Abstract
B-cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL) have been shown to promote multiple myeloma (MM) cell growth. We show that the main site of production for BAFF and APRIL is the bone marrow (BM) environment, and that production is mainly by monocytes and neutrophils. In addition, osteoclasts produce very high levels of APRIL, unlike BM stromal cells. Myeloma cells (MMCs) express TACI (transmembrane activator and calcium modulator and cyclophilin ligand interactor), the receptor of BAFF/APRIL, at varying levels. TACI expression is a good indicator of a BAFF-binding receptor. Expression data of purified MMCs from 65 newly diagnosed patients have been generated using Affymetrix microarrays and were analyzed by supervised clustering of groups with higher (TACIhi) versus lower (TACIlo) TACI expression levels. Patients in the TACIlo group had clinical parameters associated with bad prognosis. A set of 659 genes was differentially expressed between TACIhi and TACIlo MMCs. This set makes it possible to efficiently classify TACIhi and TACIlo MMCs in an independent cohort of 40 patients. TACIhi MMCs displayed a mature plasma cell gene signature, indicating dependence on the BM environment. In contrast, the TACIlo group had a gene signature of plasmablasts, suggesting an attenuated dependence on the BM environment. Taken together, our findings suggest using gene expression profiling to identify the group of patients who might benefit most from treatment with BAFF/APRIL inhibitors.
Introduction
Multiple myeloma (MM) is a universally fatal neoplasia characterized by the accumulation of malignant plasma cells (MMCs) in the bone marrow (BM).1 There is increasing evidence for a major role of interactions between MMCs and BM stroma for the support of survival and proliferation of the MMCs.2
Patients with MM can be grouped in accordance to cytogenetic aberrations identified by conventional metaphase cytogenetics or (interphase) fluorescence in situ hybridization (iFISH) in MMCs.3,4 Patients can also be grouped in accordance to gene expression profiling (GEP) with a link between cytogenetic and GEP data.4,5 A good example is the correlation between the translocation t(4;14) and the overexpression of the multiple myeloma set domain (MMSET), or between the translocation t(11;14) and a high CCND1 overexpression.6,7 GEP has the potential to identify new pathways involved in the biology of MMCs, in particular regarding communication with the tumor environment.8-10 DNA arrays permitted to show that some human myeloma cell lines (HMCLs) expressed a gene coding for heparin-binding epidermal growth factor–like (HB-EGF) growth factor, a member of the epidermal growth factor family able to bind syndecan-1.11 Further studies demonstrated that HB-EGF is a potent myeloma cell growth factor.12,13 Using Affymetrix microarrays, we also found that TACI (transmembrane activator and calcium modulator and cyclophilin ligand interactor) and BCMA (B-cell maturation antigen), each gene coding for a receptor of B-cell activating factor (BAFF)14,15 and a proliferation-inducing ligand (APRIL),16-18 were overexpressed in malignant plasma cells compared with their normal counterparts.8,9 BAFF is a tumor necrosis factor (TNF) family member involved in the survival of normal and malignant B cells as well as normal plasmablasts.18,19 APRIL is highly expressed in several tumor tissues and stimulates the growth of tumor cells.16 A third receptor specific for BAFF, BAFF-receptor (BAFF-R), was also identified.20 We and others showed that BAFF and APRIL serum levels are elevated in patients with MM and that BAFF and APRIL contribute to the survival of some HMCLs and of primary MMCs.21,22
In this study, we have shown that the tumor environment is the main source of BAFF and APRIL in patients with MM and that TACI expression yields a functional receptor. Using Affymetrix microarrays, we demonstrated that TACIhi MMCs displayed a mature plasma cell gene signature, indicating dependence on the BM environment. In contrast, the TACIlo group had a gene signature of plasmablasts, suggesting an attenuated dependence on interactions with cells in the BM environment.
Materials and methods
Cell samples
XG-1, XG-3, XG-4, XG-5, XG-6, XG-7, XG-10, XG-11, XG-12, XG-13, XG-16, XG-19, and XG-20 HMCLs were obtained as described.23 SKMM, OPM2, LP1, and RPMI8226 HMCLs were purchased from ATTC (Rockville, MD). MMCs were purified from a first series of 65 patients with myeloma at diagnosis (median age, 59 years) and a second series of 40 newly diagnosed patients after written informed consent was given according to the Declaration of Helsinki. In the first series, according to Durie-Salmon classification, 12 patients were of stage IA, 12 of stage IIA, 38 of stage IIIA, and 3 of stage IIIB. There were 11 patients who had IgAκ MM, 7 IgAλ MM, 26 IgGκ MM, 9 IgGλ MM, 7 Bence-Jones κ MM, 3 Bence-Jones λ MM, and 2 nonsecreting MM. In the second series, 6 patients were of stage IA, 5 of stage IIA, 27 of stage IIIA, and 2 of stage IIIB. There were 4 patients who had IgAκ MM, 5 IgAλ MM, 18 IgGκ MM, 7 IgGλ MM, 4 Bence-Jones κ MM, 1 Bence-Jones λ MM, and 1 nonsecreting MM. Normal BM plasma cells (BMPCs) and whole BM cells (WBMCs) were obtained from healthy donors after informed consent was given. WBMCs were collected after lysis of red blood cells with NH4Cl. After Ficoll-density gradient centrifugation, plasma cells were purified using anti-CD138 MACS microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). BM environment cells from 7 newly diagnosed patients were obtained after depletion of MMCs with anti-CD138 MACS microbeads (Miltenyi Biotech). BM culture supernatants were prepared culturing 5 × 105 BM cells depleted of MMCs for 3 days in RPMI1640 and 10% fetal calf serum (FCS). Supernatants were frozen at -20°C until use. For 3 newly diagnosed patients, BM T cells, monocytes, and polymorphonuclear neutrophils (PMNs) were purified. BM cells were labeled with a phycoerythrin (PE)–conjugated anti-CD3 monoclonal antibody (MoAb), allophycocyanin (APC)–conjugated anti-CD14 MoAb, and a fluorescein isothiocyanate (FITC)–conjugated anti-CD15 MoAb (all from Becton Dickinson, San Jose, CA). CD3+, CD14+, and CD15+ cells were sorted with a FACSAria cell sorter (Becton Dickinson). Memory B cells, polyclonal plasmablasts (PPCs), immature dendritic cells (DCs), and BM stromal cell lines (BMSCs) were generated as described previously.9,24,25 BMSC culture supernatants were prepared from 3-day culture before reaching confluence and were stored at -20°c until use.
Osteoclasts
Peripheral blood mononuclear cells were obtained from 7 patients with MM after informed consent. Cells were cultured at 2.5 × 106 cells/mL in α minimum essential medium (αMEM)–10% FCS. After 12 hours of culture, nonadherent cells were eliminated and adherent cells were cultured in αMEM–10% FCS, receptor activator of NF-κB ligand (RANKL) (50 ng/mL; PeproTech, EC Ldt, London, United Kingdom), macrophage–colony-stimulating factor (M-CSF; 25 ng/mL; Peprotech), and 10 nM dexamethasone for 14 days. Before use, osteoslasts were phenotyped by reverse transcriptase–polymerase chain reaction (RT-PCR; TRAP and cathepsin K expression), by cytometry (integrin αvβ3 expression), and by bone resorbing activity (OsteoLyse assay kit; Cambrex, Emerainville, France). Culture supernatants were prepared from mature osteoclasts cultured for 3 days with M-CSF, RANKL, and dexamethasone and were frozen at -20°C until use.
Expression of BAFF and APRIL in the BM environment cells and MMCs. (A) BAFF and APRIL expressions were determined by quantitative PCR in 4 samples of memory B cells, 7 samples of normal plasmablasts, 10 samples of HMCLs, 7 samples of primary MMCs, 7 samples of BM environment cells depleted from MMCs, and 5 samples of BM cells of healthy individuals. (B) BAFF and APRIL expressions were determined by quantitative PCR in purified BM CD3+, CD14+, or CD15+ cells. These cells were obtained from 3 newly diagnosed patients with MM (represented by differently shaded bars). (C) BAFF and APRIL expressions were determined by quantitative PCR in 7 samples of BM stromal cell lines and 7 samples of in vitro–generated osteoclasts. Primary cells were obtained from newly diagnosed patients with MM (represented by differently shaded bars). Dendritic cells (DCs) were used as positive control and assigned the expression value of 100 arbitrary units. (D) BAFF and APRIL concentrations in culture supernatants of BM cells of 10 patients with MM, 7 different BM stromal cell line samples from patients with MM, and 7 different samples of osteoclasts were determined by ELISA.
Expression of BAFF and APRIL in the BM environment cells and MMCs. (A) BAFF and APRIL expressions were determined by quantitative PCR in 4 samples of memory B cells, 7 samples of normal plasmablasts, 10 samples of HMCLs, 7 samples of primary MMCs, 7 samples of BM environment cells depleted from MMCs, and 5 samples of BM cells of healthy individuals. (B) BAFF and APRIL expressions were determined by quantitative PCR in purified BM CD3+, CD14+, or CD15+ cells. These cells were obtained from 3 newly diagnosed patients with MM (represented by differently shaded bars). (C) BAFF and APRIL expressions were determined by quantitative PCR in 7 samples of BM stromal cell lines and 7 samples of in vitro–generated osteoclasts. Primary cells were obtained from newly diagnosed patients with MM (represented by differently shaded bars). Dendritic cells (DCs) were used as positive control and assigned the expression value of 100 arbitrary units. (D) BAFF and APRIL concentrations in culture supernatants of BM cells of 10 patients with MM, 7 different BM stromal cell line samples from patients with MM, and 7 different samples of osteoclasts were determined by ELISA.
BAFF or APRIL ELISA
The concentration of BAFF was assayed using the enzyme-linked immunosorbent assay (ELISA) described previously,21 and APRIL was assayed with an ELISA purchased from Bender MedSystems (Burlingame, CA) in accordance with the manufacturer's instructions.
Modulation of the gene expression profile by addition or deprivation of BAFF/APRIL in MMCs
The modulation of gene expression by addition of BAFF and APRIL was investigated with the XG-13 HMCL.21 XG-13 cells were starved of interleukin 6 (IL-6) for 2 hours, washed, and cultured for 12 hours in RPMI1640–10% FCS. Then BAFF and APRIL (200 ng/mL each) were added in one culture group for 10 hours. We also investigated the modulation of gene expression profile in primary MMCs by BAFF and APRIL.26 BM cells of a newly diagnosed patient with TACIhi MMCs were harvested and WBMCs (5 × 105 cells/mL) were cultured for 24 hours in RPMI1640–10% FCS. Then, 20 μg/mL TACI-Fc (R&D Systems, Abington, United Kingdom) was added for 10 hours in one culture group. Cells were harvested, labeled with CD138 MACS microbeads at 4°C, and primary MMCs were purified. RNA was extracted for gene expression profiling.
Preparation of complementary RNA and microarray hybridization
RNA was extracted using the RNeasy Kit (Quiagen, Hilden, Germany). Biotinylated complementary RNA was amplified with a double in vitro transcription and hybridized to the human HG U133 A and B or U133 2:0 plus GeneChips, according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Fluorescence intensities were quantified and analyzed using the GECOS software (Affymetrix).
Conventional and real-time RT-PCR
BCMA, TACI, and BAFF-R PCR analysis was done as previously described.21 The assays-on-demand primers and probes and the TaqMan Universal Master Mix were used according to the manufacturer's instructions (Applied Biosystems, Courtaboeuf, France). Real-time RT-PCR was performed using the ABI Prism 7000 Sequence Detection System (Applied Biosystems) and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample.
Binding of BAFF to MMCs
The binding of BAFF to HMCLs and primary MMCs was determined using a human BAFF-murine CD8 biotinylated fusion protein (Ancell, Bayport, MN) and fluorescence-activated cell sorting (FACS) analysis as previously described.21
Fluorescence in situ hybridization (FISH)
Interphase FISH was performed according to our previously reported standard protocol.27 Metaphase spreads and interphase cells were evaluated using a DM RXA2 fluorescence microscope (Leica, Bensheim, Germany).
Statistical analysis and TACIlo and TACIhi group definition
Gene expression data were normalized with the MAS5 algorithm and analyzed with our bioinformatics platform (RAGE [remote analysis of microarray gene expression]) designed by Dr T Reme (INSERM U475, Montpellier, France). Hierarchical clustering was performed with Cluster and Treeview software from Eisen et al.28 We performed a 2-sided supervised clustering between a group of patients with a high (TACIhi) and one with low (TACIlo) expression of TACI. To account for the log-normal distribution of TACI expression, we performed this analysis with decreasing proportions of patients in the TACIhi and TACIlo group: 40%, 25%, and 15% of patients with the highest versus lowest TACI expression. Genes were interpreted as differentially expressed if the P value (Mann-Whitney test) was .01 and the ratio 1.5 or 0.67. A permutation test29 was performed to generate 100 repetitions of 2 arbitrary groups comprising 40%, 25%, or 15% of patients in order to get an evaluation for the number of genes differentially expressed between 2 arbitrary groups (P = .01 and ratio 1.5 or 0.67) by chance. Statistical comparisons were done with Mann-Whitney, chi-square, or Student t tests.
Results
BAFF and APRIL genes are mainly expressed by the BM environment
BAFF and APRIL gene expression levels were analyzed by real-time RT-PCR in 4 samples of memory B cells, 7 of normal plasmablasts, 10 of HMCLs, 7 of primary MMCs, 7 of BM environment cells depleted from MMCs, and 5 of BM cells of healthy individuals. These data were compared with those of dendritic cells, to which an arbitrary value of 100 was assigned (Figure 1A). BAFF and APRIL were highly expressed in the MM BM environment as well as in normal BM. No significant differences were found between MM and normal BM samples (P = .26 for BAFF expression and P = .88 for APRIL). Concerning MM BM environment samples, median expression values were 66 for BAFF and 46 for APRIL, about half of the expression in dendritic cells known to highly produce BAFF or APRIL.30 Primary MMCs weakly expressed BAFF or APRIL; the median expressions were 102-fold and 45-fold lower, respectively, than those in the tumor environment (P = .005). The majority of HMCLs and normal plasmablasts also expressed BAFF or APRIL but at a rather low level as compared with the BM environment. The 2 cell lines (XG-5 and XG-20) with the highest APRIL expression have been previously identified with conventional RT-PCR.21 BAFF and APRIL expressions in plasmablasts were 29- and 31-fold lower, respectively, than those in memory B cells from which they arose (P = .002 and P = .007).9 Thus, BAFF and APRIL genes are mainly expressed by the BM environment and only occasionally by MMCs.
In order to identify the cell type expressing BAFF or APRIL in the MM environment, BM CD3, CD14, and CD15 cells from 3 patients were purified. BAFF was mainly expressed by BM CD14 and CD15 cells and APRIL by CD14 cells. CD3 cells expressed BAFF and APRIL only at a low level (Figure 1B). To assess a broad range of populations present in the bone microenvironment, BMSCs and osteoclasts were also studied. BMSCs from 7 patients with MM weakly expressed BAFF and APRIL (Figure 1C). Of major interest, osteoclasts largely expressed the APRIL gene. Median APRIL expression was 80-fold higher than that in BM environment (P = .001). Osteoclasts also express the BAFF gene but in the same extent as the BM environment.
BAFF and APRIL production by BM cells
The production of BAFF and APRIL was investigated in 3-day culture supernatants of BM cells of 10 newly diagnosed patients, 7 osteoclasts, and 7 BMSCs. In agreement with real-time RT-PCR data, a low amount of BAFF and no APRIL could be found in BMSC supernatants. Median concentrations of 12.5 ng/mL BAFF and 24.6 ng/mLAPRIL were measured in supernatants of BM cells. Again, in agreement with RT-PCR data, osteoclasts produced large amounts of APRIL (a concentration of 206 ng/mL), ie, 8-fold higher than BM cells (P = .001). The median production of BAFF by osteoclasts was 3-fold higher than that of BM cells (P = .001).
TACI gene expression yields a functional BAFF-binding receptor in HMCLs and primary MMCs
Gene expression profiling (GEP) was performed on 17 HMCLs using Affymetrix U133 A+B microarrays. On these, no probeset for BAFF-R was available. The Affymetrix data for TACI expression perfectly matched the real-time RT-PCR and previous RT-PCR data,21 and were consistent with the HMCL ability to bind BAFF-murine CD8. Indeed, TACI had an Affymetrix “absent” call (as decided by the GECOS software) in the HMCLs that showed a lack of TACI mRNA expression by real-time or conventional RT-PCR and a lack of binding of BAFF-murine CD8. TACI had an Affymetrix “present” call in the TACI RT-PCR+ HMCLs, which also bound BAFF-murine CD8 (Table 1). The BCMA gene had a “present” call in 17 of 17 HMCLs, as had been assumed after BCMA was found in RT-PCR experiments (Table 1). These data suggest that although BCMA is expressed, the BCMA protein is not functional, possibly due to retention in the Golgi apparatus as shown for plasma cells.31 This observation holds true for purified primary MMCs. MMCs (no. 1-5) that highly expressed BCMA but weakly expressed TACI according to RT-PCR results, failed to bind BAFF-murine CD8 (Figure 2). In contrast, primary MMCs (no. 6-10) that highly expressed TACI bound human BAFF-murine CD8 (Figure 2). Thus, TACI gene expression yields a BAFF-binding receptor in HMCLs or primary MMCs.
TACI gene expression yields a BAFF/APRIL receptor with functional binding capacity in HMCLs
. | PCR . | . | . | Binding of BAFF-murine CD8 . | Real-time RT-PCR TACI . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|
| HMCLs . | BCMA . | BAFF-R . | TACI . | . | . | Affymetrix TACI . | Affymetrix BCMA . | ||
| XG-12 | + | – | + | + | + | P | P | ||
| XG-13 | +* | +* | +* | +* | + | P | P | ||
| XG-19 | + | + | + | + | + | P | P | ||
| XG-20 | +* | +* | +* | +* | + | P | P | ||
| RPM18226 | +* | +* | +* | +* | + | P | P | ||
| LP1 | +* | +* | +* | +* | + | P | P | ||
| XG-1 | +* | +* | –* | –* | – | A | P | ||
| XG-3 | + | – | – | – | – | A | P | ||
| XG-4 | + | + | – | – | – | A | P | ||
| XG-5 | +* | –* | –* | –* | – | A | P | ||
| XG-6 | +* | –* | –* | –* | – | A | P | ||
| XG-7 | +* | +* | –* | –* | – | A | P | ||
| XG-10 | + | + | – | – | – | A | P | ||
| XG-11 | +* | –* | –* | –* | – | A | P | ||
| XG-16 | + | – | – | – | – | A | P | ||
| OPM2 | + | + | – | – | – | A | P | ||
| SKMM | + | – | – | – | – | A | P | ||
. | PCR . | . | . | Binding of BAFF-murine CD8 . | Real-time RT-PCR TACI . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|
| HMCLs . | BCMA . | BAFF-R . | TACI . | . | . | Affymetrix TACI . | Affymetrix BCMA . | ||
| XG-12 | + | – | + | + | + | P | P | ||
| XG-13 | +* | +* | +* | +* | + | P | P | ||
| XG-19 | + | + | + | + | + | P | P | ||
| XG-20 | +* | +* | +* | +* | + | P | P | ||
| RPM18226 | +* | +* | +* | +* | + | P | P | ||
| LP1 | +* | +* | +* | +* | + | P | P | ||
| XG-1 | +* | +* | –* | –* | – | A | P | ||
| XG-3 | + | – | – | – | – | A | P | ||
| XG-4 | + | + | – | – | – | A | P | ||
| XG-5 | +* | –* | –* | –* | – | A | P | ||
| XG-6 | +* | –* | –* | –* | – | A | P | ||
| XG-7 | +* | +* | –* | –* | – | A | P | ||
| XG-10 | + | + | – | – | – | A | P | ||
| XG-11 | +* | –* | –* | –* | – | A | P | ||
| XG-16 | + | – | – | – | – | A | P | ||
| OPM2 | + | + | – | – | – | A | P | ||
| SKMM | + | – | – | – | – | A | P | ||
Expression of BCMA, BAFF-R, and TACI mRNA was determined by RT-PCR in 17 HMCLs. TACI and BCMA expressions were also determined with Affymetrix microarrays. The probeset for BAFF-R was not available on the Affymetrix HG U133 A + B DNA microarrays. The expression of BAFF binding was determined by flow cytometry using biotinylated human BAFF-murine CD8 fusion protein and phycoerythrin-conjugated streptavidin.
+ indicates the presence of the receptor; –, the receptor's absence; P, present cell; and A, absent cell.
These data are from our previously published data21
Clinical and genetic data of the TACIlo and TACIhi patients
Gene expression profiles of the 65 primary myeloma cell samples were determined with Affymetrix microarrays. TACI had a “present” Affymetrix call in all primary MMCs investigated, displaying a lognormal distribution. We did not find the clear-cut difference (absent or present Affymetrix call) for primary MMCs that was observed for HMCLs. One possible explanation is the presence of MMC subclones with different levels of TACI expression in the same patient. We looked for differences in the major clinical, biologic, or genetic markers in patients with a low or high TACI expression in MMCs (TACIlo or TACIhi patients). To find the best way to delimit TACIlo and TACIhi patients, we considered subgroups defined by 50%, 40%, 25%, or 15% of the patients with the highest or lowest TACI expression in MMCs. The clinical and biologic differences observed showed the clearest distinction when the 15% group was considered. Here, the TACIhi group included a higher frequency of patients within stage I MM, lambda light-chain isotype, t(4;14) translocations, and hemoglobin levels more than 10 g/dL, and a lower frequency of patients with bone lesions (Table 2). The percentage of patients with MMCs harboring a t(4;14) translocation was 14%, in agreement with published data.3,32 Most of these differences were kept when considering TACI groups representing 25% and 40% of the patients (distribution of bone lesions, t(4;14) translocations, lambda light-chain isotype). However, they were lost when considering TACI subgroups delimited by the median. Whatever method was used to define TACIlo and TACIhi subgroups, no differences in age, β2 microglobulin, C-reactive protein (CRP), albumin, lactate dehydrogenase (LDH) levels, IgG subtypes, or chromosome 13 deletion were found (Table 2).
Clinical data of the TACIhi and TACIlo patients
. | Definition of TACIlo and TACIhi subgroups, % of all patients . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 50/50 . | . | 40/40 . | . | 25/25 . | . | 15/15 . | . | |||||||
| Categories . | TACIhi . | TACIlo . | TACIhi . | TACIlo . | TACIhi . | TACIlo . | TACIhi . | TACIlo . | |||||||
| Age of at least 65 y | 30 | 24 | 28 | 28 | 20 | 40 | 20 | 40 | |||||||
| Kappa light chain | 65 | 70 | 64 | 80 | 60 | 66 | 50 | 70 | |||||||
| Lambda light chain | 35 | 26 | 36 | 16 | 40 | 16 | 50 | 20 | |||||||
| Nonsecreting | 0 | 4 | 0 | 4 | 0 | 18 | 0 | 10 | |||||||
| IgA subtype | 25 | 31 | 24 | 24 | 33 | 27 | 20 | 30 | |||||||
| β2-microglobulin at least 4 mg/L | 44 | 37 | 40 | 40 | 40 | 60 | 30 | 60 | |||||||
| C-reactive protein at least 4 mg/L | 44 | 53 | 40 | 56 | 47 | 67 | 50 | 70 | |||||||
| Lactate dehydrogenase at least 190 IU/L | 28 | 16 | 28 | 20 | 27 | 20 | 40 | 30 | |||||||
| Albumin less than 35 g/L | 37 | 37 | 40 | 36 | 47 | 33 | 60 | 40 | |||||||
| Hemoglobin less than 10 g/dL | 28 | 35 | 36 | 44 | 20 | 47 | 20 | 60 | |||||||
| Bone lesions | |||||||||||||||
| 0 lesions | 42 | 20 | 45 | 10 | 43 | 8 | 44 | 0 | |||||||
| At least 1 lesion | 58 | 80 | 55 | 90 | 57 | 92 | 56 | 100 | |||||||
| Cytogenetic abnormalities | |||||||||||||||
| Chromosome 13 deletion | 22 | 31 | 24 | 32 | 27 | 33 | 30 | 40 | |||||||
| t(4;14) translocation | 22 | 6 | 20 | 8 | 27 | 6 | 30 | 0 | |||||||
| t(11;14) translocation | 9 | 9 | 12 | 12 | 20 | 6 | 20 | 10 | |||||||
| Staging | |||||||||||||||
| I | 9 | 3 | 8 | 3 | 6 | 0 | 4 | 0 | |||||||
| II | 5 | 5 | 5 | 4 | 2 | 3 | 1 | 2 | |||||||
| III | 18 | 24 | 12 | 18 | 7 | 12 | 5 | 8 | |||||||
| P* | .1 | .02 | .02 | .08 | |||||||||||
. | Definition of TACIlo and TACIhi subgroups, % of all patients . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 50/50 . | . | 40/40 . | . | 25/25 . | . | 15/15 . | . | |||||||
| Categories . | TACIhi . | TACIlo . | TACIhi . | TACIlo . | TACIhi . | TACIlo . | TACIhi . | TACIlo . | |||||||
| Age of at least 65 y | 30 | 24 | 28 | 28 | 20 | 40 | 20 | 40 | |||||||
| Kappa light chain | 65 | 70 | 64 | 80 | 60 | 66 | 50 | 70 | |||||||
| Lambda light chain | 35 | 26 | 36 | 16 | 40 | 16 | 50 | 20 | |||||||
| Nonsecreting | 0 | 4 | 0 | 4 | 0 | 18 | 0 | 10 | |||||||
| IgA subtype | 25 | 31 | 24 | 24 | 33 | 27 | 20 | 30 | |||||||
| β2-microglobulin at least 4 mg/L | 44 | 37 | 40 | 40 | 40 | 60 | 30 | 60 | |||||||
| C-reactive protein at least 4 mg/L | 44 | 53 | 40 | 56 | 47 | 67 | 50 | 70 | |||||||
| Lactate dehydrogenase at least 190 IU/L | 28 | 16 | 28 | 20 | 27 | 20 | 40 | 30 | |||||||
| Albumin less than 35 g/L | 37 | 37 | 40 | 36 | 47 | 33 | 60 | 40 | |||||||
| Hemoglobin less than 10 g/dL | 28 | 35 | 36 | 44 | 20 | 47 | 20 | 60 | |||||||
| Bone lesions | |||||||||||||||
| 0 lesions | 42 | 20 | 45 | 10 | 43 | 8 | 44 | 0 | |||||||
| At least 1 lesion | 58 | 80 | 55 | 90 | 57 | 92 | 56 | 100 | |||||||
| Cytogenetic abnormalities | |||||||||||||||
| Chromosome 13 deletion | 22 | 31 | 24 | 32 | 27 | 33 | 30 | 40 | |||||||
| t(4;14) translocation | 22 | 6 | 20 | 8 | 27 | 6 | 30 | 0 | |||||||
| t(11;14) translocation | 9 | 9 | 12 | 12 | 20 | 6 | 20 | 10 | |||||||
| Staging | |||||||||||||||
| I | 9 | 3 | 8 | 3 | 6 | 0 | 4 | 0 | |||||||
| II | 5 | 5 | 5 | 4 | 2 | 3 | 1 | 2 | |||||||
| III | 18 | 24 | 12 | 18 | 7 | 12 | 5 | 8 | |||||||
| P* | .1 | .02 | .02 | .08 | |||||||||||
We separated 65 newly diagnosed patients with MM into 2 subgroups according to TACI gene expression in MMCs assayed with Affymetrix microarrays. The 2 subgroups represent either 50% (n = 32), 40% (n = 25), 25% (n = 15), or 15% (n = 10) of the patients with highest (TACIhi) or lowest (TACIlo) TACI expression. Data are the percentages of patients within each subgroup with the indicated clinical or biologic parameters. When the percentages were different with the chi-square test (P = .05), the data are shown in italics.
P value compares high and low subgroups in each category
Expression of the receptors for BAFF and APRIL in MMCs. (Top) Expression of BCMA, BAFF-R, TACI, and GAPDH mRNA were determined by RT-PCR in CD138+ purified primary MMCs from 10 patients with intramedullary MM. (Bottom) The ability of MMCs to bind BAFF was determined by flow cytometry using a biotinylated human BAFF-murine CD8 fusion protein and phycoerythrin-conjugated streptavidin. Dotted lines indicate Ig control; solid lines, Baff-mu CD8.
Expression of the receptors for BAFF and APRIL in MMCs. (Top) Expression of BCMA, BAFF-R, TACI, and GAPDH mRNA were determined by RT-PCR in CD138+ purified primary MMCs from 10 patients with intramedullary MM. (Bottom) The ability of MMCs to bind BAFF was determined by flow cytometry using a biotinylated human BAFF-murine CD8 fusion protein and phycoerythrin-conjugated streptavidin. Dotted lines indicate Ig control; solid lines, Baff-mu CD8.
Gene expression signature of microenvironment dependence in TACIhi MMCs and plasmablastic signature in TACIlo MMCs with Affymetrix microarrays
In order to investigate if different gene signatures could be identified comparing 2 groups with different TACI expression, we performed a supervised clustering. We considered the 3 groups with significant clinical differences (40%, 25%, and 15% of the patients). Of 49 000 probesets present on the U133 A+B DNA microarrays, we found 1612, 2709, and 1805 probesets, respectively, to be differentially expressed between the 2 groups (P = .01 with Mann-Whitney test and ratio of the mean expressions of 1.5 or 0.67). To get an impression about the number of genes differentially expressed by chance between the groups, we performed 100 permutations with 2 subgroups comprising 40%, 25%, or 15% of the patients with MM randomly sorted out of the 65 samples. Only 5% of the genes appeared to be differentially expressed, if the groups were selected by chance. A common set of 548 distinct genes and 111 distinct expressed sequence tags (ESTs) were differentially expressed between TACIhi and TACIlo MMCs independently of the definition of TACIhi and TACIlo groups. This list of 659 genes/ESTs is available on the Blood website as Supplemental Tables S1 and S2 (see the Supplemental Tables link at the top of the online article).
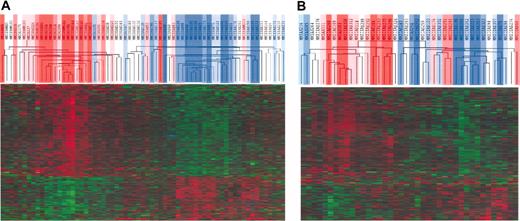
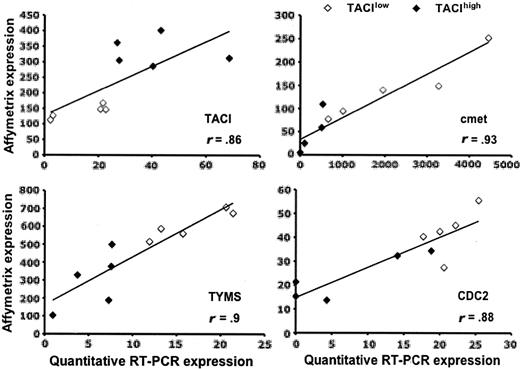
We performed an unsupervised hierarchical clustering using these 659 genes/ESTs (Figure 3A). TACIhi and TACIlo MMCs appear in different branches of the cluster analysis. The TACI40lo, TACI25lo, and TACI15lo groups were represented by graded blue color (more intense in TACI15lo) and were classified in one branch with limited misclassifications in each category. For example, the 40% groups were significantly classified (P = .01), with 5 misclassifications. The TACI40hi, TACI25hi and TACI15hi groups were represented with graded red color (more intense in TACI15hi) and classified in another branch. Patients outside the subgroups (white color) were interdispersed in the 2 clusters. The ability of this 659 gene/EST list to classify TACIhi and TACIlo MMCs was validated with another series of GEP from 40 purified MMCs, unrelated to the 65 previous ones, that was obtained with Affymetrix U133 2.0 plus microarrays (Figure 3B). Although this new series of primary MMCs was smaller and a different microarray was used, the U133 2.0 plus probesets corresponding to the 659 U133 A+B genes/ESTs were able to efficiently classify TACIhi and TACIlo MMCs. In particular, the 40% groups were significantly classified (P = .01), with 6 misclassifications. This differential gene expression in TACIhi and TACIlo MMCs observed with Affymetrix microarrays was validated for TACI and 3 other genes (CDC2, TYMS, and c-met) with real-time RT-PCR.33,34 Real-time RT-PCR and Affymetrix data showed a good correlation (r = .85, P = .01; Figure 4).
Of the 659 genes and ESTs mentioned above, 339 (52% of the 479 genes/ESTs overexpressed in TACIhi patients and 50% of the 180 genes/ESTs overexpressed in TACIlo patients) could be assigned to 8 functional categories listed in Table 3 using gene ontology terms. TACIhi MMCs expressed a higher percentage of genes coding for intercellular communication signals, cytoskeleton-associated proteins, and signal transduction (Table 3). Conversely, TACIlo MMCs overexpressed genes coding for proteins involved in cell cycle and nuclear functions (Table 3).
Intercellular communication signature in TACIhi MMCs and plasmablastic signature in TACIlo MMCs
Genes coding for protein implicated in different functions . | No. genes* . | TACIlo, % . | TACIhi, % . | P† . |
|---|---|---|---|---|
| Intercellular communication signals | 69 | 2.1 | 26.3 | < .001 |
| Cytoskeleton | 20 | 2.1 | 7.2 | .04 |
| Transduction signals | 52 | 11.2 | 17 | .05 |
| Protein synthesis and regulation | 19 | 7.7 | 4.8 | NS |
| Cell cycle | 11 | 10 | 0.8 | < .001 |
| Metabolism | 85 | 22.1 | 26.6 | NS |
| Apoptosis | 10 | 4.2 | 2.8 | NS |
| Nuclear functions | 73 | 40.6 | 15.2 | < .001 |
| Total of classified genes | 339 | 100‡ | 100§ | NS |
Genes coding for protein implicated in different functions . | No. genes* . | TACIlo, % . | TACIhi, % . | P† . |
|---|---|---|---|---|
| Intercellular communication signals | 69 | 2.1 | 26.3 | < .001 |
| Cytoskeleton | 20 | 2.1 | 7.2 | .04 |
| Transduction signals | 52 | 11.2 | 17 | .05 |
| Protein synthesis and regulation | 19 | 7.7 | 4.8 | NS |
| Cell cycle | 11 | 10 | 0.8 | < .001 |
| Metabolism | 85 | 22.1 | 26.6 | NS |
| Apoptosis | 10 | 4.2 | 2.8 | NS |
| Nuclear functions | 73 | 40.6 | 15.2 | < .001 |
| Total of classified genes | 339 | 100‡ | 100§ | NS |
Of the 659 genes differentially expressed between the TACIhi and TACIlo groups, 339 could be assigned to 8 functional categories using gene ontology terms. Data are the percentages of genes of a given category compared with the total number of TACIlo (91 genes) or TACIhi (248 genes) genes.
NS indicates not significant.
n indicates the number of genes out of the 339 in one category
The statistical comparisons of the percentages of genes belonging to one gene ontology category between the TACIlo and TACIhi group genes were performed with the chi-square test
n = 91
n = 248
Unsupervised hierarchical clustering of TACIhi and TACIlo MMCs. The 659 genes that statistically (P = .01) distinguished (ratio 1.5 or 0.67) TACIhi patients and TACIlo patients are presented graphically using hierarchical clustering. The color of each cell in the tabular image represents the expression level of each gene (red, expression higher than the mean; green, expression lower than the mean; increasing color intensity represents a higher magnitude of deviation from the mean). The TACI40lo, TACI25lo, and TACI15lo subgroups were represented by graded blue color (more intense in TACI15lo) and the TACI40hi, TACI25hi, and TACI15hi) subgroups were represented with graded red color (more intense in TACI15hi). (A) Initial set of 65 patients. (B) Validation set of 40 patients.
Unsupervised hierarchical clustering of TACIhi and TACIlo MMCs. The 659 genes that statistically (P = .01) distinguished (ratio 1.5 or 0.67) TACIhi patients and TACIlo patients are presented graphically using hierarchical clustering. The color of each cell in the tabular image represents the expression level of each gene (red, expression higher than the mean; green, expression lower than the mean; increasing color intensity represents a higher magnitude of deviation from the mean). The TACI40lo, TACI25lo, and TACI15lo subgroups were represented by graded blue color (more intense in TACI15lo) and the TACI40hi, TACI25hi, and TACI15hi) subgroups were represented with graded red color (more intense in TACI15hi). (A) Initial set of 65 patients. (B) Validation set of 40 patients.
TACIhi MMCs have a BMPC signature and TACIlo MMCs have a plasmablastic signature
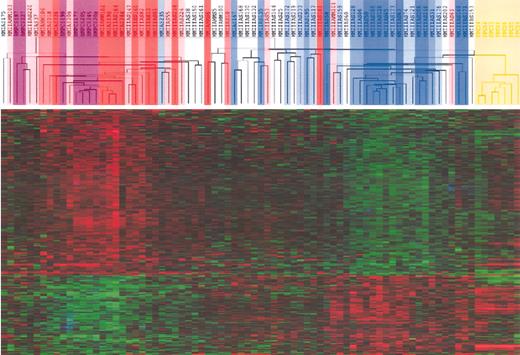
We investigated the association of plasmablasts and normal BMPCs with MMCs in the TACIhi and TACIlo groups. Using an unsupervised clustering with the above-mentioned 659 genes, the samples of patients in the TACIlo group clustered together with plasmablasts (correlation coefficient, 0.33; P = .001) whereas TACIhi samples clustered with BMPCs (correlation coefficient, 0.35; P = .001; Figure 5). When comparing plasmablasts with normal BMPCs, of the 659 genes/ESTs differentially expressed between the TACIhi and TACIlo groups, 195 also appeared as differentially expressed between the plasmablasts and BMPCs. Of these, 115 were overexpressed in BMPCs compared with plasmablasts and 80 were overexpressed in plasmablasts compared with BMPCs (ratio of the mean expressions 1.5 and 0.67; P = .01). Of 115 BMPC genes, 111 were also upregulated in TACIhi compared with TACIlo MMCs. Conversely, 77 of the 80 PPC genes were overexpressed in TACIlo MMCs (P = .01). BMPC and plasmablast genes are indicated in Tables S3 and S4.
Validation of Affymetrix data. Gene expressions of TACI, MET, TYMS, and CDC2 in TACIlo and TACIhi patients were assayed with real-time RT-PCR and normalized with GAPDH. The XG-13 HMCL was used as the standard. The coefficient of correlations between Affymetrix and real-time RT-PCR values were determined.
Validation of Affymetrix data. Gene expressions of TACI, MET, TYMS, and CDC2 in TACIlo and TACIhi patients were assayed with real-time RT-PCR and normalized with GAPDH. The XG-13 HMCL was used as the standard. The coefficient of correlations between Affymetrix and real-time RT-PCR values were determined.
Taken together, these results suggest that grouping of patients in accordance with “high” and “low” TACI expression delineates a group of patients whose MMCs share gene expression characteristics with plasmablasts (TACIlo) and a group in which the gene expression profiling resembles normal mature BM plasma cells (TACIhi). TACIhi MMCs overexpress many genes coding for potentially interesting intercellular communication or transduction signals whose biology is discussed below. In contrast, TACIlo MMCs mainly overexpress cell cycle genes.
Modulation of gene expression profile by addition or deprivation of BAFF and APRIL in MMCs
In order to determine whether the TACIhi and TACIlo gene signature could be induced by BAFF/APRIL, we first used the XG-13 HMCL, which is stimulated by BAFF and APRIL.21 After growth factor starvation, addition of BAFF and APRIL for 10 hours induced an increase of 196 genes/ESTs and a decrease of 348 genes/ESTs (P = .01 and ratio 1.5 or 0.67). However, only 19 genes were common with the TACI-related gene signature (supplemental data in Table S5). We then decided to investigate the effect of BAFF/APRIL deprivation using a whole BM culture system in which BAFF and APRIL were blocked by TACI-Fc inhibitor for 10 hours. Subsequently, MMCs were purified for gene expression profiling with Affymetrix U133 2.0 plus microarrays. There were 188 genes and ESTs that were downregulated in BAFF/APRIL-deprived MMCs (treated with TACI-Fc) and 259 that were upregulated (P = .01 and ratio 1.5 or 0.67). However, only some of these genes were common with the TACIhi of TACIlo gene signature. This list is available as supplemental data in Table S6.
Hierarchical clustering of MMCs, BMPCs, and PPCs identifies a signature of BM stroma interaction for TACIhi patients and a plasmablastic signature for TACIlo patients. Unsupervised hierarchical clustering analysis of the expression profile of MMCs of 65 patients at diagnosis, 7 PPC samples, and 7 BMPC samples show that TACIlo MMCs cluster (blue) together with PPCs (yellow) whereas TACIhi MMCs cluster (red) with normal BMPCs (purple). The clustering was performed on the 659 genes differentially expressed between TACIhi and TACIlo MMCs. The TACI40lo, TACI25lo, and TACI15lo subgroups were represented by graded blue color (more intense in TACI15lo) and the TACI40hi, TACI25hi, and TACI15hi subgroups with graded red color (more intense in TACI15hi).
Hierarchical clustering of MMCs, BMPCs, and PPCs identifies a signature of BM stroma interaction for TACIhi patients and a plasmablastic signature for TACIlo patients. Unsupervised hierarchical clustering analysis of the expression profile of MMCs of 65 patients at diagnosis, 7 PPC samples, and 7 BMPC samples show that TACIlo MMCs cluster (blue) together with PPCs (yellow) whereas TACIhi MMCs cluster (red) with normal BMPCs (purple). The clustering was performed on the 659 genes differentially expressed between TACIhi and TACIlo MMCs. The TACI40lo, TACI25lo, and TACI15lo subgroups were represented by graded blue color (more intense in TACI15lo) and the TACI40hi, TACI25hi, and TACI15hi subgroups with graded red color (more intense in TACI15hi).
Discussion
Two recent studies have shown that BAFF and APRIL are growth factors for human MMCs and that serum levels of BAFF and APRIL are increased in these patients.21,22 Using real-time PCR, we show that BAFF and APRIL genes are poorly expressed by MMCs and are expressed mainly by the BM environment of patients with MM. BM cells from healthy individuals exhibit a similar expression. The MM bone environment comprises mainly of PMNs (45%), monocytes (5%), and T cells (10%). We show here that BAFF was largely expressed by PMNs and monocytes, whereas APRIL was expressed by monocytes only. A majority of the studies about MM use BM mononuclear cells after removal of PMNs with centrifugation on a Ficoll hypaque cushion. Thus, the contribution of PMNs is in many cases ignored, whereas they are the main BM cell components in vivo and are here shown to be the main source of BAFF. Another cell component in the BM environment important for myeloma cell biology is bone cells. Primary MMCs can survive in contact with BMSCs35 or osteoclasts.36 In addition, fetal bone is critical to grow MMCs in severe combined immunodeficient (SCID) mice.37,38 As these bone cells cannot be harvested by BM aspiration, they are usually produced in vitro. We show here that cultured osteoclasts display a huge APRIL expression, about 80-fold higher than those found in the MM BM environment or in dendritic cells. The BAFF expression was in the same order of magnitude in both cases. Using ELISA, we confirmed that osteoclasts produced very large amounts of APRIL. Thus, APRIL could likely play an important role in the MMC growth stimulation by osteoclasts.36 BM stromal cells produced only a low amount of BAFF and no APRIL.
Purified MMCs also express BAFF or APRIL but in the median about 100-fold weaker than the BM environment. This BAFF or APRIL expression pattern and distribution resembles that of IL-6. A current consensus is that IL-6 is a paracrine growth factor in MM and that some MMCs may produce autocrine IL-6 and use it to escape from their dependency on the tumor environment.39,40 In particular, several HMCLs use IL-6 as an autocrine growth factor.41,42 Similarly, we have previously found that 2 HMCLs (L363 and RPMI 8226) use BAFF or APRIL as autocrine growth factors, whereas we found here that BAFF and APRIL are mainly paracrine growth factors if assessing primary MM cells. The autocrine production may have contributed to the escape of these MMCs from the tumor environment.
Given that BAFF and APRIL are paracrine growth factors, our aim was to look for a difference in the GEP of patients' MMCs expressing a high or a low level of BAFF and APRIL receptors. An important point is that TACI expression (assayed with Affymetrix microarrays or RT-PCR) is a good indicator for the presence of a receptor, able to bind BAFF. There are 3 receptors for BAFF and APRIL: BCMA, BAFF-R, and TACI. Probesets for BCMA and TACI were available on the Affymetrix U133 A+B chips, unlike BAFF-R. We could not identify an EST with BAFF-R sequence homology in any Affymetrix probeset. BCMA was probably not functional in MMCs and TACI expression was a good indicator of a functional BAFF receptor. Indeed, only 6 of the 17 HMCLs with a “present” call for TACI expression with Affymetrix microarrays were able to bind a BAFF-murine fusion protein, independent of BCMA expression. BCMA was already shown to be not functional in several cell types, in particular in human plasma cells and the U266 myeloma cell line due to retention in Golgi apparatus.31 However, it is not likely the case for murine plasma cells since BCMA-/- mice have normal B-cell function but fail to develop mature plasma cells.43
When grouping the patients in accordance with the TACI expression levels as described above, the MMCs in the TACIhi group show a signature resembling mature BMPCs with an emphasis on the expression of genes with products functioning in intercellular communication and signal transduction. In contrast, the TACIlo MMCs showed a signature sharing similarities with plasmablasts, especially with regard to the expression of cell cycle genes. The consistency of this signature was validated on an independent group of 40 MMCs using Affymetrix U133 2.0 plus microarrays. We looked for the possibility that this gene signature could be due in part to signal transduction induced by TACI activation. We used first the XG-13 HMCL, whose growth is stimulated by BAFF or APRIL.21 We also used primary MMCs from a newly diagnosed patient. In this case, as primary MMCs rapidly apoptosed as soon as they are purified,26 we looked for the effect of BAFF/APRIL deprivation on the gene expression profile of MMCs cultured together with their BM environment. In the 2 cases investigated, a large number of genes were up- or down-regulated but only a few genes belonged to the TACI signature.
This indicated that the gene signature is not linked to TACI stimulation but to the degree of dependency of myeloma cell growth on the interaction with the BM environment. As BAFF or APRIL are mainly produced by the BM environment and are potent growth factors for normal and malignant plasma cells, it seems logical that MMCs that depend more on BAFF/APRIL for their growth overexpress TACI and molecules involved in the interaction with their environment.
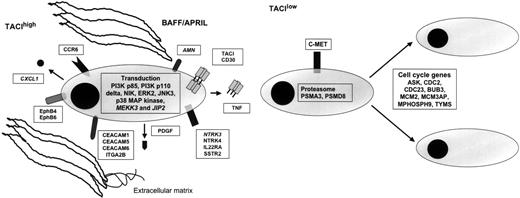
BM environment dependence signature of TACIhi patients and plasmablastic signature of TACIlo patients. (A) TACIhi MMCs had a BM environment dependence signature with overexpression of intercellular communication and transduction genes. (B) TACIlo MMCs have a plasmablastic signature with an overexpression of genes involved in the cell cycle. ITGA2B indicates integrin, alpha 2b; PDGF, platelet-derived growth factor beta polypeptide; IL22RA, interleukin 22 receptor alpha 1; AMN, amnionless homolog; PI3K, phosphoinositide-3-kinase; C-MET, met proto-oncogene (hepatocyte growth factor receptor); PSMA3, proteasome subunit, alpha type 3; PSMD8, proteasome subunit, non-ATPase, 8; ASK, activator of S phase kinase; CDC2, cell division cycle 2; CDC23, cell division cycle 23; BUB3, budding uninhibited by benzimidazoles 3; MCM2, minichromosome maintenance deficient 2; MCM3P, minichromosome maintenance deficient 3 associated protein; and MPHOSPH9, M phase phosphoprotein 9.
BM environment dependence signature of TACIhi patients and plasmablastic signature of TACIlo patients. (A) TACIhi MMCs had a BM environment dependence signature with overexpression of intercellular communication and transduction genes. (B) TACIlo MMCs have a plasmablastic signature with an overexpression of genes involved in the cell cycle. ITGA2B indicates integrin, alpha 2b; PDGF, platelet-derived growth factor beta polypeptide; IL22RA, interleukin 22 receptor alpha 1; AMN, amnionless homolog; PI3K, phosphoinositide-3-kinase; C-MET, met proto-oncogene (hepatocyte growth factor receptor); PSMA3, proteasome subunit, alpha type 3; PSMD8, proteasome subunit, non-ATPase, 8; ASK, activator of S phase kinase; CDC2, cell division cycle 2; CDC23, cell division cycle 23; BUB3, budding uninhibited by benzimidazoles 3; MCM2, minichromosome maintenance deficient 2; MCM3P, minichromosome maintenance deficient 3 associated protein; and MPHOSPH9, M phase phosphoprotein 9.
Of interest, clinical and genetic differences were found between patients with TACIlo or TACIhi MMCs. TACIlo patients had a plasmablast signature, an increase in the percentage of stage III MM, a decrease in hemoglobin level, and an increase in the percentage of bone lesions. It is of note that clinical parameters and risk factors (β2 microglobulin, CRP, LDH) were not different between TACI groups, suggesting that TACI might be an independent prognostic factor that has to be evaluated in further studies. No significant difference in event-free and overall survivals were found between TACIlo and TACIhi patients. This is understandable because the median follow-up of this series of patients was short at the time of the study (18 months). Recently, DKK gene upregulation was associated with increased bone lesions.10 Tian et al pointed out that DKK1 was not expressed in plasmablastic MMCs,10 one of the major characteristic features of TACIlo MMCs. This may explain why we found no upregulation of DKK1 in TACIlo MMCs although increased bone lesions are found in TACIlo patients. Regarding genetic alterations, we found a higher percentage of patients with a t(4;14) translocation within the TACIhi group and no increase of chromosome 13 deletion in the TACIhi group although t(4;14) was strongly associated with deletion 13.3 Actually, about half of the MMCs have deletion 13 and only 14% have a t(4;14) translocation, which may explain this discrepancy.
It was not in the scope of our work to comment on all the genes differentially expressed between TACIlo and TACIhi MMCs. Therefore, in Figure 6, we have selected for genes that appeared to us the most promising for myeloma biology understanding. First, we found an increased proportion of MMCs secreting lambda light chain in TACIhi MMCs. Given that immunoglobulin light chain rearrangement occurs in pre-B cells, we have no explanation for this observation. Focusing on growth factor–related molecules, we found that MMCs in the TACIhi group overexpress 2 neurotrophic tyrosine kinase receptors, NTRK3 which binds neurotrophin 3, and NTRK4. They also overexpress platelet-derived growth factor, CCR6, TNF, interleukin-22 receptor, 2 ephrins (B4 and B6), and the somatostatin receptor 2 (SSTR2). The IL-22 receptor complex consists of IL-10Rβ and IL-22R.44 The IL-10Rβ serves as a common receptor chain for both IL-10 and IL-22. As we previously demonstrated IL-10 to be a growth factor for human MMCs,45 the role of IL-22 in MM biology needs to be investigated. The somatostatin receptor family comprises several members and SSTR2 is expressed in different cancers.46,47 Of interest, the somatostatin analog octeotride binds SSRT2 and has been shown to induce apoptosis in MMCs.48 MMCs in the TACIhi group overexpress the carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), CEACAM5, and CEACAM6 adhesion molecules. CEACAM1 is necessary for VEGF activity.49 Given the likely role of VEGF in multiple myeloma, a role of CEACAM1 in MM demands further investigation. Furthermore, we found an overexpression of 2 ephrin receptors: EphB4 and EphB6. Ephrin receptors are overexpressed in various tumor types, suggesting that they play a role in cancer progression.50 Recently, a study demonstrated that EphB4 acts proangiogenic in breast cancer.51 It proposes that the interplay between the EphB4 ectodomain on tumor cells and the ephrin-B2 in the vasculature promotes blood vessel formation and remodeling. TACIhi MMCs overexpress the chemokine CXCL1. This is of interest because CXCL1 was recently found to bind soluble syndecan-1,52 a proteoglycan that is a hallmark of plasma cell differentiation. Actually, MMP7-deficient mice have a major defect in neutrophil attraction due to a defect in cleavage of syndecan-1 by MMP7, of binding of CXCL-1 to syndecan-1, and thus of a chemoattracting role of CXCL-1.52 MMCs produce soluble syndecan-1, the serum level of which is increased in patients with MM.53 MMP7 is expressed in all myeloma cell samples investigated by Affymetrix microarrays. CXCL1 produced by MMCs in the TACIhi group could play an important role in the biology of MMCs by recruiting cells of the microenvironment, in particular PMNs that largely express BAFF.
Regarding adhesion molecules, MMCs in the TACIhi group overexpress several of these, in particular integrin alpha 2B, also overexpressed in BMPCs compared with plasmablasts.54
Regarding the transduction elements that may be associated with TACI activation, we found an overexpression of mitogen-activated kinase kinase 3 (MKK3), mitogen-activated protein kinase (ERK2), mitogen-activated protein kinase 10 (JNK3), mitogen-activated protein kinase 14 (P38 MAP kinase), and mitogen-activated protein kinase 8-interacting protein 2 (JIP2) in MMCs belonging to the TACIhi group. The substrate of MKK3 is P38 MAP kinase, which is known to be induced by BAFF/APRIL in various cells.55 JIP2 forms complexes with MAP kinases, in particular with MKK3 and P38.56,57 The overexpression of ERK2 in MMCs of the TACIhi group is in good agreement with our previous results demonstrating that the MAPK pathway is activated, in MMCs, by BAFF and APRIL. P38 kinase is also activated in MMCs and an inhibitor of P38 induces apoptosis in MMCs.58 BAFF/APRIL trigger PI-3 kinase in MMCs,21 and in good agreement, the P85 subunit of PI-3K is overexpressed in TACIhi MMCs. PIK3CD (P110 delta polypeptide) is also overexpressed in TACIhi MMCs. P110 delta is expressed at high levels in lymphocytes of lymphoid tissues and plays a role in phosphatidylinositol 3-kinase–mediated signaling in the immune system.59 Promotion of survival of BAFF and APRIL in B cells and MMCs are consistent with NF-kappaB activation. In agreement with these data, the NF-kappaB–inducing kinase (NIK) is overexpressed in TACIhi patients. In T1 B cells, BAFF stimulation promotes B-cell survival, causing the processing of the p100 form of NFκB2 to p52 which required BAFF-R and NIK.60 Previously, we demonstrated that BAFF and APRIL induce the upregulation of the antiapoptotic protein Bcl-2.21 Confirming these results, we show here that TACIhi patients overexpress Bcl-2.
MMCs from patients belonging to the TACIlo group overexpress genes coding for proteins implicated in cell cycle and in nuclear functions. These cells overexpress genes coding for protein involved in G1/S transition and mitosis and contributing to plasmablastic signature. Among them, we validated 2 genes: thymidilate synthetase (TYMS) and cell cycle controller CDC2.61 Their overexpression is associated with chemotherapy resistance in cancer62,63 and these 2 genes are also differentially expressed between patients with or without chromosomal abnormalities.33,64 MMCs in the TACIlo group overexpress 2 genes coding for proteasome subunits and could also be an indicator of increased sensitivity to proteasome inhibitors.65 MMCs from patients belonging to the TACIlo group overexpress c-met, the receptor of HGF. HGF is a myeloma growth factor.66-68 These results suggest that c-met could be a therapeutic target in TACIlo MMCs.
In conclusion, TACI is overexpressed in patients whose MMCs have a gene signature indicative of a dependence on the tumor environment. This fits well with the production of the TACI ligands, BAFF and APRIL, by the tumor environment. TACIlo MMCs overexpress, in turn, cell cycle genes indicative of a plamablastic signature. Taken together, our findings foster a clinical use of inhibitors interfering with the BAFF/APRIL–TACI interaction like the TACI-Fc fusion protein, and at the same time suggest using gene expression profiling to identify the group of patients that might benefit most from this kind of treatment.
Prepublished online as Blood First Edition Paper, April 12, 2005; DOI 10.1182/blood-2004-11-4512.
Supported by grants from the Ligue Nationale Contre le Cancer (équipe labellisée), Paris, France.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal