4.0 software (GraphPad, San Diego, CA). A one-stage assay for hFIX:C was performed as described previously.26 Background hFIX:C levels in untreated hemophiliac mice were less than 0.03 U/mL. The baseline clotting time of citrated plasma (diluted × 100) from untreated HB mice in this assay was 87.2 ± 3 seconds, and that of normal 129/sv mice was 74.8 ± 3 seconds. Thrombin-antithrombin levels were determined using the Enzygnost TAT kit (Dade-Behring, Milton Keynes, United Kingdom). Low-molecular-weight Hirt DNA was extracted from liver at varying time points after tail-vein administration of scAAV-2/8 vector using our previously described method.27 Undigested Hirt DNA (10 μg) or DNA digested with PstI (which cuts once within the FIX expression cassette) was separated on 1% agarose gel, transferred to nitrocellulose by Southern blotting, and hybridized with an α32P-labeled 842-bp BstApI LP1 fragment at 42°C. The intensity of the hybridization was determined using the STORM phosphorimager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). To determine AAV transgene copy number, high-molecular-weight genomic DNA (10 μg) extracted from murine and macaque tissue samples, using our previously described method,13,25 was digested with either BsrDI or a combination of EcoRI and PstI and electrophoresed through a 0.8% agarose gel and transferred to a nylon membrane (Hybond-N+; Amersham Biosciences, Arlington Heights, IL and Amersham Biosciences, Amersham, United Kingdom) and then hybridized and quantitated as described earlier in this section. To evaluate the biodistribution of AAV-FIX vectors, 1 μg genomic DNA extracted from various murine and macaque tissues was subjected to PCR using primers that amplified a 617-bp region of hFIXco (5′ primer: 5′ TTTCCTGATGTGGACTATGT 3′ and 3′ primer: 5′ TCATGGAAGCCAGCACAGAACATG 3′) as described previously.25 Integrity of DNA was determined by amplifying a 604-bp region of the murine or rhesus β-actin gene using previously described primers.13 To determine which organs expressed hFIXco, total RNA was isolated and subjected to the reverse-transcription (RT) conditions in the presence or absence of reverse transcriptase as described before.13 Human FIX expression in various formalin-fixed, paraffin-embedded tissue specimens was analyzed by immunohistochemistry as described before.10
Detection of anti–human FIX antibodies and anti-AAV antibodies
Plasma samples from macaques were screened for the presence of antibodies against hFIX using an ELISA described previously.25 Additionally, the positive samples were subjected to the Bethesda assay as described before.25 An immunocapture assay was used to detect anti-AAV–specific antibodies in rhesus plasma as described before. 25 Results were expressed as the end-point titer, defined as the reciprocal of the interpolated dilution with an absorbance value equal to 5 times the mean absorbance background value. Neutralizing antibody titers were analyzed by determining the ability of rhesus serum to inhibit transduction of 293T cells by pseudotyped rAAV vector containing the CMV-GIL as previously described.25,28 The neutralizing antibody titer was arbitrarily calculated as reciprocal of the highest dilution, which inhibited transduction of 293T cells by 50%.
Epitope mapping of anti–human FIX antibodies
Antibodies were mapped using a 2-step strategy. First, domain swap mutants were used in which individual domains were exchanged by their counterparts from human FIX.29 Second, fine mapping within the protease domain was performed using purified hFIX chimeras with FX replacements in surface loops 223 to 227, 258 to 267, or 315 to 323.30 Together these loops comprise the human/rhesus differences in positions 226, 227, 261, 315 and 321.31 Mapping was performed in a standard ELISA format in which wells were coated with appropriate anti-FIX or anti-FX antibodies (0.1 μg/well). Wells were blocked with Tris-buffered saline (pH 7.4) containing 10 mM CaCl2 and 3% (wt/vol) human serum albumin prior to the addition of purified hFIX, FIXa, FX, or hFIX/X chimera (0.1 μg) in the same buffer containing 0.1% (wt/vol) albumin. After incubation (1 hour at 37°C) and washing, wells were incubated for 1 hour with various dilutions of rhesus plasma in the same buffer, and after washing, bound rhesus antibody was quantified using peroxidase-labeled antibodies against monkey IgG (product number A2054; Sigma-Aldrich, Zwijndrecht, the Netherlands).
Results
Construction and characterization of mini-hFIX expression cassettes
Our previously described 4123-bp ssAAV-HCR-hAAT-hFIX liver-restricted expression cassette was modified (Figure 1A) to create a more compact (2.1 kb) hFIX expression cassette (scAAV-LP1-hFIXco) that met the packaging requirements of scAAV, while maintaining liver-restricted expression.19,32 Key modifications included targeted deletions from the HCR and hAAT elements based on an analysis of liver transcription factor binding sites and deletion of the FIX 3′ untranslated (3′ UTR) sequences, as mutations in this region have not been described in HB patients.33 Additionally, the hFIX coding sequence was modified by using a subset of codons most frequently found in highly expressed eukaryotic genes (“codon optimization”), and then adjusted to reduce the potential for inappropriate splicing and CpG methylation to augment transgene expression. A second construct was made in which the wild-type hFIX coding sequence without the 3′ UTR was used (scAAV-LP1-hFIX). In both of these vectors, the downstream terminal resolution site (trs) was deleted to enhance the formation of tail-to-tail self-complementary dimers.15 For comparison, an almost identical single-stranded vector was made (ssAAV-LP1-hFIXcoS) in which both trs's were intact. This vector contained, in addition, 760 bp of noncoding stuffer sequence at the 3′ end and, therefore, at a total size of 2.9 kb, exceeded the packaging limit for scAAV. The yield of scAAV and the ssAAV vectors containing the LP1-hFIX cassette with or without the stuffer sequences, when pseudotyped with serotype 5 or 8 capsid protein, was in the range of 1 to 3 × 104 particles/cell, thus confirming a previous observation by others that deletion of a single trs does not compromise viral replication or packaging.15,34 Alkaline agarose gel electrophoresis confirmed that the scAAV2/8-LP1-hFIXco genome was consistently packaged as a dimer of approximately 4.6 kb (Figure 1B), while ssAAV2/8-LP1-hFIXcoS and ssAAV2/8-HCR-hAAT-hFIX were predominantly packaged as single-stranded genomes.
scAAV vectors mediate substantially higher transduction of the murine liver
Different doses of scAAV2/8-LP1-hFIXco or ssAAV2/8-HCR-hAAT-hFIX vectors were administered via tail vein to male C57Bl/6 mice, and plasma hFIX levels were assessed at 6 weeks. As shown in Figure 2A, a relatively linear vector-dose transgene-expression profile was observed with both types of vectors with no evidence of saturation kinetics at the doses examined. This is consistent with previous reports with single-stranded serotype 8 vectors.10,11 At all vector doses evaluated, steady-state hFIX levels were consistently higher for scAAV2/8-LP1-hFIXco by approximately 20-fold over the single-stranded counterparts. When 1 × 1011 scAAV2/8-LP1 hFIXco vector particles were injected into the tail vein of mice, supraphysiologic levels of hFIX (151 ± 43 μg/mL) were observed, compared with 5 ± 0.5 μg/mL with an equivalent dose of ssAAV2/8-HCR-hAAT-hFIX. We next compared transduction efficiency of our scAAV with AAV-hF.IX16, the single-stranded vector that was used in the recent clinical trial.35 This vector, which in addition to the HCR-hAAT regulatory complex also contains a 1.4-kb region from the first intron of the hFIX gene, has been shown to mediate high levels of hFIX in animal models after liver-targeted delivery.12,32,35 In our direct comparison, steady-state hFIX expression after tail-vein administration of 2.5 × 1010 serotype 8–pseudotyped AAV-hF.IX16 particles was 10 ± 0.5 μg/mL, which is almost 3-fold lower than that achieved with an equivalent dose of scAAV2/8-LP1-hFIXco (29 ± 2 μg/mL; P < .001, by Student t test). Hence, despite substantial engineering and size reduction, our scAAV-FIX vector appears to offer an advantage over the current most optimized ssAAV-FIX vector in mice.
Influence of genomic configuration and capsid proteins on transduction efficiency. (A) Plasma hFIX levels at 6 weeks after tail-vein administration of varying doses of ssAAV2/8-HCR-hAAT-FIX (▵, n = 4/dose) and scAAV2/8-LP1-hFIXco (□, n = 4/dose). Results are depicted as average together with the standard error of the mean (error bars are not visible at some time points because of the log scale). (B) Representative results of immunohistochemical staining for hFIX at 7 days and 42 days (middle 2 panels) after tail-vein administration of 1 × 1011 scAAV2/8-LP1-hFIXco compared with naive liver (far-left panel) from a control animal. The far-right panel shows hFIX expression in hepatocytes at 42 days after tail-vein administration of 1 × 1011 ssAAV2/8 vector. Magnification is × 40. (C) Influence of AAV capsid proteins on scAAV-LP1-hFIXco–mediated-hFIX expression. Plasma hFIX levels assessed at 4 weeks after tail-vein administration of 1 × 1011 vg of either scAAV2/8-LP1-hFIXco (n = 4) or scAAV2/5-LP1-hFIXco (n = 4). Results are depicted as the average together with the standard error of the mean.
Influence of genomic configuration and capsid proteins on transduction efficiency. (A) Plasma hFIX levels at 6 weeks after tail-vein administration of varying doses of ssAAV2/8-HCR-hAAT-FIX (▵, n = 4/dose) and scAAV2/8-LP1-hFIXco (□, n = 4/dose). Results are depicted as average together with the standard error of the mean (error bars are not visible at some time points because of the log scale). (B) Representative results of immunohistochemical staining for hFIX at 7 days and 42 days (middle 2 panels) after tail-vein administration of 1 × 1011 scAAV2/8-LP1-hFIXco compared with naive liver (far-left panel) from a control animal. The far-right panel shows hFIX expression in hepatocytes at 42 days after tail-vein administration of 1 × 1011 ssAAV2/8 vector. Magnification is × 40. (C) Influence of AAV capsid proteins on scAAV-LP1-hFIXco–mediated-hFIX expression. Plasma hFIX levels assessed at 4 weeks after tail-vein administration of 1 × 1011 vg of either scAAV2/8-LP1-hFIXco (n = 4) or scAAV2/5-LP1-hFIXco (n = 4). Results are depicted as the average together with the standard error of the mean.
Immunohistochemical staining of murine liver after tail-vein administration of 1 × 1011 scAAV2/8-LP1-hFIXco showed expression of hFIX in 25% of hepatocytes at day 7, which increased to more than 90% by 6 weeks (Figure 2B). This is substantially higher than that observed with an equivalent dose of ssAAV2/8 vector.10,11 Tail-vein administration of 1 × 1011 scAAV-LP1-hFIXco particles pseudotyped with serotype 5 capsid resulted in peak hFIX levels of 8.3 ± 2 μg/mL in male C57Bl/6 mice, which is about 2 logs greater than that previously reported for ssAAV2/5-HCR-hAAT-hFIX vectors,10 but approximately 20-fold lower than the levels achieved with an equivalent dose of scAAV2/8 vector (Figure 2C). Similar differences have been noted previously with ssAAV vectors, thus indicating that capsid proteins and not genomic conformation are primarily responsible for the inferior transduction observed with AAV5 vectors in murine models.10
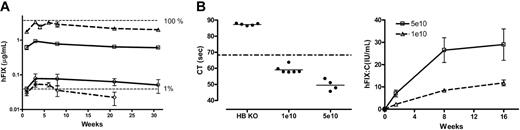
Low-dose transduction efficiency with ss and scAAV vectors in murine models. (A) Transgene expression in C57Bl/6 mice. hFIX-expression profile over 31 weeks after tail-vein administration of 2 × 109 vg/mouse (n = 3) of scAAV2/8-LP1-hFIXco (▵), scAAV2/8-LP1-hFIX (□), ssAAV2/8-LP1-hFIXcoS (○), and ssAAV2/8-HCR-hAAT-FIX (⋄) into C57Bl/6 mice. All hFIX results are depicted as the average together with the standard error of the mean. (B) Correction of clotting times and expression of hFIX:C in 129/sv HB mice. Following tail-vein injection of either 1 × 1010 (low-dose cohort, n = 6) or 5 × 1010 (high-dose cohort, n = 4) scAAV2/8-LP1-hFIXco vector particles into 129/sv HB mice, the clotting time (left-hand panel at 4 weeks) and biologically active hFIX:C levels (right-hand panel) over 16 weeks were determined.
Low-dose transduction efficiency with ss and scAAV vectors in murine models. (A) Transgene expression in C57Bl/6 mice. hFIX-expression profile over 31 weeks after tail-vein administration of 2 × 109 vg/mouse (n = 3) of scAAV2/8-LP1-hFIXco (▵), scAAV2/8-LP1-hFIX (□), ssAAV2/8-LP1-hFIXcoS (○), and ssAAV2/8-HCR-hAAT-FIX (⋄) into C57Bl/6 mice. All hFIX results are depicted as the average together with the standard error of the mean. (B) Correction of clotting times and expression of hFIX:C in 129/sv HB mice. Following tail-vein injection of either 1 × 1010 (low-dose cohort, n = 6) or 5 × 1010 (high-dose cohort, n = 4) scAAV2/8-LP1-hFIXco vector particles into 129/sv HB mice, the clotting time (left-hand panel at 4 weeks) and biologically active hFIX:C levels (right-hand panel) over 16 weeks were determined.
The transgene-expression profile was next assessed following tail-vein administration of 2 × 109 vector genomes (vg), a dose below the transduction threshold for ssAAV2 vectors.13,36 Surprisingly, there was no difference in expression kinetics between vectors containing ssAAV2/8 (HCR-hAAT-hFIX and LP1-hFIX-coS) or scAAV2/8 (LP1-hFIXco and LP1-hFIX) genomes (Figure 3A), with hFIX being detectable within a week and reaching peak levels after a lag phase of 4 weeks. There then followed a decline in transgene expression in all 4 cohorts of between 40% to 70% over a period of 20 weeks prior to stabilization, similar to that previously observed with ssAAV2/8 vectors.10,11 Peak levels of hFIX in ssAAV2/8-HCR-hAAT-hFIX–transduced mice were 0.052 ± 0.01 μg/mL, consistent with our previous report.10 Similar levels of hFIX (0.076 ± 0.03 μg/mL) were achieved in the ssAAV2/8-LP1-hFIXcoS cohort. In contrast, tail-vein administration of 2 × 109 scAAV2/8-LP1-hFIXco vector particles resulted in hFIX expression at almost 100% of physiologic levels (3.2 ± 0.15 μg/mL). This difference in transgene expression between single-stranded and scAAV was highly significant (P < .001, by one-way ANOVA analysis). Expression of hFIX in the scAAV2/8-LP1-hFIX cohort was 4-fold lower (0.77 ± 0.03 μg/mL). Transgene copy number in the liver of selected animals from the scAAV2/8-LP1-hFIXco and scAAV2/8-LP1-hFIX cohorts was similar (7.3 and 6.2 copies per diploid genome [c/dg], respectively), indicating that expression of the hFIX gene in mice appears to be influenced by codon use, when its 3′ UTR is removed.
To confirm correction of phenotype with this vector, we injected either 1 × 1010 (low-dose cohort, n = 6) or 5 × 1010 (high-dose cohort, n = 4) scAAV2/8 LP1-hFIXco into the tail vein of 129/sv HB mice that have a large deletion within exon 8 of the murine FIX gene.23 Peak hFIX:C levels, as determined by a one-stage clotting assay, were 8 ± 0.7 and 27 ± 6 IU/mL in the low- and high-dose cohorts, respectively (Figure 3B). These levels were significantly above background (untreated HB hemophiliac mice hFIX:C level = 0.03 U/mL) and significantly higher than the normal human FIX:C value, which is defined as 1 IU/mL. Complete concordance between hFIX:C and hFIX antigen levels was noted in both cohorts of mice at all time points examined. Of importance, all 6 untreated HB mice required wound cautery after tail-vein puncture to achieve hemostasis, while this was not necessary in the scAAV-treated mice. Plasma thrombin-antithrombin complexes (2.2 ± 0.2 μg/L) were not elevated, indicating that supraphysiologic levels of biologically active hFIX do not induce a noticeable hypercoagulable state in HB mice. Finally, anti-hFIX antibodies were not detected in the scAAV-treated HB mice at any stage after gene transfer.
Molecular configuration and biodistribution of scAAV vector
Southern blot analysis of Hirt DNA extracted from the liver at 24 hours showed that most of the scAAV2/8-LP1-FIXco genome was present as a double-stranded 2.1-kbp linear monomer, as confirmed by PstI digestion, which cuts approximately 500 bp from the 3′Δ ITR to release a 1.8-kbp product (Figure 4A, lane 3). Even at this early time point, aside from supercoiled circles, an array of larger molecular forms including high-molecular-weight concatamers (HMWCs, ∼ 5%) were observed. At 42 days after tail-vein administration of scAAV2/8-LP1-hFIXco, supercoiled circular and concatameric forms predominated. Digestion with PstI generated both head-to-head and head-to-tail concatamer fragments in a ratio of 3:1. The circular forms, as expected, were resistant to digestion with plasmid-safe DNase.
To distinguish between transgene expression from integrated and extrachromosomal vector, we induced hepatocellular regeneration by performing a two-thirds hepatectomy in HB mice 16 weeks after administration of 1 × 1010 scAAV2/8-LP1-hFIXco. Although hemostasis was easily achieved, there was a sharp decline (∼ 90%) in hFIX:C levels, which did not recover despite complete restoration of liver-cell mass within 4 weeks of surgery. In addition, a concomitant decline in the vector copy number (∼ 86%) was also observed after hepatectomy (Figure 4B), indicating that extrachromosomal and not integrated scAAV genomes are primarily responsible for transgene expression.
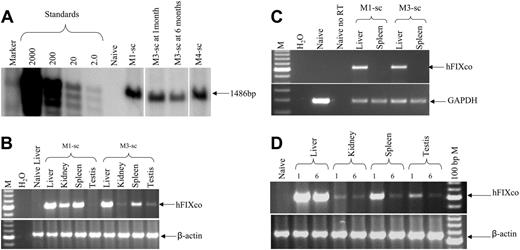
Finally, using a semiquantitative PCR assay, scAAV genomes could be detected in all tissues examined 6 weeks after tail-vein administration of scAAV2/8-LP1-hFIXco (Figure 5), with disproportionately higher levels of the transgene in the liver, as previously reported.10,11 hFIX mRNA expression, however, was detected only in the liver by RT-PCR analysis. Direct intramuscular injection of 3 × 1010 scAAV2/8-LP1-hFIXco resulted in efficient transduction but not transcription as assessed by RT-PCR analysis of skeletal muscle, suggesting that the LP1 regulatory element is not efficient at directing transgene expression in nonhepatic murine tissues.
Liver-targeted delivery of lower doses of scAAV mediates expression of hFIX at therapeutic levels in nonhuman primates
Based on extrapolation from murine studies, we have previously effected efficient transduction of the macaque liver with a standard dose of 4 × 1012 ssAAV vector/kg pseudotyped with serotype 2, 5, or 8 capsid.10,25 A similar dose of AAV-hF.IX16 mediated transient expression of hFIX following liver-directed delivery of serotype 2 vector in severe HB patients.35 Lower doses of ssAAV2 (2 × 1011 or 1 × 1012 vg/kg) were ineffective in humans and generated inconsistent expression in animals.10,25,35,37 The higher potency of scAAV in mice prompted the evaluation of 2 lower doses of scAAV2/8-LP1-hFIXco in macaques (Figure 6). The first group of macaques (M1-sc and M2-sc) received 1 × 1012 vp/kg vector, which was infused into the mesenteric vein. scAAV2/8-LP1-hFIXco was well tolerated without perturbation of serum interleukin-6 levels (< 3 pg/mL) or liver transaminases (alanine aminotransferase, < 65 U/L) over a period of 6 weeks after gene transfer. Within 24 hours of vector administration, hFIX was detectable at 0.25 and 1.2 μg/mL and continued to rise over a period of 2 weeks prior to reaching peak values of approximately 12% and 30% of normal in M1-sc and M2-sc, respectively (Figure 6A). In M1-sc, these levels were stably maintained for 9 weeks before being abrogated by anti-hFIX antibodies whose titer increased over a period of 4 weeks, reaching a stable level of 15 BIAU/mL. The coagulation screen in this animal was normal, which implies selective neutralization of hFIX and not its rhesus cognate by this antibody. Epitope mapping studies using hFIX domain swap variants demonstrated that the rhesus antihuman antibodies were exclusively directed against the serine protease domain (Figure 6B). Of the 12 amino acid differences between hFIX and its rhesus cognate, 6 are located in the catalytic domain, spread over surface loops that border the reactive site cleft. Using a panel of loop substitution mutants, antibody binding could be attributed predominantly to hFIX loop 258 to 267, which differs from its rhesus cognate by Ala instead of Thr in position 261. Treatment with rituximab and daily oral cyclosporine was commenced to eradicate the neutralizing anti-hFIX antibody, and within 4 weeks the inhibitor became undetectable. Human FIX was once again detectable in rhesus plasma at approximately 0.8 ± 0.1 μg/mL (Figure 6A) with no perturbation of serum albumin levels, suggesting the absence of nephrotic syndrome.
Molecular configuration of scAAV genome in murine liver. (A) Southern analysis of modified Hirt DNA extracted from the liver of a subset of mice at day 1 or 42. Approximately 15 μg DNA from each time point was electrophoresed uncut (uncut), or following digestion with single cutters (PstI) or plasmid safe DNase (PS-D). Shown are bands representing monomeric supercoiled circular (MSC) or dimeric supercoiled circular (DSC), ds-linear (L-ds), or high-molecular-weight concatamers (HMWCs) in the head-tail (HT) and head-to-head (HH) formation. For comparison, 5 × 106 and 2 × 107 purified vector particles were loaded directly on the gel after denaturation (last 2 lanes). Bottom panel is a schematic representation of dimeric scAAV-LP1-hFIXco in the HT (top) and HH (bottom) configuration together with the sites of cleavage by PstI. (B) Southern blot analysis of liver DNA isolated before and after partial hepatectomy at 16 and 20 weeks, respectively, after tail-vein administration of 1 × 1010 scAAV2/8-LP1-hFIXco vector particles into 129/sv HB mice (HBM1 and 2). Each lane contains 10 μg DNA digested with EcoRI and PstI, which releases a 1.1-kb fragment. Proviral copy number (shown at the bottom) was deduced from standards, which consisted of serial dilutions of vector DNA (0.13 to 13 copies) in 10 μg negative genomic DNA.
Molecular configuration of scAAV genome in murine liver. (A) Southern analysis of modified Hirt DNA extracted from the liver of a subset of mice at day 1 or 42. Approximately 15 μg DNA from each time point was electrophoresed uncut (uncut), or following digestion with single cutters (PstI) or plasmid safe DNase (PS-D). Shown are bands representing monomeric supercoiled circular (MSC) or dimeric supercoiled circular (DSC), ds-linear (L-ds), or high-molecular-weight concatamers (HMWCs) in the head-tail (HT) and head-to-head (HH) formation. For comparison, 5 × 106 and 2 × 107 purified vector particles were loaded directly on the gel after denaturation (last 2 lanes). Bottom panel is a schematic representation of dimeric scAAV-LP1-hFIXco in the HT (top) and HH (bottom) configuration together with the sites of cleavage by PstI. (B) Southern blot analysis of liver DNA isolated before and after partial hepatectomy at 16 and 20 weeks, respectively, after tail-vein administration of 1 × 1010 scAAV2/8-LP1-hFIXco vector particles into 129/sv HB mice (HBM1 and 2). Each lane contains 10 μg DNA digested with EcoRI and PstI, which releases a 1.1-kb fragment. Proviral copy number (shown at the bottom) was deduced from standards, which consisted of serial dilutions of vector DNA (0.13 to 13 copies) in 10 μg negative genomic DNA.
Two additional macaques (M3-sc and M4-sc) received 4 × 1011vp/kg scAAV2/8-LP1-hFIXco, which represents a log reduction of our standard dose. As shown in Figure 6C, steady-state hFIX levels of 0.19 μg/mL (3.75% of normal) were achieved within 2 weeks of liver-targeted administration of vector in M3-sc. Transgene expression in this animal has been stably maintained at 1% to 3% level for the duration of the study (> 6 months). The second macaque in this group (M4-sc) had a low but detectable pre-existing anti-AAV8 antibody titer (3.4 compared with between 0.6-1.0 relative units for monkeys M1-sc–M3-sc). This macaque was not successfully transduced after liver-targeted administration of 4 × 1011 vg/kg, suggesting that even modest levels of pre-existing immunity were sufficient to block successful transduction of the liver. However, switching capsid proteins resulted in successful transduction in this animal when challenged with 1 × 1012 vg/kg scAAV2/5-LP1-hFIXco, despite high titers of anti-AAV8 antibodies (> 27 relative units) at the time of vector administration (Figure 6B). Anti-AAV5 antibody titer in this animal rose from 0.83 prior to gene transfer to 33 relative units at 6 weeks after mesenteric-vein administration of scAAV2/5. The kinetics of hFIX expression was identical to that observed in M1-sc and M2-sc given scAAV-2/8, with rapid increase in expression to peak levels of 20% of normal within 2 weeks after vector administration.
Biodistribution of scAAV2/8-LP1-hFIXco following tail-vein administration of vector. (A) PCR analysis of 1 μg genomic DNA, isolated from the indicated organs 6 weeks after tail-vein (TV) administration of 1 × 1011 particles using primers unique to hFIXco designed to amplify a 617-bp product. Control consists of genomic DNA extracted from the liver of an untransduced animal. scAAV2/8-LP1-hFIXco vector particles (3 × 1010) were also administered directly into the quadriceps muscle for comparison. Genomic DNA (1 μg) extracted from the muscle tissue was subjected to PCR using identical conditions. Integrity of DNA was determined by amplifying a 604-bp region of the murine β-actin gene and is shown at the bottom panel. (B) RT-PCR analysis. Expression analysis of hFIXco mRNA by RT-PCR following tail-vein injection of 1 × 1011 or intramuscular administration of 3 × 1010 scAAV-LP1-hFIXco particles or mock-transduced mice (Naive liver). RNA samples were amplified with and without RT to exclude genomic DNA amplification. Integrity of the RNA was determined by amplifying a 295-bp region of the murine GAPDH gene and is shown at the bottom panel.
Biodistribution of scAAV2/8-LP1-hFIXco following tail-vein administration of vector. (A) PCR analysis of 1 μg genomic DNA, isolated from the indicated organs 6 weeks after tail-vein (TV) administration of 1 × 1011 particles using primers unique to hFIXco designed to amplify a 617-bp product. Control consists of genomic DNA extracted from the liver of an untransduced animal. scAAV2/8-LP1-hFIXco vector particles (3 × 1010) were also administered directly into the quadriceps muscle for comparison. Genomic DNA (1 μg) extracted from the muscle tissue was subjected to PCR using identical conditions. Integrity of DNA was determined by amplifying a 604-bp region of the murine β-actin gene and is shown at the bottom panel. (B) RT-PCR analysis. Expression analysis of hFIXco mRNA by RT-PCR following tail-vein injection of 1 × 1011 or intramuscular administration of 3 × 1010 scAAV-LP1-hFIXco particles or mock-transduced mice (Naive liver). RNA samples were amplified with and without RT to exclude genomic DNA amplification. Integrity of the RNA was determined by amplifying a 295-bp region of the murine GAPDH gene and is shown at the bottom panel.
Transduction in rhesus macaques following liver-targeted delivery of scAAV2/8 and scAAV2/5-LP1-hFIXco. (A) Human FIX concentration in rhesus plasma was determined at the indicated time points after administration of 1 × 1012 vg/kg (M1-sc [□], M2-sc [○]) scAAV2/8-LP1-hFIXco into the mesenteric vein of 2 rhesus macaques. Treatment of M1-sc with rituximab (Rit × 2 doses) and oral cyclosporine (CyA) is shown. (B) Graphic representation of the reactivity profile of the anti-hFIX antibody in M1-sc, as determined using a panel of hFIX/X chimeras with domain or surface loop substitutions derived from FX. Antibody binding to chimeras was expressed as percentage of binding to wild-type FIX. (C) Transgene expression after mesenteric-vein administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in 2 macaques (M3-sc [⋄] and M4-sc [▿]). M4-sc additionally received 1 × 1012 vg/kg scAAV2/5-LP1-hFIXco at 56 days after initial exposure to scAAV2/8. Each sample was independently evaluated on at least 3 separate occasions, and the results are depicted as an average together with the standard error of the mean.
Transduction in rhesus macaques following liver-targeted delivery of scAAV2/8 and scAAV2/5-LP1-hFIXco. (A) Human FIX concentration in rhesus plasma was determined at the indicated time points after administration of 1 × 1012 vg/kg (M1-sc [□], M2-sc [○]) scAAV2/8-LP1-hFIXco into the mesenteric vein of 2 rhesus macaques. Treatment of M1-sc with rituximab (Rit × 2 doses) and oral cyclosporine (CyA) is shown. (B) Graphic representation of the reactivity profile of the anti-hFIX antibody in M1-sc, as determined using a panel of hFIX/X chimeras with domain or surface loop substitutions derived from FX. Antibody binding to chimeras was expressed as percentage of binding to wild-type FIX. (C) Transgene expression after mesenteric-vein administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in 2 macaques (M3-sc [⋄] and M4-sc [▿]). M4-sc additionally received 1 × 1012 vg/kg scAAV2/5-LP1-hFIXco at 56 days after initial exposure to scAAV2/8. Each sample was independently evaluated on at least 3 separate occasions, and the results are depicted as an average together with the standard error of the mean.
Transgene copy number in rhesus liver at 1 month after liver-targeted delivery of 1 × 1012 vg/kg scAAV2/8-LP1-hFIXco or scAAV2/5-LP1-hFIXco was 61 and 49 c/dg, respectively. This compares favorably with our previous experience in macaques given a 4-fold higher dose (4 × 1012 vg/kg) of ssAAV where the transgene copy number ranged between 2 to 40 c/dg.10,25 In M3-sc, which was transduced with 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco, the transgene copy number in the liver was 34 c/dg (Figure 7A) at 1 month but than declined to 16 c/dg by 6 months. However, hFIX expression was stably maintained at 1% to 3% of normal over this period, showing once again the discordance between transgene expression and copy number reported by us before.10,25 Semiquantitative PCR analysis demonstrated the presence of the provirus in all the tissues examined including the testis, although there was no clear relationship between vector spillover and vector dose outside of the liver and spleen (Figure 7B). RT-PCR analysis revealed the presence of the hFIX mRNA only in the liver, again confirming the fidelity of the expression cassette (Figure 7C). Further analysis of the vector genome distribution at 6 months revealed a substantial decline in copy number in all the nonhepatic tissues including the testis in M3-sc, implying non–germ-cell transduction (Figure 7D).
Molecular analysis in rhesus macaques. (A) Southern blot analysis of DNA (10 μg) derived from liver biopsy specimens from monkeys M1-sc and M4-sc 1 month after liver-targeted delivery of 1 × 1012 vg/kg scAAV2/8-LP1-hFIXco and scAAV2/5-LP1-hFIXco, respectively, or at 1 and 6 months after administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in M3-sc. After digestion with BsrDI, the released 1486-bp fragment was probed with an LP1-specific probe. (B) Limited biodistribution analysis. Genomic DNA (top panel) isolated at 1 month from the indicated organs following liver-targeted administration of either 1 × 1012 vg/kg or 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco into monkeys M1-sc or M3-sc, respectively, was subjected to PCR amplification using primers unique to hFIXco designed to amplify a 617-bp product. Integrity of DNA was determined by amplifying a 604-bp region of the rhesus β-actin gene and is shown at the bottom of this panel. (C) RT-PCR analysis of RNA extracted from the indicated organs following mesenteric-vein injection of either 1 × 1012 vg/kg or 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco into monkeys M1-sc and M3-sc, respectively. RNA samples were amplified with (+) and without (–) RT to exclude genomic DNA amplification. Integrity of the RNA was determined by amplifying a 295-bp region of the rhesus GAPDH gene and is shown at the bottom of the panel. (D) Persistence of the scAAV transgene was assessed by subjecting genomic DNA (1 μg) isolated at 1 or 6 months from the indicated organs following liver-targeted administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in monkey M3-sc to PCR using primers unique to hFIXco designed to amplify a 617-bp product. Integrity of DNA was determined by amplifying a 604-bp region of the rhesus β-actin gene and is shown at the bottom of the panel.
Molecular analysis in rhesus macaques. (A) Southern blot analysis of DNA (10 μg) derived from liver biopsy specimens from monkeys M1-sc and M4-sc 1 month after liver-targeted delivery of 1 × 1012 vg/kg scAAV2/8-LP1-hFIXco and scAAV2/5-LP1-hFIXco, respectively, or at 1 and 6 months after administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in M3-sc. After digestion with BsrDI, the released 1486-bp fragment was probed with an LP1-specific probe. (B) Limited biodistribution analysis. Genomic DNA (top panel) isolated at 1 month from the indicated organs following liver-targeted administration of either 1 × 1012 vg/kg or 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco into monkeys M1-sc or M3-sc, respectively, was subjected to PCR amplification using primers unique to hFIXco designed to amplify a 617-bp product. Integrity of DNA was determined by amplifying a 604-bp region of the rhesus β-actin gene and is shown at the bottom of this panel. (C) RT-PCR analysis of RNA extracted from the indicated organs following mesenteric-vein injection of either 1 × 1012 vg/kg or 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco into monkeys M1-sc and M3-sc, respectively. RNA samples were amplified with (+) and without (–) RT to exclude genomic DNA amplification. Integrity of the RNA was determined by amplifying a 295-bp region of the rhesus GAPDH gene and is shown at the bottom of the panel. (D) Persistence of the scAAV transgene was assessed by subjecting genomic DNA (1 μg) isolated at 1 or 6 months from the indicated organs following liver-targeted administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in monkey M3-sc to PCR using primers unique to hFIXco designed to amplify a 617-bp product. Integrity of DNA was determined by amplifying a 604-bp region of the rhesus β-actin gene and is shown at the bottom of the panel.
Discussion
In this study, we report the design and systematic evaluation of a novel mini-hFIX expression cassette that is efficiently packaged as self-complementary dimers in a single AAV virion. In mice, scAAV vectors encoding hFIX mediated substantially higher levels of transgene than possible with ssAAV containing a variety of different carefully optimized expression cassettes. Although a relatively small number of macaques were used in these difficult and expensive studies, it is clear that scAAV vectors are also highly efficient at transducing the primate liver. Significantly, lower titers of scAAV vector were able to mediate higher transduction of the rhesus hepatocytes than previously achieved with ssAAV vectors at a molecular level. Furthermore, vector doses previously regarded as subtherapeutic in humans and animal models of hemophilia mediated therapeutic levels of hFIX (3%-30% of physiologic norm) when administered in the context of scAAV vectors. Of importance, liver-targeted delivery of scAAV in nonhuman primates was well tolerated with no evidence of toxicity. Thus the higher potency of scAAV in both murine and nonhuman primate models highlights the importance of the viral genome configuration in mediating efficient transduction of the liver.
The superiority of scAAV results from its distinct molecular fate within the liver, as after uncoating the scAAV genome rapidly forms stable double-stranded, linear molecules that are efficiently converted to HMWCs. Using a hepatocellular regeneration model, we show that the majority of the scAAV genomes, such as ssAAV, is maintained in the liver as extrachromosomal, not integrated, forms. Hence the risk of insertional mutagenesis with this vector system, which is an important consideration for all gene therapy strategies, is likely to be low. However, maintenance of hFIX expression in the therapeutic range over the lifetime of a patient with HB may require repeated administration of scAAV to compensate for loss of episomal viral genomes as a consequence of natural hepatocyte turnover. Indeed, we observed a decline in scAAV transgenes in the liver of one of our macaques (M3-sc) over a period of 6 months after liver-targeted delivery of 4 × 1011 vg/kg (Figure 7D). Repeated transduction with vectors based on the same serotype will unfortunately be blocked by capsid-specific antibodies even when using the more potent scAAV vector system. We show that this obstacle can be circumvented with scAAV vectors pseudotyped with capsid protein of an alternative serotype, thereby extending our previous observation in macaques with ssAAV.10
An important observation in this context is the similar high transduction efficiency observed with scAAV vectors pseudotyped with either serotype 5 or 8 capsid in macaques. We have previously reported similar transduction efficiency with ssAAV vector pseudotyped with 2, 5, or 8 capsid proteins in nonhuman primates.10 This contrasts markedly with the disparate transduction efficacy observed with different serotypes in mice where the selection of capsid proteins strongly influences hepatocyte gene transfer efficiencies. These species-specific differences in the biologic properties of AAV may be due to differences in distribution of AAV receptors or intracellular processing of vector, highlighting once again the importance of evaluating new vector systems in a context relevant to humans.
One of the macaques that received scAAV (M1-sc) developed neutralizing antibodies to hFIX, although this did not occur in other animals given the same dose and expressing transgene at a higher level (M2-sc and M4-sc). We have previously observed neutralizing antibodies in a few ssAAV-transduced macaques.10,25 Although it is unclear why antibodies occur in some animals and not others, epitope mapping suggests that this may be related to species-specific epitope differences. As the development of this humoral response is of some concern, we evaluated a rituximab-based regimen in M1-sc because of its efficacy at treating autoantibody-mediated diseases such as acquired hemophilia.38 The rapid decline and eventual loss of the neutralizing anti-hFIX antibody that followed rituximab/cyclosporine treatment suggests that this regimen may prove useful in treating humoral responses to the transgene product that develop after AAV-mediated gene transfer. However, further studies are required to substantiate this interesting observation.
In summary, therefore, scAAV vectors encoding hFIX represent a significant advance for gene therapy of HB and raise new hope for disorders affecting the liver.
Prepublished online as Blood First Edition Paper, December 1, 2005; DOI 10.1182/blood-2005-10-4035.
Supported by The Assisi Foundation of Memphis; the American Lebanese Syrian Associated Charities (ALSAC); National Heart, Lung, and Blood Institute (NHLBI) grant HL073838; The Katharine Dormandy Trust, UK; The National Blood Service (R&D Research grant BS02/1/RB28); and the Department of Health, UK.
A.C.N. wrote the paper, designed and directed the murine and nonhuman primate studies, and assisted in the design of the mini–human FIX expression cassette. J.T.G. designed the mini–human FIX expression cassette, created the self-complementary vectors, and generated vector particles for the experiments in mice and nonhuman primates. C.Y.C.N. determined transduction efficiency in nonhuman primates by Southern blot analysis. J.Z. performed the biodistribution studies in mice and nonhuman primates. Y.S. generated vector required for the murine and nonhuman primate studies and determined human FIX expression in mice. S.N.W. determined transduction of scAAV vectors in FIX knock-out mice. E.G.D.T. contributed to the experiments focused on abrogation of FIX inhibitor in nonhuman primates. G.K.-C. determined functional activity of human FIX in plasma from FIX knock-out mice. J.M. determined transduction efficiency with scAAV vectors in nonhuman primates and FIX knock-out mice and contributed to the study of the thrombin-antithrombin pathway. M.B.-S. conducted the rhesus antihuman FIX epitope mapping studies. K.M. designed and directed the rhesus anti–human FIX epitope mapping studies. A.M.D. wrote the paper, and designed, directed, and performed the murine and nonhuman primate studies.
A.C.N. and J.T.G. contributed equally to this study.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dorothy Bush for her assistance with immunohistochemistry. In addition, we would like to thank the staff of the Vector Core Facility at St Jude Children's Research Hospital for their assistance in generating pseudotyped rAAV vector particles required for this study. Finally, we are most grateful to Dr Mark Kay at Stanford School of Medicine (Stanford, CA) for his generous gift of AAV-hF.IX16.




![Figure 6. Transduction in rhesus macaques following liver-targeted delivery of scAAV2/8 and scAAV2/5-LP1-hFIXco. (A) Human FIX concentration in rhesus plasma was determined at the indicated time points after administration of 1 × 1012 vg/kg (M1-sc [□], M2-sc [○]) scAAV2/8-LP1-hFIXco into the mesenteric vein of 2 rhesus macaques. Treatment of M1-sc with rituximab (Rit × 2 doses) and oral cyclosporine (CyA) is shown. (B) Graphic representation of the reactivity profile of the anti-hFIX antibody in M1-sc, as determined using a panel of hFIX/X chimeras with domain or surface loop substitutions derived from FX. Antibody binding to chimeras was expressed as percentage of binding to wild-type FIX. (C) Transgene expression after mesenteric-vein administration of 4 × 1011 vg/kg scAAV2/8-LP1-hFIXco in 2 macaques (M3-sc [⋄] and M4-sc [▿]). M4-sc additionally received 1 × 1012 vg/kg scAAV2/5-LP1-hFIXco at 56 days after initial exposure to scAAV2/8. Each sample was independently evaluated on at least 3 separate occasions, and the results are depicted as an average together with the standard error of the mean.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/7/10.1182_blood-2005-10-4035/5/m_zh80070693560006.jpeg?Expires=1767710921&Signature=YKC~y17gG7hYkYM5cTRIUV5gZQjQqGp8XglfMxbrUs0YDV94oaoPDwiWzRxGT7be7qWWP-xFYQFWKJ30nGUFS05W4BiUd8KM0As~7reivRDmfUQuNe~3xkgU1~JTIMyDn7nTI0B6rHZqfXWr-m-DDoFuIB0r94S7JGOqEpH4AqHs0awn7Z4cuZ0w06V-JHaB6V3IvFM-ZB~V3v6M0GjsgN8VbyLcCTHZNMcEJshDR37CDevtMGZ5ZcCGlAeB1JMj7lPUAGq8aywwfs-CoG03atplE2JkGxQIXcp8IerAYo41ZB2Lf5WK6xy3dk9eJsyol7H~8LH~6L4ojgKZUr~WnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal