Abstract
T cells recognizing self-peptides are typically deleted in the thymus by negative selection. It is not known whether T cells against persistent viruses (eg, herpesviruses) are generated by the thymus (de novo) after the onset of the infection. Peptides from such viruses might be considered by the thymus as self-peptides, and T cells specific for these peptides might be deleted (negatively selected). Here we demonstrate in baboons infected with baboon cytomegalovirus and baboon lymphocryptovirus (Epstein-Barr virus–like virus) that after autologous transplantation of yellow fluorescent protein (YFP)–marked hematopoietic cells, YFP+ CD4 T cells against these viruses were generated de novo. Thus the thymus generates CD4 T cells against not only pathogens absent from the host but also pathogens present in the host. This finding provides a strong rationale to improve thymopoiesis in recipients of hematopoietic cell transplants and, perhaps, in other persons lacking de novo–generated CD4 T cells, such as AIDS patients and elderly persons.
Introduction
Patients who have undergone hematopoietic cell transplantation, AIDS patients, patients with congenital thymic hypoplasia, and elderly persons lack naive T cells because of insufficient generation of T cells de novo (thymopoiesis).1-6 Ways to improve de novo generation have recently been studied using prothymopoietic cytokines such as interleukin-7 (IL-7) or keratinocyte growth factor,7-9 thymic transplantation,5 or a thymic organoid (“artificial thymus”).10 It is not known whether improved thymopoiesis might result in more T cells only for pathogens typically absent from the body (eg, neopathogens such as Ebola virus or cleared recall pathogens such as measles virus) or for pathogens or malignant cells present in the body (eg, herpesviruses that infected the host before transplantation or the leukemia that necessitated hematopoietic cell transplantation). The latter T cells may be more important, as suggested by our recent study of 115 recipients of hematopoietic cell transplants (Storek et al11 and J.S., unpublished observations, 2001). Of 158 infections with a known etiologic agent that occurred after engraftment (primarily caused by T-cell deficiency), 67 of 158 (42%) infections were clearly caused by endogenous microorganisms (herpesviruses), and 34 of 158 (22%) infections were likely caused by endogenous microorganisms (digestive tract bacteria and yeasts). Relapse of the underlying hematologic malignancy occurred after transplantation in at least 15% of patients.
Human cytomegalovirus (CMV) and Epstein-Barr virus (EBV) are examples of viruses present in seropositive hosts during posttransplantation immune reconstitution. They cause serious disease in recipients of transplant (CMV pneumonia or gastroenteritis, EBV lymphoma).12,13 Seropositivity is a marker of infection because after primary infection of humans with human CMV or EBV, anti-CMV/EBV antibodies are generated for life. In addition, the virus is always present in the infected human host—CMV in myeloid cells and EBV in B cells and pharyngeal epithelial cells.14 Baboon CMV (bCMV, SA6-like virus) and baboon lymphocryptovirus (bLCV, cercopithecine herpesvirus 12, herpesvirus papio) infections of baboons closely resemble CMV and EBV infections of humans.15-17 As do humans, baboons typically become infected with bCMV and bLCV during childhood.18-20 bLCV can immortalize baboon B cells as human EBV can immortalize human B cells,21 and similar genes are expressed in the immortalized B cells.22 Baboon CMV genes are homologous and colinear with human CMV genes.19 Blewett's team at Oklahoma State University showed that baboons shedding bCMV from 1996 to 1998 still shed the virus in 2002 (E. Blewett, unpublished observations, November 2003, with permission), which is consistent with the lifetime viral persistence documented for human CMV and EBV in infected humans.14 Collectively, bCMV and bLCV infections of baboons are useful models of clinically important persistent viral infections.
To address whether CD4 T cells against persistent viruses can be generated de novo, we searched for YFP-marked CD4 T cells specific for bCMV or bLCV in bCMV/bLCV-seropositive baboons that underwent transplantation with autologous YFP-marked CD34 cells.
Materials and methods
Animals and transplantation
Four juvenile baboons (Papio cynocephalus-anubis), seropositive for bCMV and bLCV before transplantation, underwent irradiation and transplantation with YFP marker gene–transduced autologous CD34 cells, as decribed.23 Briefly, 100 μg/kg daily granulocyte–colony-stimulating factor (G-CSF) and 50 μg/kg daily stem cell factor (SCF) were given subcutaneously from day -8 to day -4 to increase the number of CD34 cells in the marrow. On day -3, bone marrow was harvested into preservative-free heparin from both humeri and femora. CD34 cells were immunomagnetically enriched to 77% to 88% purity using antibody 12.8 and goat anti–mouse immunoglobulin M (IgM)–coated microbeads (Miltenyi, Auburn, CA) and were transduced according to the stem cell transduction protocol (described in the next section). After total body irradiation (10.2 Gy in 2 fractions at 0.07 Gy/min), the stem cells were reinfused on day 0. All animals were housed and cared for at the University of Washington Regional Primate Research Center under conditions approved by the American Association for the Accreditation of Laboratory Animal Care. The study was approved by the University of Washington Animal Care and Use Committee and the Fred Hutchinson Cancer Research Center (FH-CRC) Institutional Review Board.
Stem cell transduction protocol
Transduction of CD34 cells with yellow fluorescent protein (YFP) gene was performed as described.24 Briefly, the magnetically purified CD34 cells from the marrow of G-CSF– and SCF–primed baboons were cultured in Iscove medium with 10% fetal bovine serum and 100 ng/mL each of filgrastim (G-CSF), stem cell factor (SCF), megakaryocyte growth and differentiation factor (MGDF), Flt3 ligand (Flt3L), IL-3, and IL-6 for 48 hours. All the cytokines were kindly donated by Dr Graham Molineux (Amgen, Thousand Oaks, CA). Two 4-hour incubations with viral vector were performed over the subsequent 48 hours using a medium containing Phoenix gibbon ape leukemia virus (GALV)–pseudotyped oncoretrovirus carrying the MNDMFGeYFP transgene (MNDMFG promoter and enhanced YFP gene) in flasks coated with retronectin (Takara, Shiga, Japan). T cells sorted through fluorescence-activated cell sorting (FACS) were subjected to the same treatment to determine whether T cells become transduced with the stem cell transduction protocol.
Antibodies for bCMV and bLCV
The presence of serum antibodies against bCMV and bLCV was tested at the Simian Diagnostic Laboratory (San Antonio, TX) using the rapid dot-immunobinding assay.25 In brief, virus or control antigens were spotted on the nitrocellulose sheet and allowed to absorb for 30 minutes at room temperature. Then the nitrocellulose sheet was submerged in 5% nonfat milk in phosphate-buffered saline (PBS) to block nonspecific binding for 1 hour, followed by washing with PBS-Tween (0.1%). After test sera were applied over the test antigen dots, the nitrocellulose sheet was incubated for 1 hour at 37°C and then washed 3 times in PBS-Tween. Staphylococcal protein A–conjugated horseradish peroxidase was used to detect primate IgG. After incubation and washing, the substrate, 4-chloro-1-naphthol-hydrogen peroxide, was used, and a blue spot was developed as a positive result. Controls consisted of uninfected cell culture antigens and known positive and negative sera.
Intracellular interferon-γ assay for the detection of anti-bCMV/bLCV CD4 T cells
As stimulators, we used either bCMV lysate (containing multiple bCMV antigens) or autologous bLCV-transformed B-lymphoblastoid cells (LBLCs). The bCMV lysate was prepared as follows: MRC5 human lung fibroblast cell line (ATCC CCL-171) was infected with bCMV, strain PGH-01 (ATCC VR-1530) or OCOM4-37 (kindly provided by E. Blewett, Oklahoma State University). After infection had progressed to more than 90% cytopathic effect, the cells were harvested and subjected to multiple freeze-thaw cycles. Then the cells were washed twice in glycine-buffered saline, and the pellets were sonicated on ice. After centrifugation at approximately 600g for 10 minutes, the virus-containing supernatants (from both bCMV strains) were mixed and heat inactivated at 56°C for 30 minutes. Protein concentration was determined with a BCA protein assay kit (Pierce, Rockford, IL). Aliquots of the bCMV lysate were stored in -80°C. Control lysate with the same protein concentration was prepared from uninfected MRC5 cells. LBLCs were obtained by outgrowth of bLCV-containing B cells from peripheral blood mononuclear cells in the presence of 1 μg/mL cyclosporin A (animal M00093) or by infection of blood mononuclear cells with the supernatant of M00093-derived LBLCs (filtered through a 0.45-μm filter) in the presence of 1 μg/mL cyclosporin A (animals M00117, M00227, and J00116).
Peripheral blood mononuclear cells (MNCs), 1.5 × 106, were incubated in 1 mL RPMI 1640 supplemented with 2.05 mM glutamine, 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 10% fetal bovine serum (Hyclone, Logan, UT), anti-CD28 and anti-CD49d antibodies (0.5 μg/mL each; BD Biosciences, San Jose, CA), recombinant baboon IL-7 (5 ng/mL; kindly provided by Dr Michel Morre, Cytheris, Vanves, France), and IL-15 (5 ng/mL; Peprotech, Rocky Hill, NJ) and were stimulated with bCMV lysate (5 μg protein/mL) or autologous LBLCs (3 × 105) overnight in a humidified incubator (5% CO2 and 37°C). Lysate from uninfected MRC5 cells was used as the negative control for bCMV. For bLCV, no negative control was used during cell culture. However, LBLCs without cultured MNCs were permeabilized and stained concurrently to control for nonspecific antibody binding/fluorescence signal from LBLCs. To control for nonspecific stimulation of T cells by LBLCs during cell culture, MNCs from an EBV-seronegative animal were cultured with autologous LBLCs. Brefeldin A (10 μg/mL; Sigma-Aldrich, St Louis, MO) was added 2 hours after starting the culture. After overnight incubation, cells were treated with 1 mM EDTA (ethylenediaminetetraacetic acid) for 10 minutes at room temperature (RT). Then 2.5 mL FACS Lysing solution (BD Biosciences) was added for 10 minutes at RT. After centrifugation, the cells were incubated in 0.5 mL FACS permeabilizing solution (BD Biosciences) for 10 minutes at RT and were washed with flow buffer (PBS with 1% bovine serum albumin [BSA] and 0.1% sodium azide). Cells were stained with anti–interferon (IFN)-γ–phycoerythrin (PE) (clone 45.15; Beckman Coulter [Immunotech], Miami, FL), anti-CD3-PerCPCy5.5 (clone SK34-2; BD Biosciences), anti-CD8 APCCy7 (clone SK1; BD Biosciences), and anti–CD4-Alexa647 (clone 289-14120; Molecular Probes, Eugene, OR) for 30 minutes at RT in darkness. After a final wash with flow buffer, data acquisition was performed on an LSR II cytometer (BD Biosciences). Unless otherwise noted, the number of collected events was between 5 × 105 and 4 × 106 to collect more than 100 IFN-γ+YFP+ CD4 T cells. Data were analyzed using FlowJo software (Treestar, San Carlos, CA). The percentage of virus-specific IFN-γ+ CD4 T cells was determined as percentage IFN-γ+ CD3+CD4+ cells in the stimulated sample minus percentage IFN-γ+ CD3+CD4+ cells in the negative control sample (uninfected MRC5 cell lysate–stimulated MNCs for bCMV and medium-stimulated MNCs for bLCV). The second negative control for bLCV (MNCs from a bLCV-seronegative animal stimulated with autologous EBLCs) showed consistently less than 0.05% IFN-γ+ cells among CD3+CD4+ cells.
Flow cytometric detection of YFP T cells in grafts
Cells (2 × 106 per tube) were incubated with multicolor antibody mixture containing CD10-PE (DAKO, Glostrup, Denmark), CD13-PE (Beckman Coulter [Immunotech]), CD14-PE (Beckman Coulter [Immunotech]), CD16-PE (Beckman Coulter [Immunotech]), CD19-PE (Beckman Coulter [Immunotech]), CD20-PE (BD Biosciences), CD34-PE (BD Biosciences), CD56-PE (Beckman Coulter [Immunotech]), CD3-PECy7 (clone Sp34-2, BD Biosciences), CD4-Alexa647 (clone 289-14120; Molecular Probes, Eugene, OR), and CD8 APCCy7 (clone SK1; BD Biosciences). After incubation for 30 minutes at 4°C, cells were washed and resuspended in 150 μL Accumax (Innovative Cell Technologies, San Diego, CA) and were incubated at 37°C for 30 minutes. Flow buffer and 1 μg/mL DAPI (Molecular Probes) were added, and 106 events were acquired on BD LSR II. CD4 T cells were defined as CD3+CD4+ lymphocytes that were negative for DAPI, CD10, CD13, CD14, CD16, CD19, CD20, CD34, or CD56.
Detection of bCMV and bLCV in sera by PCR
DNA was extracted from 200 μL serum using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA). The following primers were used: 5′-TACGTCATTGGTACCCTCC-3′ (forward) and 5′-GGAGTACTGCCAATGTACTA-3′ (reverse) for bCMV,15 and 5′-GCAGGAGTCTGCACTCCCTG-3′ (forward) and 5′-CTGGGACTACGTGGCCTCTT-3′ (reverse) for bLCV.21 Each reaction contained 2.5 U platinum-Taq polymerase (Invitrogen, Carlsbad, CA), 2.5 mM MgCl2, 500 μM dNTP, 500 nM each primer, 20 μL extracted DNA (of total 100 μL yielded by the QIAamp kit) in a total volume of 50 μL. The conditions of each reaction were: 1 cycle at 95°C for 9 minute; 40 cycles at 94°C for 60 seconds, 58°C for 60 seconds, and 72°C for 60 seconds; and 1 cycle at 72°C for 9 minute. Polymerase chain reaction (PCR) products were separated on 2% agarose gel and visualized by ethidium bromide staining. As a positive control for bCMV and a negative control for bLCV, we used DNA extracted from the bCMV lysate (derived from bCMV-infected MRC5 cells). As a positive control for bLCV and a negative control for bCMV, we used DNA extracted from LBLCs obtained by the outgrowth of bLCV-containing B cells from animal M00093.
Detection of bCMV and bLCV in thymi by immunohistochemistry
Thymic tissue of animals M00117, M00227, and F00262 (another bCMV- and bLCV-seropositive baboon undergoing autologous CD34 cell transplantation) was obtained by biopsy under general anesthesia at 3 months after transplantation and by necropsy at 6 months after transplantation. (Thymic tissue of animals M00093 and J00116 was unavailable.) The tissue was fixed in buffered formalin and embedded in paraffin. Baboon CMV was detected using 2 monoclonal antibodies, 35C7 (1:1000) and 37E12.1D1 (1:10), generated against bCMV-infected cell proteins by Dr E. Blewett, as described.26 Baboon LCV was detected using a monoclonal antibody directed against the BZLF1 protein of EBV (1:50, M7005, clone BZ; DAKO Cytomation, Carpinteria, CA).1 To ensure that the tissue could be immunostained, sections were also stained for vimentin (1:100, M7020, clone Vim 3B4; DAKO Cytomation). Control tissue was made by fixing cell pellets of bCMV-infected MRC5 cells, noninfected MRC5 cells, and bLCV-infected LBLCs with formalin and embedding the cells in paraffin. Formalin-fixed, paraffin-embedded tissue was sectioned at 5 μm and was deparaffinized. Tissue stained for bCMV and vimentin was treated with citrate buffer pH 6.0 (Target Retrieval Solution; DAKO Cytomation) under steam for 20 minutes, then allowed to cool in buffer for 25 minutes. Tissue stained for bLCV was treated with proteinase K (DAKO Cytomation) at RT for 10 minutes. Endogenous peroxidase activity was blocked by incubating the sections in 3% hydrogen peroxide (Medi-Pak; McKesson, Richmond, VA) at RT for 8 minutes. Protein blocking was performed at RT for 10 minutes with 15% normal swine serum and 5% normal human serum (Jackson ImmunoResearch, Bar Harbor, ME) with 1% BSA in TBS with Tween (Wash Buffer; DAKO Cytomation). Primary antibodies were applied to the tissue and incubated at RT for 30 minutes. Immunoglobulin concentration–matched isotype controls were included for each antibody. Bound antibody was detected using EnVision+ HRP for mouse antibodies and dimethylaminoazobenzene (DAB) (DAKO Cytomation). Sections were counterstained with hematoxylin (DAKO Cytomation). Baboon CMV– and bLCV–infected cell pellets demonstrated positive staining for the bCMV and bLCV antibodies, respectively. Noninfected cell pellets and isotype controls showed no staining. All cell pellets and thymus tissue had positive vimentin staining.
Results
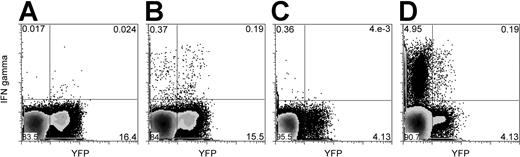
We detected de novo–generated (YFP+) and functional (IFN-γ–producing) virus-specific CD4 T cells in 4 of 4 baboons for bCMV and in 3 of 4 baboons for bLCV (Table 1; Figure 1). These cells were likely generated by the thymus because an extrathymic pathway for T-cell development from hematopoietic cells, though potentially existing for CD8 T cells, does not result in the generation of functional CD4 T cells.27-29
Percentages of YFP+ bCMV/bLCV-specific CD4 T cells of total YFP+ CD4 T cells and, for comparison, percentages of YFP- bCMV/bLCV-specific CD4 T cells of total YFP- CD4 T cells
. | bCMV-specific CD4 T cells, % . | . | bLCV-specific CD4 T cells, % . | . | Absolute YFP+ and YFP- CD4 T cell counts, ×106/L . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Animal and time after transplantation, mo . | YFP+ . | YFP- . | YFP+ . | YFP- . | YFP+ . | YFP- . | |||
| M00117 | |||||||||
| 6 | 2.55 | 1.82 | 0.89 | 0.68 | 83 | 941 | |||
| M00227 | |||||||||
| 6 | 0.21 | 8.45 | 1.81 | 0.27 | 56 | 586 | |||
| J00116 | |||||||||
| 16 | 3.03 | 3.18 | ND | ND | 82 | 1618 | |||
| M00093 | |||||||||
| 3 | 0.75 | 0.02 | ND | ND | 137 | 532 | |||
| 5 | 0.34 | 1.05 | 1.19 | 0.43 | 143 | 334 | |||
| 6 | 0.47 | 2.84 | 1.56 | 0.75 | 192 | 578 | |||
| 15 | 1.01 | 2.11 | 0.29 | 0.34 | 659 | 1466 | |||
. | bCMV-specific CD4 T cells, % . | . | bLCV-specific CD4 T cells, % . | . | Absolute YFP+ and YFP- CD4 T cell counts, ×106/L . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Animal and time after transplantation, mo . | YFP+ . | YFP- . | YFP+ . | YFP- . | YFP+ . | YFP- . | |||
| M00117 | |||||||||
| 6 | 2.55 | 1.82 | 0.89 | 0.68 | 83 | 941 | |||
| M00227 | |||||||||
| 6 | 0.21 | 8.45 | 1.81 | 0.27 | 56 | 586 | |||
| J00116 | |||||||||
| 16 | 3.03 | 3.18 | ND | ND | 82 | 1618 | |||
| M00093 | |||||||||
| 3 | 0.75 | 0.02 | ND | ND | 137 | 532 | |||
| 5 | 0.34 | 1.05 | 1.19 | 0.43 | 143 | 334 | |||
| 6 | 0.47 | 2.84 | 1.56 | 0.75 | 192 | 578 | |||
| 15 | 1.01 | 2.11 | 0.29 | 0.34 | 659 | 1466 | |||
Percentages shown are averages of 2 independent experiments, except for EBV-specific cells in M00117, which were determined only once. Percentages were determined as percentage of IFN-γ+ YFP+ (YFP-) CD3+CD4+ cells in the stimulated sample minus percentage of IFN-γ+ YFP+ (YFP-) CD3+CD4+ cells in the negative control sample. ND indicates none detected.
Baboon LCV- and bCMV-specific (IFN-γ–producing) CD4 T cells include YFP+ cells originating from grafted hematopoietic cells. Blood mononuclear cells were stimulated with autologous bLCV-transformed B cells (B) or bCMV lysate (D). As controls, the mononuclear cells were stimulated with medium only (A) or with uninfected MRC5 cell lysate (C). In this example, cells from animal M00093 were stimulated with medium only or bLCV (A-B), and cells from animal J00116 were stimulated with MRC5 or bCMV lysate (C-D). Dot plots shown here were gated on CD3+CD4+ lymphocytes. The same number of dots is displayed for the stimulated condition and the control condition. Numbers displayed represent percentages of cells in respective quadrants.
Baboon LCV- and bCMV-specific (IFN-γ–producing) CD4 T cells include YFP+ cells originating from grafted hematopoietic cells. Blood mononuclear cells were stimulated with autologous bLCV-transformed B cells (B) or bCMV lysate (D). As controls, the mononuclear cells were stimulated with medium only (A) or with uninfected MRC5 cell lysate (C). In this example, cells from animal M00093 were stimulated with medium only or bLCV (A-B), and cells from animal J00116 were stimulated with MRC5 or bCMV lysate (C-D). Dot plots shown here were gated on CD3+CD4+ lymphocytes. The same number of dots is displayed for the stimulated condition and the control condition. Numbers displayed represent percentages of cells in respective quadrants.
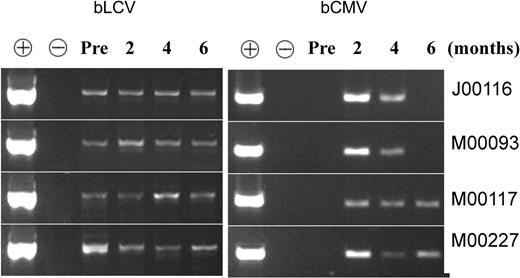
The presence of bCMV and bLCV in the animals during immune reconstitution was documented by PCR. The viruses were detectable in the serum on at least 2 of 3 determinations between 2 and 6 months after transplantation (Figure 2).
Both bLCV and bCMV were present in all 4 animals during immune reconstitution. Serum was obtained before transplantation (“Pre”) and at approximately 2, 4, and 6 months after transplantation and were subjected to PCR, as described in “Materials and methods.” DNA extracted from bCMV lysate (derived from bCMV-infected MRC5 cells) was used as a positive control (+) for bCMV and as a negative control (-) for bLCV. DNA extracted from bLCV-transformed B lymphoblastoid cells was used as a positive control for bLCV and a negative control for bCMV. The fact that bLCV was detected in all 4 animals before transplantation is consistent with the observation of Dittmer et al (D. Dittmer, unpublished information, April 2004, with permission), who detected bLCV in the blood of 71 of 90 normal seropositive baboons. The fact that bCMV was undetectable before transplantation and detectable at 2 to 4 months after transplantation is consistent with the expected viral reactivation, leading to increased viral load early after transplantation.
Both bLCV and bCMV were present in all 4 animals during immune reconstitution. Serum was obtained before transplantation (“Pre”) and at approximately 2, 4, and 6 months after transplantation and were subjected to PCR, as described in “Materials and methods.” DNA extracted from bCMV lysate (derived from bCMV-infected MRC5 cells) was used as a positive control (+) for bCMV and as a negative control (-) for bLCV. DNA extracted from bLCV-transformed B lymphoblastoid cells was used as a positive control for bLCV and a negative control for bCMV. The fact that bLCV was detected in all 4 animals before transplantation is consistent with the observation of Dittmer et al (D. Dittmer, unpublished information, April 2004, with permission), who detected bLCV in the blood of 71 of 90 normal seropositive baboons. The fact that bCMV was undetectable before transplantation and detectable at 2 to 4 months after transplantation is consistent with the expected viral reactivation, leading to increased viral load early after transplantation.
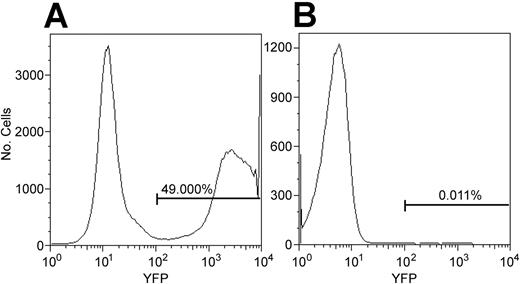
The conclusion that anti-bCMV/bLCV CD4 T cells were generated de novo could be incorrect if anti-bCMV/bLCV CD4 T cells contaminating the CD34 cell graft were YFP marked. To address this possibility, we determined whether CD4 T cells might become YFP marked during 3-day CD34 cell transduction—2 days of culturing the cells with G-CSF, SCF, MGDF, Flt3L, IL-3, and IL-6 and 1 day of exposing them to the YFP gene–containing oncoretrovirus. Seven aliquots of magnetically enriched marrow CD34 cells from the 4 animals studied were marked with YFP using the stem cell transduction protocol (see “Materials and methods”). By flow cytometry, the percentage of YFP+ T cells among all viable cells at 1, 3, 8, and 14 days after the last exposure to the oncoretrovirus was not greater than background (ie, not more than 0.05%) (data not shown). However, only approximately 1% of cells were T cells, suggesting that we would have detected YFP+ T cells only if more than 5% T cells contaminating the graft expressed YFP. To increase the sensitivity of the determination of whether T cells may be marked using the stem cell transduction protocol, we used blood mononuclear cells, enriched them for CD3+ cells by FACS sorting, cultured them with G-CSF, Flt3L, MGDF, IL-3, IL-6, and SCF, and exposed them to the oncoretrovirus per the stem cell transduction protocol. CD3+ cells from the blood of the same animal that were cultured with the same cytokines and were not exposed to the oncoretrovirus served as negative control. Bone marrow CD34 cells that were cultured with the same cytokines and exposed to the same oncoretrovirus were used as positive control. YFP expression was checked by flow cytometry 24 hours after the last exposure to the oncoretrovirus because, in our experience, all transduced hematopoietic cells become YFP+ by flow cytometry by 24 hours after the last exposure to the virus. In 4 of 4 experiments, the percentage of YFP+ cells among T cells was not greater than the detection limit established using the negative controls (ie, not more than 0.05%) (Figure 3). Theoretically, the transgene might not be expressed in vitro but could become expressed in vivo after transplantation. Thus, it was important to determine whether unexpressed transgene might be present in CD4 T cells subjected to the stem cell transduction protocol. Using real-time PCR that can detect 1 transgene copy in 10 000 cells,30 in 2 experiments we found no YFP gene among 100 000 FACS-sorted CD3+CD4+ cells of a normal baboon that were treated per the stem cell transduction protocol. This was expected because oncoretroviruses can mark only proliferating cells,31 and none of the 6 cytokines we used to induce proliferation of CD34 cells is known to induce the proliferation of T cells.32-37 Thus, the YFP+ CD4 T cells detected after transplantation originated from grafted hematopoietic cells and not from T cells contaminating the graft.
CD34 cells, but not CD4 T cells, were transduced using the stem cell transduction protocol. The stem cell transduction protocol is described in “Materials and methods.” Cultured CD34 cells were exposed to the YFP transgene-containing oncoretrovirus (A). In parallel, cultured T cells were exposed to the same oncoretrovirus (B). Flow cytometry was performed 24 hours after the last exposure to the oncoretrovirus. The histogram on the right is gated on CD3+CD4+ lymphocytes. In 4 of 4 experiments, the percentage of YFP+ CD4 T cells was not greater than our detection limit of 0.05%, determined using negative controls (not shown).
CD34 cells, but not CD4 T cells, were transduced using the stem cell transduction protocol. The stem cell transduction protocol is described in “Materials and methods.” Cultured CD34 cells were exposed to the YFP transgene-containing oncoretrovirus (A). In parallel, cultured T cells were exposed to the same oncoretrovirus (B). Flow cytometry was performed 24 hours after the last exposure to the oncoretrovirus. The histogram on the right is gated on CD3+CD4+ lymphocytes. In 4 of 4 experiments, the percentage of YFP+ CD4 T cells was not greater than our detection limit of 0.05%, determined using negative controls (not shown).
Discussion
The reason de novo–generated anti-bCMV/bLCV CD4 T cells were not intrathymically deleted remains to be elucidated. Possibly, the viruses or the virus-infected cells might not have entered the thymopoietic tissue because of the putative blood–thymus barrier.38 Alternatively, the amount of the virus or viral peptides entering the thymopoietic tissue might have been too small to trigger negative selection—human CMV is harbored by 1 of 104 to 105 CD33+ cells, and human EBV is harbored by 1 of 104 to 106 B cells.14 Possibly, the cells that harbored the virus and circulated through the thymus expressed only a limited number of viral antigens (only antigens expressed during latency). Thus, T cells against peptides expressed during latency might have been deleted in the thymus, whereas T cells against peptides expressed during lytic infection and detected by the intracellular IFN-γ assay might not have been deleted. By immunohistology, thymi from animals M00117, M00227, and F00262 at 3 and 6 months after transplantation were negative for bCMV and bLCV proteins expressed during lytic infection (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article).
The generation of antiviral T cells de novo after hematopoietic cell transplantation is consistent with clinical observations. Anti-CMV CD8 T cells developed in 6 of 7 CMV-seropositive patients receiving T cell–depleted grafts from CMV-seronegative donors.39,40 The T cells were of donor origin (documented in 3 patients). Although the T cells might have been derived from donor naive T cells remaining in the graft, it is possible that they were generated after transplantation from donor hematopoietic progenitors. In agreement with that, in 2 of 4 recipients of CMV-seropositive transplants from CMV-seropositive donors, new anti-CMV CD8 T-cell clones (that had been undetectable in the donors) were detected in the recipients.40 Similar indirect clinical information on de novo generation of antiviral CD4 T cells has not been published.
The finding that hematopoietic cell–derived, herpesvirus-specific CD4 T cells can be generated de novo provides a rationale to improve thymopoiesis of patients recovering from lymphopenia and, perhaps, of patients with chronic infections (hepatitis C, HIV) or of elderly persons.
Prepublished online as Blood First Edition Paper, October 12, 2004; DOI 10.1182/blood-2004-01-0348.
T.K. and H.L. contributed equally to this study.
Supported by grants HL69710, AI46108, CA18221, HL53750, NCRR00166, DK56465, and DK47754 from National Institutes of Health and by Research Project No. 00000064203 from the Ministry of Health, Czech Republic.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Bobbie M. Thomasson, Julie Morris, Roxanne Velez, and Renee Kendall for excellent technical assistance. We appreciate the valuable input of Dr Marian G. Michaels (Children's Hospital of Pittsburgh) on bCMV detection by PCR. The study could not have been completed without the dedicated work of the staff of the University of Washington National Primate Research Center, in particular Michael Gough, Leslie Falch, Ray Angeles, Sarah Kirkbride, Ed Novak, Dr Judy Johnson, Dr David Anderson, Dr Steven Kelley, and Dr Maggie Gillen. We also thank Amgen and Cytheris for providing most of the cytokines used. Immunohistochemistry was performed at the FHCRC Experimental Histopathology Shared Resource.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal