Abstract
We examined expression of B cell–activating factor of the tumor necrosis factor (TNF) family (BAFF) and a proliferation-inducing ligand (APRIL) on chronic lymphocytic leukemia (CLL) B cells and nurselike cells (NLCs), which differentiate from CD14+ cells when cultured with CLL B cells. NLCs expressed significantly higher levels of APRIL than monocytes and significantly higher levels of BAFF and APRIL than CLL B cells. Also, the viability of CLL B cells cultured with NLCs was significantly reduced when CLL B cells were cultured with decoy receptor of B-cell maturation antigen (BCMA), which can bind both BAFF and APRIL, but not with BAFF receptor:Fc (BAFF-R:Fc), which binds only to BAFF. The effect(s) of BAFF or APRIL on leukemia cell survival appeared additive and distinct from that of stromal cell–derived factor-1α (SDF-1α), which in contrast to BAFF or APRIL induced leukemia cell phosphorylation of p44/42 mitogen-activated protein kinase (extracellular signal-regulated kinase-1/2 [ERK1/2]) and AKT. Conversely, BAFF and APRIL, but not SDF-1α, induced CLL-cell activation of the nuclear factor–κB1 (NF-κB1) and enhanced CLL-cell expression of the antiapoptotic protein Mcl-1. However, BAFF, but not APRIL, also induced CLL-cell activation of NF-κB2. We conclude that BAFF and APRIL from NLCs can function in a paracrine manner to support leukemia cell survival via mechanisms that are distinct from those of SDF-1α, indicating that NLCs use multiple distinct pathways to support CLL-cell survival.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is characterized by the accumulation of monoclonal B cells in the blood, secondary lymphoid tissues, and marrow.1 The leukemia cells primarily are arrested in the G0/G1 phase of the cell cycle and appear resistant to programmed cell death.2,3 Despite their apparent longevity in vivo, CLL cells typically undergo spontaneous apoptosis under conditions that support the growth of human B-cell lines in vitro.4-7 This implies that the factors essential for survival are not intrinsic to the CLL B cell.4-6,8
In vitro, a subset of blood mononuclear cells (BMCs) from patients with CLL can differentiate into large, round, adherent cells that can attract leukemia cells and protect them from undergoing apoptosis.9 When removed from these cells, the CLL B cells experience a rapid decline in viability. Because these cells attract CLL B cells, share features in common with thymic nurse cells, and support CLL B-cell survival, the adherent cells are termed nurselike cells, or NLCs.
Subsequent studies found that NLCs differentiated from CD14+ blood mononuclear cells upon coculture with leukemia cells in vitro. Nevertheless, despite expressing myelomonocytic antigens, NLCs were found to have an expression profile of surface and cytoplasmic antigens (CD14lo, CD68hi, CD83-, CD106-) that is distinct from those of monocytes, macrophages, or blood-derived dendritic cells.10 Abundant cells with the morphology and phenotype of NLCs are present in secondary lymphoid tissues of patients with CLL,10 suggesting they might also function to promote leukemia cell survival in vivo.
The mechanisms whereby NLCs promote CLL-cell survival are not resolved. NLCs express high levels of stromal-derived factor-1α (SDF-1α),9 a CXC chemokine capable of inducing chemotaxis, phosphorylation of mitogen-activated protein kinases (MAPKs), and improved survival of CLL cells in vitro.9 Nevertheless, the viability of CLL B cells cultured with even high concentrations of SDF-1α is not as high as that achieved by coculture with NLCs, indicating that factors other than SDF-1α also might be responsible for promoting CLL B-cell survival by NLCs in vitro.
Investigators have reported that CLL cells express B-lymphocyte stimulator (BLyS), otherwise known as B cell–activating factor of the tumor necrosis factor (TNF) family (BAFF).11-13 BAFF is a type II transmembrane protein that can act in a membranebound or soluble form to promote B-cell survival (reviewed by Mackay and colleagues14 ). Moreover, in mice, disruptive mutations of either BAFF or its receptor, BAFF-R, cause profound loss of mature B cells, indicating that BAFF–BAFF-R interactions are critical for the differentiation and/or survival of mature B cells.15-17 CLL B cells also were found to express the primary BAFF receptor (BAFF-R) as well as 2 other receptors that can interact with BAFF: B-cell maturation antigen (BCMA) and transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI).13 Kern and colleagues also detected expression of BAFF on the surface of CLL cells, implying that BAFF may function in an autocrine manner to support CLL B-cell survival.11
Two of the BAFF receptors, BCMA and TACI, also can bind a proliferation-inducing ligand (APRIL), a factor that also can contribute to B-cell survival.14 The third receptor for BAFF, BAFF-R, is specific for BAFF and cannot bind to APRIL. APRIL originally was found in tumor cells and supposedly is expressed primarily as a secreted soluble molecule through the action of furin proteases present in the Golgi.18 However, Kern and colleagues reported that CLL cells also can express surface APRIL11 and suggested that this factor also may function as an autocrine survival factor in this disease.
Whether the expression of BAFF and/or APRIL on CLL cells is sufficient for optimal leukemia cell survival is not known. Of note, addition of recombinant BAFF could significantly enhance leukemia cell viability, suggesting that the amount of BAFF expressed on isolated CLL cells may be insufficient to support leukemia cell survival, at least in vitro.11,13 Because of the noted dependency of leukemia B cells on accessory cells such as NLCs for survival in vitro, and presumably in vivo, we examined the blood mononuclear cells, NLCs, and isolated leukemia cells of patients with CLL for their relative expression of BAFF and APRIL.
Materials and methods
Cell preparation
After informed consent was obtained per the Declaration of Helsinki, blood samples were collected from patients at the University of California, San Diego (UCSD) Medical Center who satisfied diagnostic and immunophenotypic criteria for common B-cell CLL.1 Blood mononuclear cells were isolated via density-gradient centrifugation with Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). Cells were suspended in fetal calf serum (FCS) containing 5% dimethyl sulfoxide for storage in liquid nitrogen. The viability of the CLL cells was at least 85% at the initiation of cell culture, as assessed by their capacity to exclude propidium iodide (PI) (Molecular Probes, Eugene, OR). All CLL mononuclear cell samples contained more than 95% CD19+/CD5+/CD3- CLL B cells, as assessed by flow cytometry using fluorochrome-conjugated monoclonal antibodies (mAbs) specific for CD19, CD5, or CD3 (BD PharMingen, La Jolla, CA). CLL cells were cultured in RPMI 1640 (Gibco, Rockville, MD) supplemented with 10% FCS and penicillin-streptomycin-glutamine (culture medium) in 5% CO2 in air at 37°C.
CD14+ blood mononuclear cells or CD19+ B cells of healthy donors were isolated from the buffy coat of blood samples collected from adult volunteers at the San Diego Blood Bank (San Diego, CA), as described.10,19 CD14+ cells were cultured with isolated CLL B cells in culture medium at respective cell densities of 1 × 105/mL and 1 × 107/mL. After 10 to 14 days, the plates were rinsed free of the nonadherent CLL cells. The adherent NLCs were then removed for analyses, as described.10
Reagents
Anti–human BAFF mAb was purchased from RDI (Flanders, NJ). Isotype control mouse immunoglobulin G1 (IgG1) (MOPC-21) and fluorescein isothiocyanate (FITC)–conjugated anti–mouse IgG1 were purchased from BD PharMingen. Phycoerythrin (PE)–conjugated anti–human BAFF mAb was purchased from R&D Systems (Minneapolis, MN). Goat anti–human APRIL (R15) polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). FITC-conjugated anti–goat IgG was from Rockland (Gilbertsville, PA). Recombinant human BAFF (rhBAFF) was a kind gift from Dr G. Zhang (National Jewish Medical and Research Center, Denver, CO). Recombinant human APRIL MegaLigand and BCMA-Fc were purchased from Alexis Biochemicals (San Diego, CA). BAFF-R:Fc and control Ig were purchased from R&D Systems. We received the CXCR4 antagonist 4F-benzoyl-TE14011 (4F), which specifically can inhibit the activity of SDF-1α,20 as a gift from Dr N. Fujii (Graduate School of Pharmaceutical Sciences, Kyoto University, Japan).
Cell isolation
Isolated blood mononuclear cells of patients with CLL were incubated with saturating amounts of Dynabeads coated with anti-CD2 or anti-CD14 mAbs (Dynal, Oslo, Norway). Beadbound cells were removed with a strong magnetic field. Following depletion, less than 0.5% of cells were CD2+ or CD14+, whereas more than 99% were CD19+, as assessed via flow cytometry (data not shown). Peripheral normal CD19+ B cells were purified from the buffy coat of blood samples collected from adult volunteers at the San Diego Blood Bank using CD19 Dynabeads and Detatch A Bead (Dynal), following the manufacturer's instruction. The purity of the isolated B cells was more than 95%, as assessed by flow cytometry using a fluorochrome-conjugated anti-CD19 mAb that does not compete with the anti-CD19 mAb used for prior positive selection.
Real-time quantitative RT-PCR
Total RNA was isolated from normal CD14+ cells, NLCs, normal peripheral B cells, and CLL cells before or after depletion of CD14+ cells, using RNeasy Mini Kit (QIAGEN, Valencia, CA). In other experiments, CD14+ monocytes were added to isolated CLL B cells at the indicated ratio, and total RNA was made from each sample. To remove contaminating DNA, the isolated RNA was treated with RQ1 RNase-Free DNase (Promega, Madison, WI) according to the manufacturer's instructions. First-strand cDNA synthesis was performed with SuperScript First-Strand Synthesis System for reverse transcriptase–polymerase chain reaction (RT-PCR) (Invitrogen, Carlsbad, CA). For real-time PCR, SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) was used with 300 nM forward and reverse primers in a final volume of 50 μL for each reaction. Amplification primers were as follows: human BAFF, 5′-ACCGCGGGACTGAAAATCT-3′ and 5′-CACGCTTATTTCTGCTGTTCTGA-3′; human APRIL, 5′-CTGCACCTGGTTCCCATTAAC-3′ and 5′-AAGAGCTGGTTGCCACATCA-3′; human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-ACGGATTTGGTCGTATTGGGC-3′ and 5′-TTGACGGTGCCATGGAATTTG-3′. Each sample was run in duplicate. The polymerase chain reactions were performed using GeneAmp 5700 Detection System (Applied Biosystems) with an initial incubation at 95°C for 10 minutes, followed by 40 cycles, each cycle consisting of a 1-minute incubation at 60°C, followed by a 15-second denaturation step at 95°C. For each run, serially diluted cDNA of U937 cells was used in samples run in parallel to standardize the assay. We determined the cell equivalence (CE) numbers of BAFF, APRIL, and GAPDH mRNA in each sample using the 7700 sequence detector (Applied Biosystems) using the standard curve generated from the diluted U937 cells. The unit number showing relative BAFF or APRIL mRNA level in each sample was determined as a value of BAFF or APRIL CE normalized with GAPDH CE. Melting curve analysis was performed to assess the specificity of PCR product. Following 40 cycles of PCR, samples were heated to 95°C for 30 seconds and 60°C for 20 seconds and then heated to 95°C at a ramp rate of 0.2°C per second. Melting curves for each sample were drawn with 5700 sequence detector software (Applied Biosystems).
Flow cytometry
The cells were stained with saturating amounts of antibodies for 30 minutes at 4°C in deficient RPMI 1640 supplemented with 0.5% bovine serum albumin (fluorescence-activated cell sorter [FACS] buffer), washed 2 times, and then analyzed on a FACSCalibur (Becton Dickinson, Mountain View, CA). Flow cytometry data were analyzed using FlowJo software (Tree Star, San Carlos, CA).
Immunofluorescence staining
CD14+ monocytes were cultured with CLL B cells on Lab-Tek chambered cover glass (Nalge Nunc International, Naperville, IL) for immunofluorescence staining, as described.10 After 14 days, the cells were prepared for immunofluorescence staining using the Cytofix/Cytoperm Kit (BD PharMingen), as per the manufacturer's instructions. The fixed and permeabilized cells were incubated with control antibodies, PE-conjugated anti-BAFF mAb and FITC–anti-CD19 (BD PharMingen), or goat anti-APRIL IgG and PE–anti-CD19 (BD PharMingen). The latter was counterstained with FITC-conjugated anti–goat IgG to detect cellbound goat antibody. Hoechst 33342 (Molecular Probes, Eugene, OR) was used to stain the nuclei. Cover slips were mounted with ProLong Gold antifade reagent (Molecular Probes). Images of fluorochrome-labeled cells were captured with a Delta Vision Restoration microscope system (Applied Precision, Issaquah, WA), using a Photometrics Sony Coolsnap HQ charged-coupled device camera system attached to an inverted, wide-field fluorescent microscope (Nikon TE-200; Nikon, Tokyo, Japan) at the Digital Imaging Core of the Moores Cancer Center of the University of California–San Diego. Optical sections were acquired using a 40×/1.3 NA oil immersion objective in 0.2 μm steps in the z-axis using the attached Applied Precision motorized stage. Images were saved, processed, and analyzed on Silicon Graphics workstations (O2, Octane; Silicon Graphics, Mountain View, CA), using the Delta Vision software package Soft Worx (Version 2.50).
Immunoblot analysis
Cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [tris(hydroxymethyl)aminomethane] [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 5 mM EDTA [ethylenediaminetetraacetic acid]) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.28 TIU (typsin inhibitor units)/mL aprotinin, 50 μg/mL leupeptin, 1 mM benzamidine, and 0.7 μg/mL pepstatin. Lysates were normalized for total protein (20 μg) and subjected to SDS–polyacrylamide gel electrophoresis (PAGE) (4% to 15% gradient gels; Bio-Rad, Hercules, CA) and immunoblot assay. We incubated the blots with secondary antibodies that were conjugated with horseradish peroxidase. Blots then were prepared for enhanced chemiluminescence (ECL) detection system (Amersham, Little Chalfont, United Kingdom) and subsequent autoradiography with Super RX film (Fuji, Tokyo, Japan). The mouse mAb against APRIL (APRIL8) was from Alexis Biochemicals. The mouse mAb against inhibitor of kappa B-α (IκBα) was from Imgenex (San Diego, CA). The antibodies against antiphospho-MAPK extracellular signal-related kinase-1/2 (ERK1/2) and antibody specific for the carboxy-terminal 35 amino acids of MAPK 2/ERK2 (designated “anti-MAP kinase 1/2 [Erk 1/2-CT]” by Upstate Biotechnology) were purchased from Upstate Biotechnology (Lake Placid, NY). Antibodies against Akt or phospho-Akt (Ser473) were from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal antibodies (Mcl-1, Bcl-2, and Bax) were raised against synthetic peptides.21 Also, primary antibodies included β-actin (Sigma Immunochemicals, St Louis, MO). Anti-p52 and anti-p65 antibodies were purchased from Upstate Biotechnology.
Subcellular fractionation and detection of cytoplasmic or nuclear NF-κB
For fractionation experiments, cells were collected by centrifugation and washed with phosphate-buffered saline (PBS). The cell pellet containing 5 × 106 cells was suspended in 100 μL hypotonic buffer (50 mM Tris [pH 7.4], 5 mM EDTA, 10 mM NaCl, 0.05% Nonidet P-40 [NP-40], 1 mM PMSF, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 10 μg/mL pepstatin, 10 mM β-glycerophosphate, 1 mM sodium vanadate, and 1 mM NaF). After 10 minutes, the lysate was spun and the supernatant was collected as cytoplasmic lysates. The pellet was washed 5 times in hypotonic buffer containing 0.1% NP-40. The remaining pellet was suspended in 100 μL RIPA buffer containing protease and phosphatase inhibitors. After an appropriate amount of 3 × sample buffer (200 mM Tris [pH 6.8], 30 mM EDTA, 30% glycerol, 6% SDS) was added, the sample was boiled for 10 minutes, spun for 10 minutes, and the supernatant was recovered as nucleus lysates. Anti–nuclear factor-κB (anti–NF-κB) p52 and p65 were purchased from Upstate Biotechnology. Anti–SP-1 was purchased from Santa Cruz Biotechnology.
Measurement of cell viability
Freshly thawed CLL B cells were cultured at the concentration of 1 × 106/mL under various conditions. Determination of CLL-cell viability in this study was based on the analysis of mitochondrial transmembrane potential (ΔΨm) using 3,3′-dihexyloxacarbocyanine iodine (DiOC6) and cell membrane permeability to PI, as described.22 For viability assays, 100 μL of the cell culture was collected at the indicated time points and transferred to polypropylene tubes containing 100 μL of 60 nM DiOC6 (Molecular Probes) and 10 μg/mL PI in FACS buffer. The cells then were incubated at 37°C for 15 minutes and analyzed within 30 minutes by flow cytometry using a FACSCalibur (Becton Dickinson). Fluorescence was recorded at 525 nm (FL-1) for DiOC6 and at 600 nm (FL-3) for PI.
Statistical analysis
Results are shown as mean plus or minus SD of at least 3 samples each. For statistical comparison between groups, the Student t test or the Bonferroni t test was used. Analyses were performed using Glanzman “Primer of Biostatistics” software (McGraw-Hill, New York, NY).
Results
Expression of BAFF mRNA and protein on CLL cells and NLCs
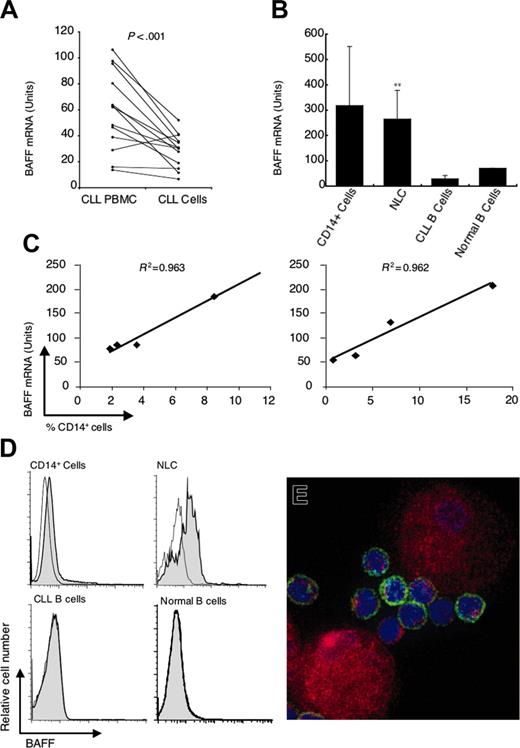
We examined the BMCs of patients with CLL for expression of BAFF mRNA by real-time RT-PCR. In each case, we detected expression of BAFF mRNA, consistent with earlier reports.11-13 Moreover, we found that rigorous depletion of CD14+ cells from the BMCs significantly lowered the amount of BAFF mRNA detected in each sample (59 ± 30 units in BMCs and 29 ± 13 units in isolated CLL B cells, n = 12, P < .001, paired t test; Figure 1A). Furthermore, the amount of BAFF mRNA detected in CD14+ cells (320 ± 230, n = 4; or NLCs 270 ± 110, n = 12) was significantly greater than that noted in the isolated leukemia B cells (P < .001; Figure 1B) or isolated CD19+ blood B cells of healthy donors.
Small numbers of CD14+ cells present in the BMCs isolated from patients with CLL potentially could contribute a large proportion of the BAFF mRNA detected by real-time RT-PCR assay, which uses GAPDH mRNA to normalize the assay. To evaluate this possibility we added small numbers of CD14+ blood mononuclear cells to purified CD19+ CLL B cells and examined how this affected the amount of BAFF mRNA detected in each sample (Figure 1C). For each 1% of added CD14+ cells there was an increase in the detected amount of BAFF mRNA of 10 to 13 units. At the y-intercept of each graph (Figure 1C) when the proportion of CD14+ cells was extrapolated to 0%, we detected 30 to 40 units of BAFF mRNA. We attribute this to the amount of BAFF mRNA expressed by CLL B cells themselves, because this is the amount we detected in the isolated leukemia B cells (Figure 1B; eg, 29 ± 13 units). This implies that on a cell-per-cell basis, CD14+ cells apparently contain approximately 30-fold more BAFF mRNA than CLL B cells.
We next examined CLL cells and NLCs for surface expression of BAFF by flow cytometry. In contrast to CLL B cells or purified normal B cells, NLCs expressed high levels of BAFF that were easily detected by flow cytometry (Figure 1D) or immunofluorescence microscopy (Figure 1E). These data indicate that NLCs express large amounts of BAFF protein relative to that expressed by CLL B cells.
Expression of APRIL mRNA and protein on CLL cells and NLCs
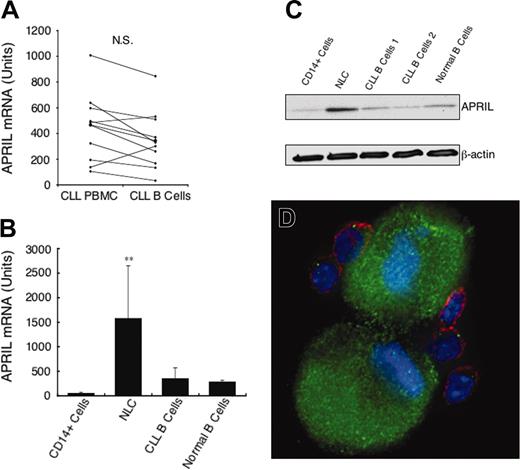
We also examined the BMCs of patients with CLL for expression of APRIL mRNA with the same techniques used for evaluating the expression of BAFF. In contrast to our studies on BAFF mRNA, we found that rigorous depletion of CD14+ cells from the BMCs did not lower the amount of APRIL mRNA detected in each sample tested (440 ± 308 units in BMCs and 348 ± 228 units in isolated CLL B cells, n = 11, not significant [NS], paired t test; Figure 2A). This indicates that CD14+ blood mononuclear cells do not contribute significantly to the amounts of APRIL mRNA found in CLL blood mononuclear cells. Consistent with this, we found that isolated CD14+ cells had very low amounts of APRIL mRNA (52 ± 20, n = 5).
In contrast, the amount of APRIL mRNA detected in differentiated NLCs was significantly higher (Figure 2B; 1595 ± 1090, n = 11) than that of nondifferentiated CD14+ blood mononuclear cells. Moreover, NLCs had a significantly greater amount of APRIL mRNA than that noted in the isolated leukemia B cells or isolated CD19+ blood B cells of healthy donors (P < .01, Bonferroni t test; Figure 2B).
Expression of BAFF mRNA and protein. (A) Quantitative real-time RT-PCR was performed on RNA samples isolated from the blood mononuclear cells of individual patients with CLL before (left) and after (right) depletion of CD2+ and CD14+ cells. The lines connect the preisolation and postisolation levels of BAFF mRNA detected in each sample. The amount of BAFF mRNA detected is indicated in arbitrary units. The amount of BAFF mRNA detected in an equivalent number of U937 cells is 1000 units (data not shown). (B) Quantitative real-time RT-PCR measurement of the average amount of BAFF mRNA detected in CD14+ cells (n = 4), NLCs (n = 12), purified CLL B cells (n = 12), and isolated CD19+ blood B cells of healthy donors (n = 2), as indicated at the bottom of the panel, ± SD (**the level of BAFF mRNA detected in NLCs was significantly greater than that found in isolated CLL B cells, P < .001). (C) Reconstitution experiments in which small numbers of CD14+ blood mononuclear cells are added to 5 × 106 isolated CLL B cells that subsequently were evaluated for BAFF mRNA in 2 representative patients. On the x-axis is the percent of CD14+ cells detected by FACS in the reconstituted cell population prior to extraction of RNA. The y-axis indicates the level of BAFF mRNA detected in units as defined in the description for panel A. (D) Representative histograms depicting surface BAFF detected by flow cytometry on CD14+ cells, NLCs, CD19+ CLL B cells, or CD19+ blood B cells of healthy donors, as indicated at the top of each graph. Shaded histograms represent the fluorescence of cells stained with a fluorochrome-labeled anti-BAFF mAb, whereas the open histograms depict the fluorescence of cells stained with an isotype control mAb. (E) An immunofluorescence picture of NLCs and CLL cells stained with fluorescein-labeled anti-CD19 mAb (green) and a PE-labeled anti-BAFF mAb (red). The nuclei are labeled blue with Hoechst 33342.
Expression of BAFF mRNA and protein. (A) Quantitative real-time RT-PCR was performed on RNA samples isolated from the blood mononuclear cells of individual patients with CLL before (left) and after (right) depletion of CD2+ and CD14+ cells. The lines connect the preisolation and postisolation levels of BAFF mRNA detected in each sample. The amount of BAFF mRNA detected is indicated in arbitrary units. The amount of BAFF mRNA detected in an equivalent number of U937 cells is 1000 units (data not shown). (B) Quantitative real-time RT-PCR measurement of the average amount of BAFF mRNA detected in CD14+ cells (n = 4), NLCs (n = 12), purified CLL B cells (n = 12), and isolated CD19+ blood B cells of healthy donors (n = 2), as indicated at the bottom of the panel, ± SD (**the level of BAFF mRNA detected in NLCs was significantly greater than that found in isolated CLL B cells, P < .001). (C) Reconstitution experiments in which small numbers of CD14+ blood mononuclear cells are added to 5 × 106 isolated CLL B cells that subsequently were evaluated for BAFF mRNA in 2 representative patients. On the x-axis is the percent of CD14+ cells detected by FACS in the reconstituted cell population prior to extraction of RNA. The y-axis indicates the level of BAFF mRNA detected in units as defined in the description for panel A. (D) Representative histograms depicting surface BAFF detected by flow cytometry on CD14+ cells, NLCs, CD19+ CLL B cells, or CD19+ blood B cells of healthy donors, as indicated at the top of each graph. Shaded histograms represent the fluorescence of cells stained with a fluorochrome-labeled anti-BAFF mAb, whereas the open histograms depict the fluorescence of cells stained with an isotype control mAb. (E) An immunofluorescence picture of NLCs and CLL cells stained with fluorescein-labeled anti-CD19 mAb (green) and a PE-labeled anti-BAFF mAb (red). The nuclei are labeled blue with Hoechst 33342.
Expression of APRIL mRNA and protein. (A) Quantitative real-time RT-PCR was performed on RNA samples isolated from the blood mononuclear cells of patients with CLL before (left) and after (right) depletion of CD2+ and CD14+ cells. The lines connect the preisolation and postisolation levels of APRIL mRNA in each sample. The amount of APRIL mRNA detected is indicated in arbitrary units. The amount of APRIL mRNA detected in an equivalent number of U937 cells is 30 units (data not shown). (B) Quantitative real-time RT-PCR measurement of the average amount of APRIL mRNA detected in CD14+ cells (n = 4), NLCs (n = 11), purified CLL B cells (n = 11), or isolated CD19+ blood B cells of healthy donors (n = 3), as indicated at the bottom of the histogram, ± SD (**the mean level of APRIL mRNA detected in NLCs was significantly greater than that found in isolated CLL B cells, P < .01). (C) Representative immunoblot data showing the expression of APRIL by NLCs, CD14+ blood mononuclear cells, CLL B cells, or isolated CD19+ blood B cells of healthy donors. Whole cell lysates were prepared as described in “Materials and methods.” The protein content was normalized to 20 μg and subjected to immunoblot analysis with antibodies specific for APRIL or β-actin using ECL-based detection. (D) An immunofluorescence picture of NLCs and CLL cells stained with phycoerythrin-labeled anti-CD19 mAb (red) and goat IgG anti-APRIL polyclonal antibody that was detected using a fluorescein-labeled anti–goat IgG (green). The nuclei are labeled blue with Hoechst 33342.
Expression of APRIL mRNA and protein. (A) Quantitative real-time RT-PCR was performed on RNA samples isolated from the blood mononuclear cells of patients with CLL before (left) and after (right) depletion of CD2+ and CD14+ cells. The lines connect the preisolation and postisolation levels of APRIL mRNA in each sample. The amount of APRIL mRNA detected is indicated in arbitrary units. The amount of APRIL mRNA detected in an equivalent number of U937 cells is 30 units (data not shown). (B) Quantitative real-time RT-PCR measurement of the average amount of APRIL mRNA detected in CD14+ cells (n = 4), NLCs (n = 11), purified CLL B cells (n = 11), or isolated CD19+ blood B cells of healthy donors (n = 3), as indicated at the bottom of the histogram, ± SD (**the mean level of APRIL mRNA detected in NLCs was significantly greater than that found in isolated CLL B cells, P < .01). (C) Representative immunoblot data showing the expression of APRIL by NLCs, CD14+ blood mononuclear cells, CLL B cells, or isolated CD19+ blood B cells of healthy donors. Whole cell lysates were prepared as described in “Materials and methods.” The protein content was normalized to 20 μg and subjected to immunoblot analysis with antibodies specific for APRIL or β-actin using ECL-based detection. (D) An immunofluorescence picture of NLCs and CLL cells stained with phycoerythrin-labeled anti-CD19 mAb (red) and goat IgG anti-APRIL polyclonal antibody that was detected using a fluorescein-labeled anti–goat IgG (green). The nuclei are labeled blue with Hoechst 33342.
We evaluated for expression of APRIL by immunoblot analysis. As seen in Figure 2C, total lysates from NLCs had higher amounts of APRIL than did CD14+ blood mononuclear cells, purified CLL B cells, or isolated CD19+ blood B cells of healthy donors. NLCs also were found to express high levels of APRIL relative to CLL B cells by immunofluorescence staining (Figure 2D).
Effect of BCMA-Fc or BAFF-R:Fc on the viability of CLL cells cultured with NLCs
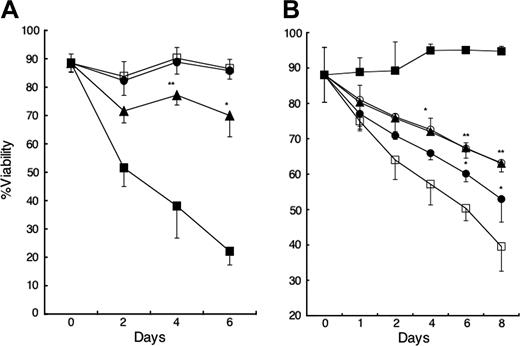
Because NLCs express both BAFF and APRIL, we examined whether these factors contributed to the capacity of NLCs to sustain CLL-cell survival in vitro. We cultured CLL B cells with decoy receptors of BCMA (BCMA-Fc), which can bind to both BAFF and APRIL, and BAFF-R (BAFF-R:Fc), which binds only to BAFF, and compared the viability of the leukemia cells with that of such cells cultured with control Ig. We observed that addition of BCMA-Fc to cocultures of CLL cells and NLCs significantly reduced the viability of the CLL cells relative to that of cocultures treated with control Ig (Figure 3A). In contrast, there was no decline in leukemia cell viability in such cocultures when we added saturating amounts of BAFF-R:Fc (Figure 3A), which in parallel studies were found capable of inhibiting B-cell survival in cocultures with rhBAFF or fibroblastlike synoviocytes that expressed BAFF, but not APRIL (data not shown).19
CLL-cell survival with or without NLCs. (A) Inhibition of CLL-cell survival on NLCs by BCMA-Fc but not BAFF-R:Fc. CLL B cells were cultured with (□) or without (▪) NLCs and 1 μg/mL control Ig. BCMA-Fc (▴) or BAFF-R:Fc (•) at 1 μg/mL was added to the wells of CLL B cells cultured with NLCs at day 0. Viability was subsequently determined for each time point, as indicated on the horizontal axis. Displayed is the mean percent viability ± SD (error bars) of samples from each of 5 patients. The percent viability of BCMA-Fc–treated cultures was significantly less than that of control Ig–treated cultures (*P < .05; **P < .01; Bonferroni t test). (B) Enhanced CLL-cell survival with NLCs or rhBAFF or rhAPRIL. A total of 1 × 106/mL isolated CD19+ CLL B cells was cultured alone (□), with 50 ng/mL rhBAFF (▴), 500 ng/mL rhAPRIL (•), both rhBAFF and rhAPRIL (○), or NLCs (▪) and evaluated over time. Displayed is the mean percent viability ± SD of samples from each of 3 patients. The percent viability of rhBAFF-treated CLL cells or rhAPRIL-treated CLL cells was significantly greater than that of control-treated CLL cells (*P < .05; **P < .01; Bonferroni t test).
CLL-cell survival with or without NLCs. (A) Inhibition of CLL-cell survival on NLCs by BCMA-Fc but not BAFF-R:Fc. CLL B cells were cultured with (□) or without (▪) NLCs and 1 μg/mL control Ig. BCMA-Fc (▴) or BAFF-R:Fc (•) at 1 μg/mL was added to the wells of CLL B cells cultured with NLCs at day 0. Viability was subsequently determined for each time point, as indicated on the horizontal axis. Displayed is the mean percent viability ± SD (error bars) of samples from each of 5 patients. The percent viability of BCMA-Fc–treated cultures was significantly less than that of control Ig–treated cultures (*P < .05; **P < .01; Bonferroni t test). (B) Enhanced CLL-cell survival with NLCs or rhBAFF or rhAPRIL. A total of 1 × 106/mL isolated CD19+ CLL B cells was cultured alone (□), with 50 ng/mL rhBAFF (▴), 500 ng/mL rhAPRIL (•), both rhBAFF and rhAPRIL (○), or NLCs (▪) and evaluated over time. Displayed is the mean percent viability ± SD of samples from each of 3 patients. The percent viability of rhBAFF-treated CLL cells or rhAPRIL-treated CLL cells was significantly greater than that of control-treated CLL cells (*P < .05; **P < .01; Bonferroni t test).
Additive effects of SDF-1α and BAFF or APRIL on CLL B-cell survival
Next we examined whether NLCs or exogenous BAFF or APRIL could enhance the viability of CLL B cells in vitro. For this, we monitored the viability of CLL B cells over time when cultured with or without NLCs or with or without rhBAFF or rhAPRIL. Consistent with prior studies,9,10 CLL cells cultured alone had less viability than leukemia cells cultured with NLCs. The addition of rhBAFF or rhAPRIL significantly improved the viability of CLL cells cultured without NLCs (Figure 3B). The viability of the CLL cells cocultured with either rhBAFF or rhAPRIL alone was not enhanced further by the addition of rhAPRIL or rhBAFF, respectively.
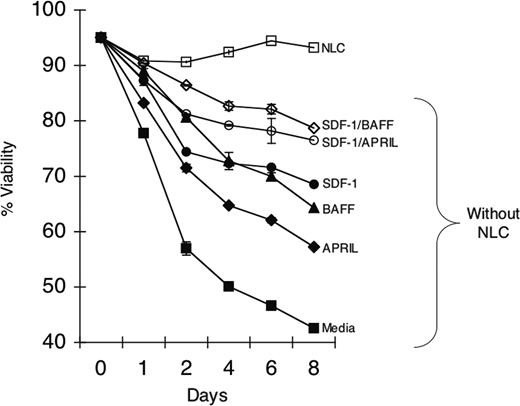
Because NLCs express BAFF, APRIL, and SDF-1α, we examined whether these factors together could support CLL B-cell survival better than either factor alone. The viability of isolated CLL B cells was highest when cocultured with NLCs (Figure 4). However, isolated CLL B cells cocultured with rhBAFF plus SDF-1α, or rhAPRIL plus SDF-1α, had a significantly greater viability than that of CLL B cells cultured with any one factor alone (Figure 4). Collectively, these data support the notion that BAFF or APRIL promotes leukemia cell survival via a mechanism(s) independent of that used by SDF-1α.
Effects of rhBAFF, rhAPRIL, or SDF-1α on signaling pathways in CLL B cells
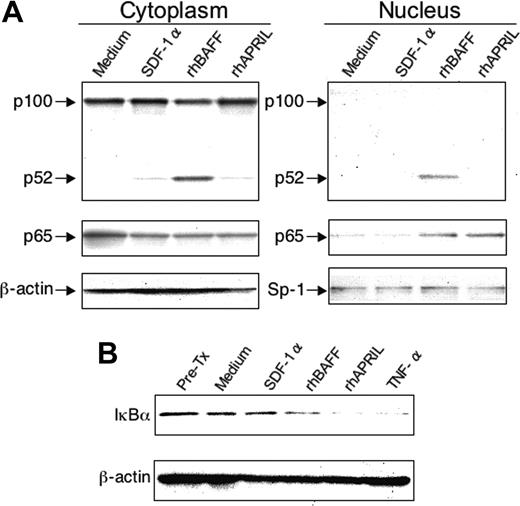
We examined the intracellular signaling pathways stimulated by rhBAFF, rhAPRIL, or SDF-1α at concentrations that can promote CLL B-cell survival in vitro. Prior studies indicated that BAFF could induce activation of the NF-κB2 in normal B cells,17,23 a pathway that appears critical for the growth and/or survival of normal B cells.24 Such activation involves processing of p100 to p52 with subsequent translocation of p52 to the nucleus. We found that rhBAFF could induce activation of NF-κB2 also in CLL B cells (Figure 5A). In contrast, we did not observe activation of NF-κB2 in CLL cells treated with rhAPRIL or SDF-1α, even at concentrations that could support CLL-cell survival in vitro. Both rhBAFF and rhAPRIL, however, induced degradation of the inhibitor of kappa B (IκBα) and translocation of p65 to the nuclear fraction, indicating activation of the classical NF-κB pathway (Figure 4B). SDF-1α, on the other hand, did not have this activity (Figure 4).
Effect of rhBAFF, rhAPRIL, and/or SDF-1α on CLL-cell survival. CLL B cells were cultured with (□) or without (▪) NLCs. SDF-1α (•) rhAPRIL (♦) at 500 ng/mL, rhBAFF (▴) at 50 ng/mL, or both (⋄) were added to wells without NLCs at day 0. Also SDF-1α and rhBAFF (⋄) or SDF-1α and rhAPRIL (○) were added to the cultures without NLCs. The mean viability ± SE of replicate wells was determined for each time point indicated on the horizontal axis. A representative example of 3 different CLL patients is presented.
Effect of rhBAFF, rhAPRIL, and/or SDF-1α on CLL-cell survival. CLL B cells were cultured with (□) or without (▪) NLCs. SDF-1α (•) rhAPRIL (♦) at 500 ng/mL, rhBAFF (▴) at 50 ng/mL, or both (⋄) were added to wells without NLCs at day 0. Also SDF-1α and rhBAFF (⋄) or SDF-1α and rhAPRIL (○) were added to the cultures without NLCs. The mean viability ± SE of replicate wells was determined for each time point indicated on the horizontal axis. A representative example of 3 different CLL patients is presented.
Activation of NF-κB in CLL B cells by rhBAFF or rhAPRIL. (A) Processing of p100 and nuclear translocation of p52 or p65. CLL B cells were cultured with or without SDF-1α (500 ng/mL), rhBAFF (50 ng/mL), and rhAPRIL (500 ng/mL) for 24 hours. Cytoplasmic and nuclear extracts were prepared as described in “Materials and methods” for immunoblot analysis with anti-p100 or anti-p65 antibodies as indicated on the left side of each panel. The agent used to treat the CLL cells is indicated at the top of each panel under the label indicating whether the extract was derived from cytoplasmic (left panel) or nuclear (right panel) cell fractions. We evaluated for equal loading in each lane by stripping the blot and probing it again with antibodies specific for β-actin (for cytoplasmic extracts) or SP-1 (for nuclear extracts), as indicated on the left side of each panel. (B) Degradation of IκBα. Extracts of CLL cells were prepared for immunoblot analysis prior to treatment (Pre-Tx) or after a 30-minute incubation with culture medium alone (Medium) or medium supplemented with SDF-1α (500 ng/mL), rhBAFF (50 ng/mL), rhAPRIL (500 ng/mL), or TNFα (50 ng/mL), as indicated at the top of each lane. The immunoblot was probed with antibodies specific for IκBα (top blot). We evaluated for equal loading in each lane by stripping the blot and probing it again with antibodies specific for β-actin (bottom blot).
Activation of NF-κB in CLL B cells by rhBAFF or rhAPRIL. (A) Processing of p100 and nuclear translocation of p52 or p65. CLL B cells were cultured with or without SDF-1α (500 ng/mL), rhBAFF (50 ng/mL), and rhAPRIL (500 ng/mL) for 24 hours. Cytoplasmic and nuclear extracts were prepared as described in “Materials and methods” for immunoblot analysis with anti-p100 or anti-p65 antibodies as indicated on the left side of each panel. The agent used to treat the CLL cells is indicated at the top of each panel under the label indicating whether the extract was derived from cytoplasmic (left panel) or nuclear (right panel) cell fractions. We evaluated for equal loading in each lane by stripping the blot and probing it again with antibodies specific for β-actin (for cytoplasmic extracts) or SP-1 (for nuclear extracts), as indicated on the left side of each panel. (B) Degradation of IκBα. Extracts of CLL cells were prepared for immunoblot analysis prior to treatment (Pre-Tx) or after a 30-minute incubation with culture medium alone (Medium) or medium supplemented with SDF-1α (500 ng/mL), rhBAFF (50 ng/mL), rhAPRIL (500 ng/mL), or TNFα (50 ng/mL), as indicated at the top of each lane. The immunoblot was probed with antibodies specific for IκBα (top blot). We evaluated for equal loading in each lane by stripping the blot and probing it again with antibodies specific for β-actin (bottom blot).
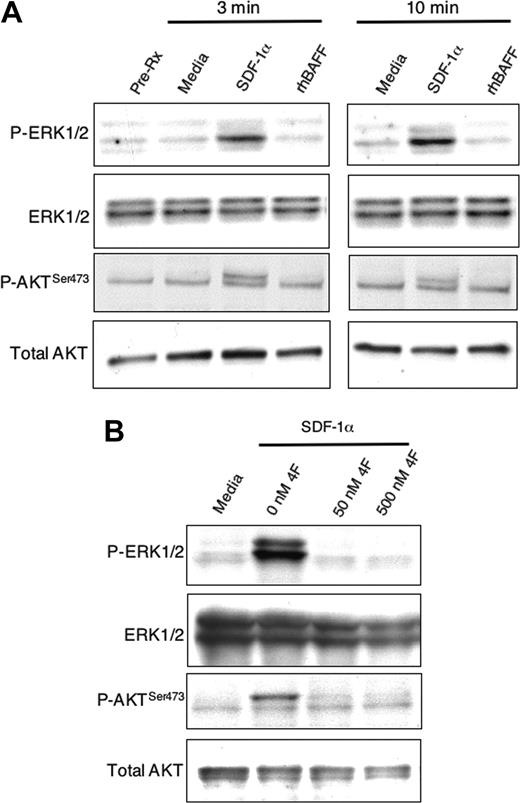
Activation of MAPK (ERK1/2) and AKT in CLL cells. (A) CLL B cells were cultured for 3 or 10 minutes with SDF-1α (200 ng/mL), rhBAFF (50 ng/mL), or media, as indicated above the sample lanes. Cell lysates were prepared and analyzed by immunoblot using antibodies specific for phosphorylated ERK1/2 (P-ERK1/2), ERK1/2, phosphorylated AKT (P-AKTSer473), or AKT, as indicated on the left margin. Equal loading in the lanes was evaluated by stripping the blot and probing again with anti-ERK1/2 and an anti-AKT antibody. Five different CLL B cells gave similar results. (B) The CLL cells were stimulated for 3 minutes with either media (far left lane) or SDF-1α (200 ng/mL) (right 3 lanes). For samples treated with SDF-1α we included the CXCR4 antagonist 4F-benzoyl-TE1401120 (4F) at 0 nM, 50 nM, or 500 nM. The samples were analyzed and the results presented as noted in panel A.
Activation of MAPK (ERK1/2) and AKT in CLL cells. (A) CLL B cells were cultured for 3 or 10 minutes with SDF-1α (200 ng/mL), rhBAFF (50 ng/mL), or media, as indicated above the sample lanes. Cell lysates were prepared and analyzed by immunoblot using antibodies specific for phosphorylated ERK1/2 (P-ERK1/2), ERK1/2, phosphorylated AKT (P-AKTSer473), or AKT, as indicated on the left margin. Equal loading in the lanes was evaluated by stripping the blot and probing again with anti-ERK1/2 and an anti-AKT antibody. Five different CLL B cells gave similar results. (B) The CLL cells were stimulated for 3 minutes with either media (far left lane) or SDF-1α (200 ng/mL) (right 3 lanes). For samples treated with SDF-1α we included the CXCR4 antagonist 4F-benzoyl-TE1401120 (4F) at 0 nM, 50 nM, or 500 nM. The samples were analyzed and the results presented as noted in panel A.
We also examined for phosphorylation and activation of AKT, which prior studies found also could enhance CLL B-cell survival.25,26 In contrast to SDF-1α,9 we found that rhBAFF or rhAPRIL could not induce phosphorylation of p44/42 mitogen-activated phosphokinase (MAPK ERK1/2) or activation of AKT in CLL B cells, even at concentrations that could promote CLL B-cell survival in vitro (Figure 6 and data not shown).
However, SDF-1α not only induced phosphorylation of ERK1/2, as noted previously,9 but also induced phosphorylation of AKT at Ser473 in isolated CLL B cells (Figure 6A). The capacity of SDF-1α to induce CLL-cell phosphorylation of ERK1/2 and AKT at Ser473 could be blocked by 4F-benzoyl-TE14011 (4F), a specific CXCR4 antagonist (Figure 6B)20
NLCs, BAFF, or APRIL, but not SDF-1α, can induce CLL-cell expression of Mcl-1
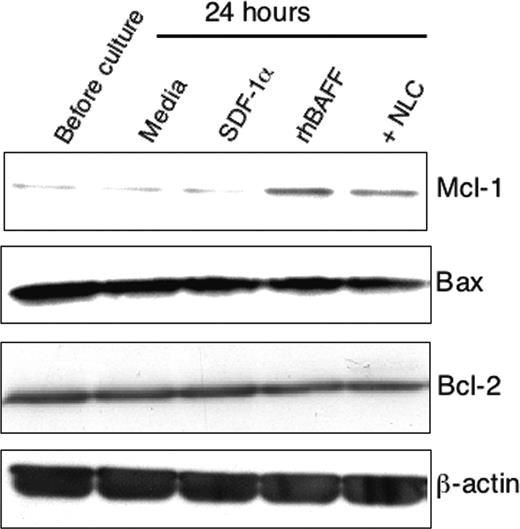
To examine mechanisms that might account for the effects on leukemia cell survival, we evaluated for the expression of proapoptotic and antiapoptotic proteins in CLL B cells following culture with or without NLCs or with either rhBAFF or SDF-1α. We did not observe significant changes in the levels of Bcl-2, Bax, or Bcl-xL expressed by isolated CLL B cells in any of the short-term culture conditions used (Figure 7 and data not shown). On the other hand, CLL B cells cocultured with NLCs, rhBAFF, or rhAPRIL were induced to express increased levels of Mcl-1 (Figure 7 and data not shown). In contrast, SDF-1α could not induce isolated CLL B cells to express higher levels of Mcl-1, even at concentrations that could protect CLL-cell survival in vitro (Figure 7 and data not shown).
Discussion
Increasing attention is being focused on cells and factors of different microenvironments that contribute to CLL-cell survival.27,28 Such accessory cells include marrow stromal cells,6,7,29,30 follicular dendritic cells,31 and NLCs.9 Defining the mechanisms whereby these cells contribute to the survival of CLL cells potentially could identify novel targets for treatment of this disease.
In this study, we found that NLCs express high levels of BAFF and APRIL, 2 factors of the TNF family that play an important role in maintaining the survival of mature B cells.15-17 Because NLCs are derived from CD14+ cells,10 expression of BAFF by NLCs was anticipated, because this factor originally was found expressed by myeloid lineage cells, such as monocytes, macrophages, or dendritic cells.32,33 Moreover, we found that CD14+ cells accounted for most of the BAFF mRNA found in the blood mononuclear cells of patients with CLL and, on a cell-per cell basis, contained approximately 30-fold more BAFF mRNA than did CLL B cells, which prior studies found could also express this B-cell survival factor.11,13 From the studies reported here, it appears that such CD14+ cells maintain high-level expression of BAFF, even after they differentiate into NLCs upon coculture with CLL B cells in vitro.
In contrast, NLCs expressed significantly more APRIL than newly isolated CD14+ blood cells, which in turn contributed little to the APRIL mRNA detected in the blood mononuclear cells of patients with CLL. Moreover, the low to negligible amount of APRIL mRNA detected in CD14+ blood mononuclear cells appeared less than that expressed by CLL B cells or even normal B cells. In contrast, CD14+ myeloid cells in the secondary lymphoid tissues of patients with non-Hodgkin lymphomas, including CLL, apparently express high levels of BAFF and APRIL.12 Conceivably, such cells may include CD14+ cells that already have differentiated into NLCs in vivo.10
Expression of Mcl-1 in CLL B cells by immunoblot analysis. Representative immunoblot data showing up-regulation of Mcl-1 by NLCs or rhBAFF. CLL B cells were cultured with or without NLCs or with SDF-1α (500 ng/mL) or rhBAFF (50 ng/mL) for 24 hours. Whole cell lysates were then prepared. The protein content was normalized to 12.5 μg and analyzed by immunoblot analysis with antibodies specific for Mcl-1, Bax, Bcl-2, or β-actin using ECL-based detection.
Expression of Mcl-1 in CLL B cells by immunoblot analysis. Representative immunoblot data showing up-regulation of Mcl-1 by NLCs or rhBAFF. CLL B cells were cultured with or without NLCs or with SDF-1α (500 ng/mL) or rhBAFF (50 ng/mL) for 24 hours. Whole cell lysates were then prepared. The protein content was normalized to 12.5 μg and analyzed by immunoblot analysis with antibodies specific for Mcl-1, Bax, Bcl-2, or β-actin using ECL-based detection.
We investigated whether BAFF and/or APRIL on NLCs could contribute to their capacity to promote leukemia cell survival in vitro. Previous studies showed BCMA-Fc could impair leukemia cell viability over time when this decoy receptor was added to isolated leukemia cells.11,13 However, we did not observe this effect on the viability of CLL B cells cultured without NLCs, even at concentrations of BCMA-Fc of 30 μg/mL (data not shown). The reason for the discrepancy between our data and others is not clear. Instead, BCMA-Fc significantly impaired the viability of CLL B cells cultured with NLCs (Figure 3A). However, BAFF-R:Fc, which can only inhibit BAFF interactions with BAFF-R, failed to impair the viability of CLL cells that were cultured either with or without NLCs, implying that APRIL may play an important role in the protective effect(s) of NLCs on CLL-cell survival. Although the studies in knock-out mice showed that APRIL appeared to be dispensable for developing normal immune systems,34 a recent study by Planelles et al found that APRIL may play a role in the pathogenesis of B1-cell malignancies, namely CLL.35 In this light, strategies that interfere only with BAFF–BAFF-R interactions may not be sufficient to affect CLL-cell viability in vivo.
Previously, we reported that NLCs also express SDF-1α, a chemokine that can trigger phosphorylation of p44/42 MAPK ERK1/2 and enhance CLL-cell survival in vitro.9 Although some studies have suggested that the ERK pathway might not be involved in preventing spontaneous apoptosis of CLL B cells,36 suppression of ERK activity is seen in CLL B cells undergoing drug-induced apoptosis,37,38 suggesting that this pathway is important for survival of CLL B cells.
Because SDF-1α had an additive effect on the viability of isolated CLL cells cultured with BAFF and/or APRIL (Figure 5), we reasoned that BAFF or APRIL might promote CLL-cell survival via a pathway(s) that is distinct from that of SDF-1α. Consistent with this notion, we found that SDF-1α, in addition to its noted capacity to induce phosphorylation of MAPK ERK1/2, could induce CLL B-cell activation of phosphatidylinositol 3-kinase (PI3K) AKT (Figure 6), a pathway that is essential for the survival of CLL B cells.25,26 These findings are consistent with those of others who found that SDF-1α could induce activation of AKT in other types of cells besides leukemia B cells.39-42 Recently, Moreaux and colleagues reported that addition of exogenous BAFF to myeloma cells induced late activation of both ERK1/2 and AKT,42 but the direct influence of BAFF on these 2 pathways was not resolved. In the study presented here, it appears that neither pathway is activated in CLL cells by rhBAFF or rhAPRIL, indicating that these factors must use other mechanisms to protect CLL B cells from spontaneous apoptosis.
Some TNF superfamily proteins like BAFF trigger their functions by activating NF-κB. Two main pathways—the canonical and alternative pathways—regulate the activity of NF-κB.24 Activation of the canonical pathway results from degradation of the inhibitor of NF-κBα (IκBα), which is induced upon its phosphorylation by the beta subunit of the IκB kinase (IKK) complex, IKKβ.43 This leads to nuclear translocation of active NF-κB heterodimers (that are composed of p65, c-Rel, or p50) where they can effect changes in gene expression. As noted for lymphoma or CLL B cells,11,12 concentrations of rhBAFF or rhAPRIL required for optimal enhancement of CLL-cell survival also induced degradation of IκBα and translocation of p65 to the nucleus, indicating that either factor can activate the canonical NF-κB pathway. Activation of the canonical NF-κB pathway in normal B cells appears secondary to the capacity of BAFF or APRIL to interact with BCMA, or BCMA and/or TACI, respectively.44
Alternative pathway activation results from processing of NF-κB2 p100 to p52, which is triggered by the phosphorylation of NF-κB2 p100 by the alpha subunit of the IKK complex, namely IKKα.43 This allows for nuclear translocation of p52 along with RelB, where this complex can influence expression of genes that are distinct from those regulated by the canonical NF-κB pathway. We noted that rhBAFF, but not rhAPRIL or SDF-1α, could induce degradation of p100 to p52 and translocation of p52 to the nucleus. Because the BAFF-R interacts with BAFF, but not APRIL, the selective activation of p100 processing by BAFF suggests that the BAFF-R may be distinct from BCMA or TACI in its capacity to activate the alternative NF-κB pathway in CLL B cells. This is similar to the interaction of BAFF with its receptor on normal B cells, which also promotes processing of NF-κB2.17,45 Moreover, studies have shown that IKKα is required for B-cell maturation and formation of secondary lymphoid organs.46,47 However, because treatment of cocultures of CLL cells and NLCs with BAFF-R:Fc failed to inhibit the protective effect of NLCs on leukemia cell survival, it appears that activation of the canonical pathway may obviate the requirement for activation of the alternative NF-κB pathway in CLL to promote leukemia cell survival, at least in the in vitro culture conditions used in this study.
Finally, we evaluated for expression of Bcl-2 family member proteins that can influence the resistance or sensitivity of CLL cells to apoptosis. Prior studies found that BAFF can upregulate expression of Bcl-2 in most B cells.12,14 BAFF-induced up-regulation of Bcl-2 was less apparent in CLL B cells, possibly secondary to the constitutive high-level expression of this antiapoptotic protein in this leukemia.48 However, we found that rhBAFF, rhAPRIL, or NLCs could induce CLL B cells to express high levels of Mcl-1 (Figure 7 and data not shown). Like Bcl-2, Mcl-1 also appears to play a role in the resistance of CLL B cells to drug-induced apoptosis,49,50 and patients with CLL who fail to achieve complete remission after chemotherapy tend to have high levels of Mcl-1.3,51 There are several reports that AKT or ERK1/2 regulate the expression of Mcl-1 in various types of cells.52-57 On the other hand, O'Connor et al reported that the persistence of plasma cells in mice was associated with a BAFF-mediated up-regulation of Mcl-1.58 In the present study, we found that rhBAFF or rhAPRIL, which did not activate AKT or ERK1/2, up-regulated Mcl-1 in CLL B cells. However, saturating amounts of BCMA-Fc or BAFF-R:Fc that could inhibit rhBAFF-induced expression of Mcl-1 failed to block the capacity of NLCs to enhance expression of Mcl-1 in CLL B cells (data not shown), suggesting that NLC-associated factors other than BAFF and APRIL also may induce expression of this antiapoptotic protein in CLL cells. In any case, we found that SDF-1α, which can activate AKT or ERK1/2 in CLL cells, was unable to induce CLL cells to express Mcl-1 (Figure 7). As such, these data suggest that BAFF up-regulates expression of Mcl-1 in CLL B cells via a pathway(s) distinct from that involving activation of MAPK or AKT.
Whereas isolated CLL B cells undergo apoptosis when cultured alone, the addition of rhBAFF, rhAPRIL, and/or SDF-1α to the CLL B cells significantly enhanced their viability (Figure 5), as noted previously.9,11 Nevertheless, the viability of CLL cells cultured with SDF-1α and rhBAFF and/or rhAPRIL still was not as high as that seen when CLL B cells were cultured with NLCs, suggesting that yet additional NLC factors are involved in promoting leukemia cell survival. In this regard, it is noteworthy that Deaglio and colleagues recently found that NLCs also express high levels of CD31 and plexin-B1, which also can contribute in part to the capacity of NLCs to sustain CLL-cell viability.59 Conceivably, strategies that can target one or more of the mechanisms whereby NLCs sustain CLL-cell survival could have therapeutic potential for patients with this disease.
Prepublished online as Blood First Edition Paper, April 28, 2005; DOI 10.1182/blood-2004-03-0889.
Supported in part by National Institutes of Health (NIH) grant PO1-CA81534 for the CLL Research Consortium and by The Leukemia & Lymphoma Society (6217-04; T.J.K.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Dr James R. Feramisco, Arrate Mallabiabarrena, and Monika K. Szeszel (UCSD School of Medicine) for their excellent assistance with immunofluorescence microscopic analysis. We also greatly appreciate the kind gift of 4F-benzoyl-TE14011 from Dr N. Fujii (Graduate School of Pharmaceutical Sciences, Kyoto University, Japan).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal