Abstract
Peptides containing the Arg-Gly-Asp (RGD) motif inhibit cell adhesion and exhibit a variety of other biologic effects including anticoagulant and antimetastatic activities. The aim of the present study was to examine the anchorage-independent effects of an RGD-containing peptide, Arg-Gly-Asp-Ser (RGDS), on human umbilical vein endothelial cells (HUVECs). Assays were performed on HUVECs seeded onto collagen IV; under these experimental conditions RGDS did not exert antiadhesive effects but significantly reduced FGF-2-dependent chemotaxis after 4 hours of treatment and reduced proliferation after 24 hours of treatment. Experiments carried out with caspase-specific inhibitors indicated that the observed antichemotactic effects required caspase 8 and caspase 9 activation. RGDS activated both caspase 8 and caspase 9 after 4 hours of treatment and caspase 3 after 24 hours of treatment, and markedly enhanced HUVEC apoptosis by transferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL)/Hoechst staining and fluorescence-activated cell sorting (FACS) analysis. Finally, confocal microscopy showed that RGDS localizes in the cytoplasm of live HUVECs within 4 hours and in vitro experiments showed that RGDS directly interacts with recombinant caspases 8 and 9 in a specific way. In summary, these results indicate that RGDS directly binds and activates caspases 8 and 9, inhibits chemotaxis, and induces apoptosis of HUVECs with a mechanism independent from its antiadhesive effect.
Introduction
Integrins mediate cell-cell and cell-extracellular matrix (ECM) interactions1,2 and recognize several ECM proteins including fibronectin, vitronectin, laminin, fibrinogen, von Willebrand factor, osteopontin, thrombospondin, and collagen. Endothelial cell adhesion to ECM is mediated by α1β1, α2β1, α3β1, α5β1, α6β1, αvβ3, and αvβ5 integrins3 and such interaction triggers mechanical as well as chemical signals,4,5 which promote the assembly of actin stress fibers6 and modulate adherent cell growth and survival.2,7
One of the major sites mediating integrin action is the Arg-Gly-Asp (RGD) motif, present in ECM proteins and in other proteins such as disintegrins.8,9 Integrins interact with ECM in both an RGD-dependent and RGD-independent manner.10,11 Peptides containing the RGD motif have been shown to inhibit cell adhesion by competing for integrins/matrix interaction; further, these peptides exert a variety of biologic effects including anti-inflammatory, anticoagulant, and antimetastatic activities.12-16 RGD-containing peptides and disintegrins have an antiangiogenic effect due at least in part to inhibition of αvβ3- and αvβ5-dependent adhesion,8,9 and prevent vascular lumen formation.17 RGD peptidomimetics have also been shown to inhibit neointima formation and lumen restenosis following coronary artery injury.18,19
Previous studies indicated that integrins regulate cell viability in different cell types; anti-integrins monoclonal antibody or RGD peptides reduce cell growth by inhibiting integrin ligation, therefore detaching cells from ECM and inducing an anchorage-dependent apoptosis, named anoikis.20-28 Hence, integrin-mediated apoptosis has been attributed to the onset of an antiadhesive effect; in fact, anchorage loss of adherent cells is known to trigger different proapoptotic signals, either by altering integrin expression,29-31 blocking integrin signaling,32-35 or inducing integrin degradation.36,37
Recent evidence shows that RGD-containing peptides may induce an anchorage-independent apoptosis of T cells and cardiomyocytes, by entering into the cell and directly promoting caspase 3 activation.38-40 We focused our interest on the anchorage-independent effects of the RGD motif, which are still poorly investigated. We set experimental conditions to avoid any RGDS antiadhesive action and examined the effect on HUVEC chemotaxis and on the activity of initiator caspases, namely caspases 8 and 9.
Materials and methods
Materials
Arg-Gly-Asp-Ser (RGDS) peptide was obtained from Bachem (Bubendorf, Switzerland). Trypsin-EDTA (ethylenediaminetetraacetic acid), basic fibroblast growth factor (FGF-2), human plasma fibronectin, phosphate-buffered saline (PBS), sodium pyruvate, minimum essential medium (MEM) amino acids without l-glutamine, l-glutamine, penicillin-streptomycin, MEM nonessential amino acids, HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), and RPMI medium were from Gibco (Paisley, Scotland). Human plasma vitronectin was obtained from Calbiochem (San Diego, CA). Mouse collagen IV was from Becton Dickinson (Bedford, MA). Bovin serum albumin (BSA), Arg-Gly-Glu-Ser (RGES) peptide, protease inhibitors, and the fluorescein isothiocyanate (FITC)-conjugated phalloidin were from Sigma (St Louis, MO). General caspase inhibitor (Z-VAD-FMK), caspase 1 inhibitor (Z-WEHD-FMK), caspase 3 inhibitor (Z-DEVD-FMK), caspase 8 inhibitor (Z-IEDT-FMK), and caspase 9 inhibitor (Z-LEHD-FMK) were from R&D Systems (Minneapolis, MN) and Calbiochem. Caspase substrates FITC-VAD-FMK and Z-DEVD-R110 were from Promega (Southampton, United Kingdom). Substrates for caspases 1, 8, and 9 (DEVD-AFC, AEVD-AFC, and IETD-AFC, respectively) were from Calbiochem. Polyclonal rabbit antihuman caspase 3 antibody was from Santa Cruz Biotechnology (Alexa, CA), antihuman caspase 8 antibody was from Zymed Laboratories (San Francesco, CA), and antihuman caspase 9 antibody was from Pharmingen (San Diego, CA). Amino terminal-biotinylated RGDS was purchased from Sigma-Genosys (Cambridge, United Kingdom). Anti-αvβ3 (clone LM609), anti-αvβ5 (clone P1F6), anti-α2β1 (clone BHA2.1) blocking antibodies and human caspases 8 and 9 active recombinant proteins were purchased from Chemicon International (Temecula, CA).
Cell culture
Human umbilical vein endothelial cells (HUVECs; Clonetics, Walkersville, MD) were maintained in EBM-2 medium (Clonetics) supplemented with endothelial growth medium 2 (EGM-2) kit (Clonetics) containing fetal calf serum (FCS; 2%), hydrocortisone (0.04%), human fibroblast growth factor B (hFGF-B; 0.4%), vascular endothelial growth factor (VEGF; 0.1%), R3-insulin-like growth factor 1 (R3-IGF-1; 0.1%), ascorbic acid (0.1%), human epidermal growth factor (hEGF; 0.1%), gentamicin sulfate amphotericin-B (GA-1000; 0.1%), heparin (0.1%) (complete medium), according to manufacturer's instructions. For all experiments, cells were used up to the sixth passage and harvested at 80% confluence. For all experiments, cells were seeded for 24 hours, then starved overnight and used.
Cell adhesion assay
Cell adhesion was quantified as reported.11 Ninety-six-well tissue-culture plates were coated overnight at 4°C with ECM proteins (fibronectin, vitronectin, or collagen IV, diluted in PBS, pH 7.4, to the final concentration of 50 μg/mL). Wells were then blocked for 1 hour at 37°C with 5 mg/mL BSA in EBM-2 medium and washed 3 times with PBS, pH 7.4, before cell seeding.
HUVECs harvested by trypsin-EDTA were suspended in EBM-2 medium plus FCS 10% with serial dilutions of either peptide (RGDS or RGES) ranging from 500 μg/mL to 62.5 μg/mL, corresponding to 1.1 mM to 0.1 mM. Cells were incubated with peptides at 37°C for 15 minutes before seeding, then 15 000 cells per well were added, and the plate was incubated at 37°C for one hour. Nonadherent cells were discarded by washing 3 times, then adherent cells were fixed with 4% formaldehyde in PBS, pH 7.4, for 10 minutes at room temperature (RT) and stained with 0.5% toluidine blue (Merck KgaA, Darmstadt, Germany) in 4% formaldehyde for 10 minutes at RT. Plates were then rinsed extensively with water and stain was extracted by incubation with sodium dodecyl sulfate (SDS) 1% in PBS, pH 7.4, for 30 minutes at RT. Cell adhesion was then quantified as optical density (OD) at 595 nm.
Immunofluorescence
HUVECs were seeded on coverslips coated with poly-l-lysine (PLL), vitronectin, or collagen IV (50 μg/mL in PBS, pH 7.4) and incubated at 37°C for 24 hours; then complete medium was replaced with unsupplemented medium (EBM-2) overnight at 37°C. After treatment with RGDS in EBM-2 0.1% BSA for 4 hours, cells were fixed with 3% paraformaldehyde in PBS, pH 7.4, for 10 minutes, permeabilized with 0.1% Triton X-100 in PBS, pH 7.4, for 5 minutes at RT, and blocked for 30 minutes with BSA 2% in PBS, pH 7.4, at RT.41 Cells were stained with FITC-conjugated phalloidin (1:1000) in PBS, pH 7.4, containing BSA 1% for one hour at RT. Coverslips were mounted with a fluorescent mounting medium (Dako, Carpinteria, CA) and cells were visualized using a Zeiss Axioplan fluorescence microscope (Zeiss, Heidelberg, Germany). Images were acquired with a Zeiss Axiocam digital camera (Zeiss).
Proliferation assay
HUVEC proliferation was evaluated as previously reported.42 Briefly, HUVECs (8 × 104 cells/well) in EBM-2 medium supplemented with EGM-2 kit were allowed to grow onto 6-well plates coated with collagen IV (50 μg/mL) (Becton Dickinson, Bedford, MA) for 24 hours at 37°C in 5% CO2. Medium was then replaced with EBM-2 overnight. Subsequently, cells were treated with serial dilutions of RGDS (from 500 μg/mL to 125 μg/mL) dissolved in EBM-2 medium plus BSA 0.1% containing basic fibroblast growth factor (FGF-2; 10 ng/mL) for 24 hours at 37°C. Then, cells were harvested by trypsin-EDTA and counted using a hemacytometer.
Cell migration assay
FGF-2-induced HUVEC migration was measured in modified Boyden chambers as previously reported.43 Briefly, 8-μm pore-size polycarbonate filters (Costar, Cambridge, MA) were coated with murine collagen type IV (10 μg/mL) for one hour. HUVECs were incubated for 48 hours in RPMI 15% FBS, 1% sodium pyruvate, 2% MEM amino acids without l-glutamine, 1% l-glutamine, 1% penicillin-streptomycin, and 1% MEM nonessential amino acids. Cells were harvested by trypsinization, resuspended in RPMI 25 mM HEPES, 0.1% BSA, and 200 μL was added to the upper portion of the chambers at 1 × 106 cells/mL with serial dilutions of RGDS (from 500 μg/mL to 125 μg/mL); the lower portion of the Boyden chamber contained FGF-2 (10 ng/mL) as a chemoattractant. After 4 hours at 37°C in 5% CO2, cells were fixed in 95% ethanol and stained with 0.5% toluidine blue in PBS, pH 7.4, for 10 minutes. The number of migrated cells was evaluated by counting 15 fields at × 400 magnification. In other experiments, cells were pretreated for 20 minutes at 37°C with caspase inhibitors, namely Z-VAD-FMK (general caspase inhibitor), Z-DEVD-FMK (caspase 3 inhibitor), Z-WEHD-FMK (caspase 1 inhibitor), Z-IEDT-FMK (caspase 8 inhibitor), and Z-LEHD-FMK (caspase 9 inhibitor) (50 μM) before treatment with RGDS.
In additional experiments, HUVECs were preincubated with RGDS (500 μg/mL, 20 minutes, 37°C); then cells were washed with 2 M NaCl, 20 mM HEPES, pH 7.4, and twice with 2 M NaCl, 20 mM acetic acid, pH 7.4, to wash out RGDS free in the medium or bound to the membranes.44 Cells treated in this way were then used for migration on collagen IV-coated filters as reported above.
Caspase activity assay
Caspase activity was measured as follows: HUVECs seeded on collagen IV were incubated at 37°C for 24 hours; then medium was replaced with EBM-2 overnight before treatment with RGDS (500 μg/mL in EBM-2, 0.1% BSA for 4 hours). Before harvesting by trypsinization, cells were incubated for 20 minutes at 37°C with the fluorescent generic caspase substrate (FITC-VAD-FMK; 10 μM). Subsequently, cells were washed twice with PBS, pH 7.4, and fixed with 2% paraformaldehyde in PBS, pH 7.4, for 30 minutes at 4°C. Cell fluorescence was then measured on a Coulter Epics XL flow cytometer (Beckman Coulter), with automatic gating. Data were analyzed with WinMDI software (Scripps Research Institute, La Jolla, CA).
Caspase 3 and 7 enzymatic activity was measured by monitoring the fluorescence of the Z-DEVD-R110 substrate according to the manufacturer's instructions. HUVECs seeded on collagen IV were treated as described above and incubated with RGDS 500 μg/mL in EBM-2 0.1% BSA for 4 hours and 24 hours at 37°C. The substrate was then added and cells were harvested by trypsinization and resuspended in PBS, pH 7.4. Fluorescence was then measured (485-nm excitation wavelength and 530-nm emission wavelength) and expressed as fold increase versus control.
Caspase 1, caspase 8, and caspase 9 activity was measured as previously reported.45,46 Briefly, cells seeded on collagen IV were incubated in the presence or absence of RGDS (as reported in “Cell culture”), cells were then harvested with trypsin, washed, and resuspended in caspase buffer (100 μL; 20 mM HEPES pH 7.6, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA [ethylene glycol tetraacetic acid], 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 50 μM of the fluorogenic substrate for caspases 1, 8, and 9 (DEVD-AFC [DEVD-7-amino-4trifluoromethylcoumarin], AEVD-AFC, and IETD-AFC, respectively). The suspension was then passed through a 21.5-gauge needle, on ice for 15 minutes, and centrifuged at 25 000g for 15 minutes. The supernatant was collected and centrifuged at 100 000g at 4°C for 75 minutes. Protein concentration in the supernatant was determined by the micro-bicinchoninic acid method (micro-BCA) method (Pierce, Tattenhall, Cheshire, United Kingdom). Caspase activity was allowed to proceed for 2 hours at 37°C. Under these conditions, caspase activity is proportional to the release of AFC from the substrate; fluorescence (emission at 505 nm and excitation at 400 nm) was then measured with a Perkin-Elmer LS50 fluorometer (Perkin-Elmer Instruments, Beaconsfield, United Kingdom). Caspase activation was then reported as fold increase versus control.
Caspase activation by Western blotting
Activation of caspases 3, 8, and 9 was also followed as function of procaspase cleaveage.38 HUVECs were incubated at 37°C for 24 hours, then medium was replaced with EBM-2 overnight at 37°C, cells were then treated with RGDS 500 μg/mL in EBM-2 0.1% BSA for 4 hours (caspase 3 activation was analyzed at 4, 16, and 24 hours) at 37°C. Cells were washed in PBS, pH 7.4, and lysed in buffer containing 150 mM NaCl, 25 mM HEPES, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EGTA, EDTA, and PMSF, 10 μg/mL aprotinin, leupeptin, and pepstatin. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. Membranes were then blocked with 5% milk (Bio-Rad Laboratories, Hercules, CA) in TPBS (0.1% Tween 20 in PBS, pH 7.4). After washing 3 times with TPBS, membranes were incubated with antibody to caspase 3 (1:200), caspase 8 (1:250), and caspase 9 (1:1000) for one hour to visualize both pro-caspase and cleaved-caspase bands. For detection, secondary antibody goat antirabbit conjugate to horseradish peroxidase (Pierce) was used, 1:5000 in 2% BSA in TPBS, followed by chemiluminescence (ECL; Amersham, Buckinghamshire, United Kingdom) and autoradiography. Bands were quantified using a calibrated imaging densitometer (GS 710; Bio-Rad) and analyzed using “Quantity one” software (Bio-Rad).
Apoptosis assays
HUVECs (5 × 104) in EBM-2 medium supplemented with EGM-2 kit were seeded onto coverslips coated with collagen IV as reported above. After 24 hours at 37°C, medium was replaced with EBM-2 overnight and then cells were incubated with RGDS 500 μg/mL diluted in EBM-2, 0.1% BSA for 24 hours. Then cells were fixed with 3% paraformaldehyde for 10 minutes at RT, permeabilized with 0.1% Triton X-100, 0.1% sodium citrate for 2 minutes on ice, washed twice with PBS, pH 7.4, and incubated for one hour at 37°C in the dark with a TUNEL reaction mixture (Boehringer Mannheim, Indianapolis, IN), for in situ detection of cell death. After washing twice with PBS, pH 7.4, cells were incubated at RT with the Hoechst solution for 5 minutes. All Hoechst-positive nuclei as well as TUNEL-positive nuclei were visualized using a Zeiss Axioplan fluorescence microscope. Then, apoptosis was expressed as a percent of fragmented Hoechst-positive nuclei versus total Hoechst-positive nuclei, and as a percent of TUNEL-positive nuclei versus total Hoechst-positive nuclei and was reported as fold increase versus control.
To further analyze the cell death-inducing effect of RGDS, FACS analysis was carried out on HUVECs grown as reported in the previous paragraph and treated with FGF-2 (10 ng/mL) or BSA alone in the presence or absence of RGDS (500 μg/mL). In additional experiments, RGDS-treated HUVECs were pretreated for 20 minutes with blocking antibodies anti-αvβ3 (10 μg/mL), anti-αvβ5 (0.1 μg/mL), or anti-α2β1 (10 μg/mL) according to manufacturer's instructions. Then, propidium iodide (PI) staining (1 μg/mL; Sigma Chemical) was performed according to standard protocols on ethanol-fixed cells. Cell cycle analysis was performed within 72 hours of the staining, on a Coulter Epics XL flow cytometer (Beckman Coulter). Data were then analyzed with WinMDI software.
Confocal microscopy
HUVECs seeded on coverslips and grown as described in “Proliferation assay” were treated for 4 hours with biotinylated RGDS (500 μg/mL) at 4°C and 37°C. Then, cells were fixed and permeabilized as described in the immunofluorescence paragraph, and blocked for 30 minutes with 10% goat serum in PBS, followed by incubation with fluorescein avidin (1:40; Vector Laboratories, Peterborough, United Kingdom) in PBS, pH 7.4, for one hour at RT. After washing in 0.3% Triton X-100 in PBS, HUVECs were incubated with PI at a final concentration of 5 μg/mL to visualize nuclei. Cells were then analyzed using a Zeiss LSM 510 meta confocal microscope (Zeiss). Laser power, beam splitters, filter settings, pinhole diameters, and scan mode were the same for all examined samples. Fields reported in Figure 7A are representative of all examined fields.
RGDS cell internalization and molecular interaction. (A) Biotinylated RGDS (500 μg/mL) penetration in live HUVECs was examined after 4 hours of incubation at 4°C and 37°C. Biotinylated RGDS internalized is shown as green stain (fluorescein-avidin staining), and nuclei are shown as red stain (PI staining, 5 μg/mL), by confocal microscopy (original magnification × 110). A short incubation (4 hours) was sufficient for RGDS to enter cells at 37°C incubation, whereas it did not penetrate at 4°C incubation. (B) Overlay assays were carried out with HUVEC membranes and cytoplasmic extracts immobilized onto nitrocellulose and incubated with biotinylated RGDS. Total binding was shown in the presence of labeled RGDS, whereas nonspecific binding was shown in the presence of an excess of unlabeled RGDS. This experiment was performed 3 times with similar results. A representative experiment is shown. (C) The binding was quantified by densitometry; the nonspecific binding was subtracted from the total binding, giving the values reported as specific binding. Data are expressed as mean ± SE of 3 experiments. (D) Overlay assays were carried out with biotinylated RGDS spotted onto immobilized proteins. Labeled RGDS specifically interacted with human recombinant active caspase 9 immobilized onto nitrocellulose (0.1 to 5 units/spot) and with human recombinant active caspase 8 (1 to 10 units/spot), after 4 hours of incubation at RT, whereas no interaction was observed with immobilized control proteins (BSA and fibronectin, 10 μg/spot). Total binding (ie, binding observed in the presence of labeled RGDS only) and nonspecific binding (ie, binding observed in the presence of labeled RGDS and an excess of unlabeled RGDS) are reported. This experiment was performed 2 times with similar results. A representative experiment is shown.
RGDS cell internalization and molecular interaction. (A) Biotinylated RGDS (500 μg/mL) penetration in live HUVECs was examined after 4 hours of incubation at 4°C and 37°C. Biotinylated RGDS internalized is shown as green stain (fluorescein-avidin staining), and nuclei are shown as red stain (PI staining, 5 μg/mL), by confocal microscopy (original magnification × 110). A short incubation (4 hours) was sufficient for RGDS to enter cells at 37°C incubation, whereas it did not penetrate at 4°C incubation. (B) Overlay assays were carried out with HUVEC membranes and cytoplasmic extracts immobilized onto nitrocellulose and incubated with biotinylated RGDS. Total binding was shown in the presence of labeled RGDS, whereas nonspecific binding was shown in the presence of an excess of unlabeled RGDS. This experiment was performed 3 times with similar results. A representative experiment is shown. (C) The binding was quantified by densitometry; the nonspecific binding was subtracted from the total binding, giving the values reported as specific binding. Data are expressed as mean ± SE of 3 experiments. (D) Overlay assays were carried out with biotinylated RGDS spotted onto immobilized proteins. Labeled RGDS specifically interacted with human recombinant active caspase 9 immobilized onto nitrocellulose (0.1 to 5 units/spot) and with human recombinant active caspase 8 (1 to 10 units/spot), after 4 hours of incubation at RT, whereas no interaction was observed with immobilized control proteins (BSA and fibronectin, 10 μg/spot). Total binding (ie, binding observed in the presence of labeled RGDS only) and nonspecific binding (ie, binding observed in the presence of labeled RGDS and an excess of unlabeled RGDS) are reported. This experiment was performed 2 times with similar results. A representative experiment is shown.
RGDS direct interaction with membranes, cytoplasm, and caspases
A partial subfractionation to obtain membrane and nonmembrane (here referred to as “cytoplasmic”) extracts was carried out as reported38,47 with some modification. Briefly, HUVEC pellet was resuspended in lysis buffer (4 mM HEPES, pH 7.4, 10 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol [DTT], 5 mM EGTA, 25 μg/mL leupeptin, 5 μg/mL pepstatin, 40 mM β-glycerophosphate, 1 mM phenylmethylsulphonyl fluoride [PMSF]) and kept for 25 minutes at 4°C. After freezing and thawing 3 times, membranes were separated by centrifugation at 15 493g (13 000 rpm) at 4°C for 20 minutes. Membranes and the corresponding cytoplasmic extracts were spotted onto nitrocellulose, as previously reported.48 In other experiments, human caspases 8 and 9 active recombinant proteins (0 to 10 units) or control proteins (BSA, fibronectin; 10 μg) were spotted onto nitrocellulose and blocked with 5% milk in TPBS. After washing 3 times with TPBS, nitrocellulose membranes were incubated for 4 hours at RT with 1 mg/mL amino terminal-biotinylated RGDS (Sigma-Genosys). Some experiments were carried out with 1 mg/mL biotinylated RGDS in the presence of 10 mg/mL not-biotinylated RGDS diluted in TPBS, as a specific competitor. After extensive washing with TPBS, nitrocellulose was incubated for 1 hour at RT with an avidin/biotinylated horseradish peroxidase kit (Vecstatin ABC; Vector) followed by chemiluminescence reaction and exposure to Kodak film (Eastman Kodak, Rochester, NY). Binding in the presence of labeled RGDS was considered “total” binding, while binding in the presence of an excess of unlabeled RGDS was considered “nonspecific” binding. Interaction was measured by densitometry (GS 710; Bio-Rad), analyzed using “Quantity one” software (Bio-Rad), and expressed as a percent of total binding.
Results
HUVEC adhesion
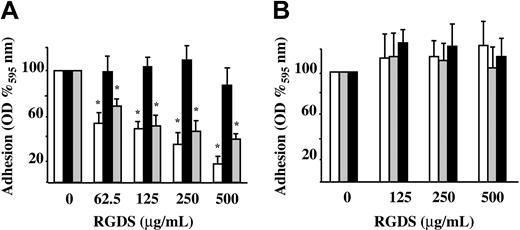
Preliminary experiments showed that HUVEC adhesion to vitronectin, fibronectin, and collagen IV (50 μg/mL) is proportional to the number of seeded cells with the midpoint set to 15 000 cells/well. Therefore, this number of cells was seeded either in the presence or absence of increasing concentrations of RGDS. In these experiments, RGDS exhibited a dose-dependent inhibitory effect on HUVEC adhesion to fibronectin and vitronectin (61% ± 4.3% and 84% ± 6.8% inhibition; P < .001, respectively, at 500 μg/mL) while it had no effect on HUVEC adhesion to collagen IV (Figure 1A). The control peptide (RGES) had no effect on HUVEC adhesion to ECM glycoproteins at any tested doses, as expected (Figure 1B).
HUVEC adhesion to different matrix glycoproteins. HUVEC adhesion to fibronectin (▦), vitronectin (□), and collagen IV (50 μg/mL diluted in PBS; ▪) was measured. Cells were seeded in the presence of 10% FCS and treated for one hour at 37°C with serial dilutions of RGDS or RGES. Cell adhesion was then quantified as optical density (OD) at 595 nm and expressed as OD percent versus control. (A) RGDS reduced in a dose-dependent manner HUVEC adhesion to fibronectin and vitronectin, but it did not inhibit adhesion to collagen IV (*P < .001). (B) Control peptide RGES had no effect on HUVEC adhesion to extracellular matrix glycoproteins. Data are expressed as mean ± SE of 3 experiments carried out in triplicate.
HUVEC adhesion to different matrix glycoproteins. HUVEC adhesion to fibronectin (▦), vitronectin (□), and collagen IV (50 μg/mL diluted in PBS; ▪) was measured. Cells were seeded in the presence of 10% FCS and treated for one hour at 37°C with serial dilutions of RGDS or RGES. Cell adhesion was then quantified as optical density (OD) at 595 nm and expressed as OD percent versus control. (A) RGDS reduced in a dose-dependent manner HUVEC adhesion to fibronectin and vitronectin, but it did not inhibit adhesion to collagen IV (*P < .001). (B) Control peptide RGES had no effect on HUVEC adhesion to extracellular matrix glycoproteins. Data are expressed as mean ± SE of 3 experiments carried out in triplicate.
Additional studies were carried out to investigate the antiadhesive effect of RGDS peptide on collagen IV. HUVECs were seeded on PLL glass, on vitronectin, and on collagen IV, and treated with RGDS (500 μg/mL) for 4 hours. RGDS induced retraction of cells seeded on vitronectin, while it had no effect on HUVECs seeded on PLL glass or on collagen IV (Figure 2). These results confirmed that, under such experimental conditions, RGDS did not exert any evident antiadhesive effect on HUVECs seeded on collagen IV. Therefore, the following studies were all carried out on cells seeded on collagen IV and on migration filters coated with collagen IV, in order to minimize the contribution of an antiadhesive action to the biologic effects observed.
HUVEC spreading on matrix glycoproteins. Cell morphology on coverslips coated with PLL, vitronectin, or collagen IV (50 μg/mL) was examined. Cells were treated for 4 hours with RGDS (500 μg/mL) and stained with FITC-conjugated phalloidin (original magnification, × 40). In all cases cells showed a similar spreading and actin stress fibers morphology, indicating that while RGDS had no antiadhesive effect on collagen IV and poly-L-lysine, it had an antiadhesive effect on vitronectin, as expected. Experiments were carried out in triplicate. The images shown refer to a representative experiment.
HUVEC spreading on matrix glycoproteins. Cell morphology on coverslips coated with PLL, vitronectin, or collagen IV (50 μg/mL) was examined. Cells were treated for 4 hours with RGDS (500 μg/mL) and stained with FITC-conjugated phalloidin (original magnification, × 40). In all cases cells showed a similar spreading and actin stress fibers morphology, indicating that while RGDS had no antiadhesive effect on collagen IV and poly-L-lysine, it had an antiadhesive effect on vitronectin, as expected. Experiments were carried out in triplicate. The images shown refer to a representative experiment.
HUVEC proliferation and migration in the presence of RGDS
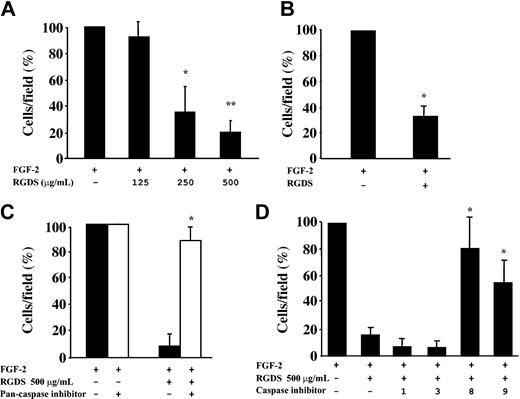
The effect of RGDS on proliferation of HUVECs seeded on collagen IV was examined in the presence of FGF-2 (Figure 3). RGDS significantly reduced in a concentration-dependent way FGF-2-induced proliferation (50% ± 4% inhibition; P < .05) at 500 μg/mL, indicating that RGDS exerts a potent antiproliferative effect independently of its antiadhesive properties.
FGF-2-induced HUVEC proliferation. FGF-2 (10 ng/mL)-induced HUVEC proliferation on collagen IV (50 μg/mL) was evaluated. Cells were treated for 24 hours at 37°C with increasing RGDS concentrations. RGDS significantly reduced endothelial cell proliferation (*P < .05 at 500 μg/mL RGDS). Data are expressed as mean ± SE of 3 experiments carried out in duplicate.
FGF-2-induced HUVEC proliferation. FGF-2 (10 ng/mL)-induced HUVEC proliferation on collagen IV (50 μg/mL) was evaluated. Cells were treated for 24 hours at 37°C with increasing RGDS concentrations. RGDS significantly reduced endothelial cell proliferation (*P < .05 at 500 μg/mL RGDS). Data are expressed as mean ± SE of 3 experiments carried out in duplicate.
RGDS effect on cell migration was investigated in FGF-2-induced chemotaxis through collagen IV-coated polycarbonate filters (Figure 4). The peptide inhibited FGF-2-induced chemotaxis achieving 65% ± 11% (P < .05) and 81% ± 8.9% (P < .01) inhibition at 250 μg/mL and 500 μg/mL, respectively (Figure 4A). This antichemotactic effect was maintained when RGDS was preincubated for 20 minutes and then washed away (67% ± 8.4% inhibition, P < .01 at 500 μg/mL; Figure 4B), showing that a short pretreatment followed by an extensive washing out is sufficient to achieve the inhibitory action observed. It should be noted that all these effects were anchorage independent, since under these experimental conditions RGDS does not induce cell detachment (Figure 2) nor does it affect cell adhesion (Figure 1).
HUVEC migration in the presence of RGDS. FGF-2-induced HUVEC migration across collagen IV (10 μg/mL) was examined in the presence of increasing doses of RGDS. Data are expressed as mean ± SE of 3 experiments carried out in duplicate. (A) RGDS reduced in a dose-dependent manner cell migration (*P < .05 and **P < .01 at 250 μg/mL or 500 μg/mL, respectively). (B) The antichemotactic effect was maintained on HUVECs pretreated with the peptide (500 μg/mL) for 20 minutes at 37°C and then washed with high ionic strength (*P < .01). (C) HUVEC migration across collagen IV (10 μg/mL) in the presence of RGDS (500 μg/mL) was examined. Migration was induced by FGF-2 (10 ng/mL) and cells were pretreated for 20 minutes at 37°C with caspase inhibitors. A pan-caspase inhibitor (Z-VAD-FMK, 50 μM) treatment abolished the RGDS inhibitory effect (*P < .01). (D) Specific caspase 8 and 9 inhibitors (named “8” and “9,” respectively) reverted the inhibitory effect of RGDS, whereas caspase 1 and 3 inhibitors (named “1” and “3”) had no effect (*P < .05).
HUVEC migration in the presence of RGDS. FGF-2-induced HUVEC migration across collagen IV (10 μg/mL) was examined in the presence of increasing doses of RGDS. Data are expressed as mean ± SE of 3 experiments carried out in duplicate. (A) RGDS reduced in a dose-dependent manner cell migration (*P < .05 and **P < .01 at 250 μg/mL or 500 μg/mL, respectively). (B) The antichemotactic effect was maintained on HUVECs pretreated with the peptide (500 μg/mL) for 20 minutes at 37°C and then washed with high ionic strength (*P < .01). (C) HUVEC migration across collagen IV (10 μg/mL) in the presence of RGDS (500 μg/mL) was examined. Migration was induced by FGF-2 (10 ng/mL) and cells were pretreated for 20 minutes at 37°C with caspase inhibitors. A pan-caspase inhibitor (Z-VAD-FMK, 50 μM) treatment abolished the RGDS inhibitory effect (*P < .01). (D) Specific caspase 8 and 9 inhibitors (named “8” and “9,” respectively) reverted the inhibitory effect of RGDS, whereas caspase 1 and 3 inhibitors (named “1” and “3”) had no effect (*P < .05).
It is known that RGD-containing peptides have a direct caspase-activating effect,44 thus we investigated the role caspases may play in such antichemotactic effect. As a first approach, it was investigated whether caspase inhibitors reverted the RGDS antichemotactic effect. RGDS inhibitory effect was completely blocked when endothelial cells were pretreated with the pan-caspase inhibitor Z-VAD-FMK (Figure 4C). Further experiments showed that specific caspase 8 and caspase 9 inhibitors Z-IEDT-FMK and Z-LEHD-FMK abolished RGDS inhibitory effect, while caspase 1 and caspase 3 inhibitors Z-WEHD-FMK and Z-DEVD-FMK had no effect (Figure 4D). These results indicated that caspase 8 and caspase 9 activity may account, at least in part, for the antichemotactic effect of RGDS.
Caspase activity and apoptosis induction
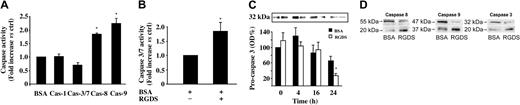
Additional studies were carried out to further investigate the caspases' role. HUVECs seeded onto collagen IV were preincubated with RGDS for 4 hours, then treated with the fluorescent pan-caspase substrate FITC-VAD-FMK. Under these conditions, cells remained adherent and a 30% fluorescence increase was revealed as compared with untreated cells, indicating that RGDS short treatment leads to an anchorage-independent caspase activation (data not shown). Caspase activation was then investigated, under similar experimental conditions, by specific enzymatic assays. It was found that RGDS treatment induced caspase 8 and caspase 9 activation (1.8 ± 0.02 fold versus control, P < .01; and 2.2 ± 0.2 fold versus control, P < .05, respectively), while it had no effect on caspase 1 and caspase 3/7 activity (Figure 5A). Conversely, it induced caspase 3 and caspase 7 activation only after 24 hours (1.8 ± 0.3 fold versus control, P < .05; Figure 5B).
Caspase activity. Effect of RGDS treatment on caspase activity. (A) HUVECs seeded on collagen IV (50 μg/mL) were treated with RGDS (500 μg/mL) for 4 hours at 37°C. Caspase 3/7 enzymatic activity was evaluated by measuring the fluorescence of the Z-DEVD-R110 substrate. Caspase 1, caspase 8, and caspase 9 activity were quantified by the release of AFC from the substrates (DEVD-AFC, AEVD-AFC, and IETD-AFC, respectively). RGDS significantly activated caspase 8 (Cas-8; *P < .01) and caspase 9 (Cas-9; *P < .05), whereas it had no effect on caspase 3/7 (Cas-3/7) and caspase 1 (Cas-1) activation. Results represent the average ± SD of 3 experiments performed in duplicate. (B) Caspase 3/7 enzymatic activity was measured as reported for panel A. RGDS increased caspase 3 activity after 24 hours (P < .05). These experiments were carried out in triplicate. (C) Cells were incubated at 37°C with RGDS for 4, 16, and 24 hours. Caspase 3 activation was followed as function of pro-caspase cleavage by Western blotting, incubating with a polyclonal rabbit antihuman caspase 3 antibody. RGDS reduced the inactive form of caspase 3 (pro-caspase 3 MW 32 kDa) after 24 hours of treatment (*P < .01), whereas at 4 hours and 16 hours it was ineffective. (D) Pro-caspases 8 and 9 (55 kDa and 47 kDa, respectively) were cleaved to the active forms (20 kDa and 37 kDa, respectively) after 4 hours of treatment, whereas the 20-kDa active subunit of caspase 3 was generated after 24 hours of treatment; the increase of the cleaved active forms was always paralleled by a corresponding decrease of the pro-caspase inactive forms. The equal loading was confirmed by Ponceau S solution staining. The figure refers to one representative experiment, whereas the quantification refers to 4 experiments.
Caspase activity. Effect of RGDS treatment on caspase activity. (A) HUVECs seeded on collagen IV (50 μg/mL) were treated with RGDS (500 μg/mL) for 4 hours at 37°C. Caspase 3/7 enzymatic activity was evaluated by measuring the fluorescence of the Z-DEVD-R110 substrate. Caspase 1, caspase 8, and caspase 9 activity were quantified by the release of AFC from the substrates (DEVD-AFC, AEVD-AFC, and IETD-AFC, respectively). RGDS significantly activated caspase 8 (Cas-8; *P < .01) and caspase 9 (Cas-9; *P < .05), whereas it had no effect on caspase 3/7 (Cas-3/7) and caspase 1 (Cas-1) activation. Results represent the average ± SD of 3 experiments performed in duplicate. (B) Caspase 3/7 enzymatic activity was measured as reported for panel A. RGDS increased caspase 3 activity after 24 hours (P < .05). These experiments were carried out in triplicate. (C) Cells were incubated at 37°C with RGDS for 4, 16, and 24 hours. Caspase 3 activation was followed as function of pro-caspase cleavage by Western blotting, incubating with a polyclonal rabbit antihuman caspase 3 antibody. RGDS reduced the inactive form of caspase 3 (pro-caspase 3 MW 32 kDa) after 24 hours of treatment (*P < .01), whereas at 4 hours and 16 hours it was ineffective. (D) Pro-caspases 8 and 9 (55 kDa and 47 kDa, respectively) were cleaved to the active forms (20 kDa and 37 kDa, respectively) after 4 hours of treatment, whereas the 20-kDa active subunit of caspase 3 was generated after 24 hours of treatment; the increase of the cleaved active forms was always paralleled by a corresponding decrease of the pro-caspase inactive forms. The equal loading was confirmed by Ponceau S solution staining. The figure refers to one representative experiment, whereas the quantification refers to 4 experiments.
An additional approach allowed us to follow cleavage of pro-caspases 8, 9, and 3, which leads to their activation. RGDS treatment reduced expression of caspase 3 inactive precursor (molecular weight [MW] 32 kDa) by 70% ± 5.5% (P < .01) after 24 hours while it was ineffective at shorter time points (Figure 5C). The inactive precursor was proteolytically cleaved, generating a 20-kDa subunit (Figure 5D). On the other hand, at a shorter incubation time (namely, 4 hours of RGDS treatment), the procaspases 8 and 9 (55 kDa and 47 kDa, respectively) were cleaved to the corresponding active forms (20 kDa and 37 kDa, respectively; Figure 5D).
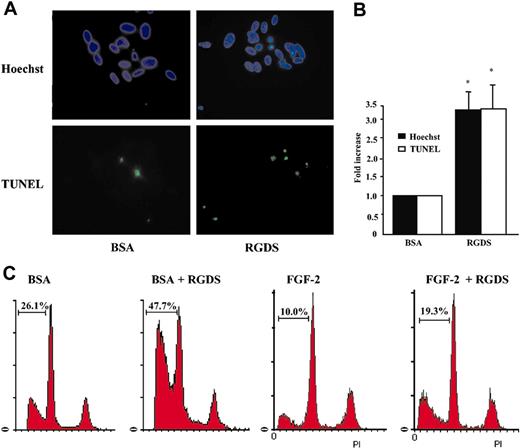
These results indicated that RGDS treatment induces an early activation of caspases 8 and 9 (4-hour treatment) and a later activation of caspase 3 (24-hour treatment). Therefore, RGDS-induced HUVEC apoptosis was examined. Cells, seeded on collagen IV, were treated with RGDS for 24 hours and apoptosis was measured by TUNEL and Hoechst staining. Both tests showed that RGDS induced a significant increase of apoptosis as compared with control (3.3 ± 0.6 fold versus control, P < .05, at 500 μg/mL; Figure 6A-B).
Apoptosis induction. Apoptosis of HUVECs seeded on collagen IV (50 μg/mL) was evaluated after 24 hours of RGDS (500 μg/mL) treatment at 37°C. (A) The apoptosis was visualized by TUNEL staining and by nuclear fragmentation (Hoechst staining). Nuclei stained with Hoechst as well as TUNEL-positive nuclei were identified by means of a Zeiss Axioplan fluorescence microscope (original magnification, × 40). The images shown refer to representative experiments. (B) Apoptosis was expressed as a percent of fragmented Hoechst-positive nuclei versus total Hoechst-positive nuclei, and as a percent of TUNEL-positive nuclei versus total Hoechst-positive nuclei and was reported as fold increase versus control. Quantification performed with the 2 staining methods gave similar results. (*P < .05). Experiments were carried out in triplicate. (C) Apoptosis was quantified by FACS analysis according to the PI staining in sub-G1-phase cells. Three independent experiments were performed with similar results; one representative experiment is shown.
Apoptosis induction. Apoptosis of HUVECs seeded on collagen IV (50 μg/mL) was evaluated after 24 hours of RGDS (500 μg/mL) treatment at 37°C. (A) The apoptosis was visualized by TUNEL staining and by nuclear fragmentation (Hoechst staining). Nuclei stained with Hoechst as well as TUNEL-positive nuclei were identified by means of a Zeiss Axioplan fluorescence microscope (original magnification, × 40). The images shown refer to representative experiments. (B) Apoptosis was expressed as a percent of fragmented Hoechst-positive nuclei versus total Hoechst-positive nuclei, and as a percent of TUNEL-positive nuclei versus total Hoechst-positive nuclei and was reported as fold increase versus control. Quantification performed with the 2 staining methods gave similar results. (*P < .05). Experiments were carried out in triplicate. (C) Apoptosis was quantified by FACS analysis according to the PI staining in sub-G1-phase cells. Three independent experiments were performed with similar results; one representative experiment is shown.
To further analyze the RGDS effect, FACS analysis was carried out on HUVECs treated for 24 hours with RGDS (500 μg/mL). Quantification of sub-G1-phase cells was considered an apoptosis marker. Data reported in Figure 6C show that RGDS treatment significantly increased percentage of cells in sub-G1 phase either in the presence of BSA only, or in the presence of FGF-2. Apoptosis of 26.1% under BSA treatment was expected, due to the 24-hour-long serum deprivation, and was significantly increased to 47.7% by RGDS treatment. Similarly, 10% apoptosis under FGF-2 treatment was significantly increased to 19.3% by RGDS treatment. Pretreatment with antibodies blocking α2β1, αvβ3, or αvβ5 did not significantly change RGDS proapoptotic activity in the presence of BSA (44.6%, 43.5%, and 42.4% respectively, versus 47.7% RGDS alone; not shown).
RGDS cell internalization and molecular interaction
It has already been reported that RGD-containing peptides enter the cells (lymphocytes, cardiomyocytes, melanoma cells) leading to apoptosis by direct interaction with—and activation of—caspase 3.44,45 We observed that RGDS is able to specifically penetrate live HUVECs under our experimental set-up (Figure 7A). Immunofluorescent confocal microscopy showed that RGDS entered the cells within 4 hours at 37°C incubation, while it was not able to penetrate at 4°C incubation, strongly suggesting a specific internalization event.
We also observed that RGDS specifically interacts with HUVEC membranes as well as cytoplasmic extracts immobilized onto nitrocellulose. In both cases the interaction was inhibited in the presence of an excess of unlabeled RGDS (Figure 7B-C), indicating that such interaction is a specific event. The specific binding corresponded to at least 50% of the total binding. Since RGDS short-treatment of HUVECs led to RGDS internalization (Figure 7A) and caspase 8 and caspase 9 activation (Figure 5), we hypothesized that such activation may be due, at least in part, to a direct interaction of RGDS with caspase 8 or caspase 9. Thus, different quantities of human active recombinant caspases 8 and 9 were immobilized and were incubated with biotinylated RGDS (1 mg/mL) in the presence or absence of unlabeled RGDS (10 mg/mL). Labeled RGDS directly interacted with caspases 8 and 9 and this binding was specifically inhibited, at least in part, in the presence of an excess of unlabeled RGDS. Under similar experimental conditions RGDS did not interact with BSA or with fibronectin (Figure 7D). The interaction with caspase 9 became evident at one immobilized unit while interaction with caspase 8 was evident at 10 immobilized units. These data indicated for the first time a direct and specific interaction of RGDS with human recombinant active caspases 8 and 9.
Discussion
Cell adhesion to ECM is a requisite for endothelial cells to grow and survive. The RGD motif in the ECM proteins (namely, fibronectin, laminin, vitronectin, collagen, osteopontin, trombospondin, bone sialoproteins, entactin, tenascin) is known to play a crucial role in integrin-mediated cell adhesion. However, integrins also interact in an RGD-independent way with collagens. Namely, this has been shown for α2β1, αvβ3, and α1β1 integrins.49-53 αvβ3 integrin has been shown to bind noncollagenous (NC1) domains of the alpha2(IV), alpha3(IV), and alpha6(IV) chains of human collagen type IV in an RGD-independent manner and such domains have shown antiangiogenic and antitumoral properties.10,54,55
The RGD motif is involved in a number of key biologic activities related to cell adhesion. More specifically, it is known to mediate proteins and virus entry into target cells and appears to play a key role in cell targeting35,56-59 and membrane receptors clustering in T lymphocytes.60 Besides the antiadhesive-dependent apoptotic effect, RGD-containing peptides have also shown an anchorage-independent apoptotic effect in lymphocytes and cardiomyocyte cells.38,39 In these cells, caspase 3 activation followed direct interaction with RGD-containing peptides, indicating the ability of the RGD motif to penetrate cells and act intracellularly. To further investigate this field, we explored the RGDS effect on HUVECs, under experimental conditions set to minimize its antiadhesive activity. Cell adhesion to collagen IV is known to be RGD independent.53-55 In fact, RGDS did not inhibit the ability of HUVECs to adhere to the matrix (Figure 1) and, on the other hand, did not induce detachment of adherent cells from collagen IV (Figure 2). Under such experimental conditions, RGDS strongly inhibited HUVEC migration and proliferation induced by FGF-2. This antichemotactic effect is related to the activity of initiator caspases (namely caspases 8 and 9) rather than to effector caspase 3. In fact, caspase 8 and caspase 9 inhibitors abolished the antichemotactic effect, while caspase 3 and caspase 1 inhibitors were ineffective. Furthermore, caspase 8 and caspase 9 activation was observed as soon as after 4 hours of RGDS treatment, suggesting that caspase 3 activation, apoptosis, and reduced proliferation, all observed at 24 hours, may be late cascade effects. The early activation of caspases 8 and 9 may account at least in part for the observed antichemotactic effect since known cell-migration modulators, namely cytoskeletal proteins, focal adhesion kinase, and focal adhesion components, are caspase substrates.61-63 RGD-dependent activation of caspases 8, 9, and 3 will be further investigated; at this stage it is possible to speculate on different mechanisms and signaling pathways potentially involved, such as phosphatidylinositol 3-kinase (PI3-kinase)64 and mitogen-activated protein kinase (MAPK).65-69 For instance, the MAPK pathway may play a role, based on previously published data.65-69 In fact, ERK has been recently shown to directly induce Thr-125 phosphorylation on caspase 9, leading to its inactivation,65 as well as to directly control caspase 8 activation state66 or to phosphorylate myosin light chain kinase (MLCK) and therefore regulate migration.67 Furthermore, the role of MAPK pathway in controlling caspase 3 activity has been also reported.68,69 We hypothesized that RGDS may either act onto the membrane surface, through an integrin-mediated mechanism,70 or it may enter HUVECs and trigger caspase activation through an intracellular mechanism, according to other evidence.38,39 At this stage we cannot exclude either hypothesis; however, we observed that RGDS penetrates live HUVECs within 4 hours at 37°C and directly interacts with the cytoplasmic extract and with caspases 8 and 9. Moreover, we observed that the proapoptotic activity of RGDS was not affected when α2β1, αvβ3, or αvβ5 integrins were neutralized by the specific antibodies, excluding an integrin-mediated effect, according to a previous report.40 All together, these findings support the hypothesis that RGDS may directly trigger the caspase cascade at an early level. Therefore, the RGDS direct interaction with caspases and their activation at different levels may represent a novel mechanism of apoptosis induction, in addition to the known death receptor- and mithocondria-mediated pathways.
Prepublished online as Blood First Edition Paper, February 24, 2004; DOI 10.1182/blood-2003-06-2144.
Supported in part by an Agenzia Spaziale Italiana (ASI) grant (ref I/R/367/02 no. 717).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal