Abstract
Histone deacetylase (HDAC) inhibitors are emerging as a promising new treatment strategy in hematologic malignancies. Here we show that NVP-LAQ824, a novel hydroxamic acid derivative, induces apoptosis at physiologically achievable concentrations (median inhibitory concentration [IC50] of 100 nM at 24 hours) in multiple myeloma (MM) cell lines resistant to conventional therapies. MM.1S myeloma cell proliferation was also inhibited when cocultured with bone marrow stromal cells, demonstrating ability to overcome the stimulatory effects of the bone marrow microenvironment. Importantly, NVP-LAQ824 also inhibited patient MM cell growth in a dose- and time-dependent manner. NVP-LAQ824-induced apoptotic signaling includes up-regulation of p21, caspase cascade activation, and poly (adenosine diphosphate [ADP]) ribose (PARP) cleavage. Apoptosis was confirmed with cell cycle analysis and annexin-propidium iodide staining. Interestingly, treatment of MM cells with NVPLAQ824 also led to proteasome inhibition, as determined by reduced proteasome chymotrypsin-like activity and increased levels of cellular polyubiquitin conjugates. Finally, a study using NVP-LAQ824 in a preclinical murine myeloma model provides in vivo relevance to our in vitro studies. Taken together, these findings provide the framework for NVP-LAQ824 as a novel therapeutic in MM. (Blood. 2003;102:2615-2622)
Introduction
Despite many recent advances in the knowledge and treatment of multiple myeloma (MM), it currently remains incurable without an allogeneic hematopoietic cell transplantation. Incomplete response rates and the eventual emergence of drug resistance have been major obstacles to treatment with conventional therapies. The recent development of new treatment strategies based upon molecular targeting, such as proteasome inhibitors and thalidomide analogs, have made great progress in the treatment of MM. In the search for more potential molecular targets, the enzyme histone deacetylase (HDAC) is emerging as a key molecule. Aberrant histone acetylation has been observed in the development of numerous malignancies,1-4 and inhibitors of HDAC are a promising new treatment strategy for malignant disease.5,6
For decades, the short-chain fatty acid sodium butyrate and the hydroxamic acid trichostatin A (TSA) have been known as differentiation inducers and HDAC inhibitors, but clinical activity has been limited by low potency (butyrate) or high reactivity and instability (TSA).7-9 In 1998, hexamethylene bisacetamide (HMBA), a hybrid polar compound (HPC) that induced terminal differentiation of transformed cells without HDAC inhibitory properties, was shown to inhibit the growth of several human myeloma cell lines and patient myeloma cells.10 Second-generation HPCs suberoylanilide hydroxamic acid (SAHA) and m-carboxycinnamic acid bishydroxamide (CBHA) were found to be more potent inducers of differentiation on a molar basis than HMBA, with activity in low micromolar concentrations, and furthermore were HDAC inhibitors.11 X-ray crystallographic studies have shown that hydroxamic acid-containing HDAC inhibitors interact with the catalytic site of HDACs, thereby blocking substrate access to the active zinc ion at its base.12 Other classes of HDAC inhibitors include cyclic peptides, benzamides such as MS-275, and the recently reported class of sulfonamide anilides.13 Over a dozen HDAC inhibitors have now been developed for clinical use, several of which have entered into clinical trials.14 In general, hydroxamic acid derivatives such as SAHA11 and pyroxamide15 are potent at micromolar concentrations, as are the sulfonamide anilides; whereas the cyclic peptides, such as FK22816 and the hybrid cyclic hydroxamic acid peptide analogs,17 are active at nanomolar concentrations. Impressive ability to overcome drug resistance in patients has been reported with FK288 for cutaneous T-cell lymphoma.18 The combination of phenylbutyrate and all-trans retinoic acid has induced a complete clinical remission in a patient with acute promyelocytic leukemia resistant to all-trans retinoic acid alone.19
NVP-LAQ824 is a structurally novel hydroxamic acid derivative, which inhibits HDAC at 0.15 μM or less, and colon cancer cell line H1299 monolayer cell growth inhibition at median inhibitory concentration (IC50) less than 1 μM. NVP-LAQ824 is a derivative of 4-aminomethylcinnamic hydroxamic acid, with a chemical formula C22H25O3 (chemical structure in press, Atadja et al20 ). NVP-LAQ824 is water soluble at a concentration of more than 20 mg/mL at 37°C and is stable for at least a month at room temperature. NVP-LAQ824 inhibits HDAC at IC50 0.03 μM, and up-regulates transcriptional activation of the p21 promoter at 0.3 μM. Activity has been demonstrated against tumor cell lines resistant to paclitaxel due to overexpression of p-glycoprotein, as well as human leukemia cell lines in vitro.20 Monolayer cell growth inhibition has been achieved at an IC50 less than 1 μM in colon cancer cell lines H1299 and HCT116, breast cancer MDA435 cells, prostate cancer DU145 and PC3 cells, and non-small cell lung cancer A549 cells. Apoptosis induced by NVP-LAQ824 has been reported in human breast cancer SKBR-3, BT-474, and MB-468 cells.21 Early preclinical evaluation in murine models showed significant in vivo activity against the growth of established human lung, colon, or breast tumor xenografts in doses ranging from 5 to 100 mg/kg.
HDAC inhibitors have previously been shown to be effective in MM,10 and we therefore evaluated the effects of NVP-LAQ824 on patient MM cells and human myeloma cell lines (HMCLs). In this study, we demonstrated in vitro activity of NVP-LAQ824 tested against freshly isolated MM patient cells from relapsed and refractory patients, as well as MM cell lines both sensitive and resistant to conventional chemotherapy. Furthermore, NVPLAQ824 has efficacy in vivo in a murine xenograft myeloma model. The molecular sequelae of NVP-LAQ824 antitumor activity in MM cells are also explored to further characterize the mechanisms of action of this novel HDAC inhibitor. These studies provide the preclinical rationale for clinical protocols using NVPLAQ824 to improve patient outcome in MM.
Materials and methods
HDAC inhibitor
NVP-LAQ824 (Novartis Pharma, Basel, Switzerland) was dissolved in deiodinized water and stored at -20°C, then thawed and diluted in media for cell culture experiments. For animal experiments, the drug was dissolved in sterile water prior to intraperitoneal injection.
MM-derived cell lines and patient cells
Dexamethasone (Dex)-sensitive (MM.1S) and Dex-resistant (MM.1R) human MM cell lines, as well as RPMI 8226 cells resistant to doxorubicin (DOX; Dox40), mitoxantrone (Mit; MR20), and melphalan (Mel; LR5), were cultured in RPMI 1640 media (Cellgro, Mediatech, VA) with 10% fetal bovine serum, 2 mM l-glutamine (GIBCO, Grand Island, NY), 100 U/mL penicillin, and 100 mg/mL streptomycin (GIBCO). Drug-resistant cell lines were cultured with either Dox, Mit, Mel, or Dex to confirm their lack of drug sensitivity. Multiple myeloma patient cells were obtained from bone marrow samples after informed consent for the use of samples for the purpose of research. The bone marrow mononuclear cells were separated using Ficoll-Hypaque density sedimentation, and plasma cells were purified (> 95% CD138+) by positive selection with anti-CD138 magnetic-activated cell separation (MACS) microbeads (Miltenyi, San Diego, CA). B cells and T cells were obtained by Ficoll-Hypaque density sedimentation of normal donor peripheral blood, and separation was obtained using B-cell isolation kit and anti-CD3 MACS antibodies, respectively. Cells were incubated in 96-well plates with or without NVP-LAQ824. B cells were stimulated with interleukin-4 (IL-4, 10 ng/mL) and anti-CD40 (G28.5, 10 μg/mL); T cells were stimulated with phytohemagglutinin (10 μg/mL).
DNA synthesis
MM cells (3 × 104 cells/well) were incubated in 96-well culture plates (Costar, Cambridge, MA) in the presence of media, NVP-LAQ824, and/or dexamethasone or recombinant IL-6 (Genetics Institute, Cambridge, MA) for 48 hours at 37°C. DNA synthesis was measured by 3H-thymidine (NEN Products, Boston, MA) uptake. Cells were pulsed with 3H-thymidine (0.5 μCi/well [0.0185 MBq/well]) during the last 8 hours of 48-hour cultures. All experiments were performed in triplicate.
Growth inhibition assay
The inhibitory effect of NVP-LAQ824 on MM cell growth was assessed by MTS ([3-(4,5-dimethyl thiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium], inner salt) assay (Promega, Madison, WI). Cells from 24-, 48-, or 72-hour cultures in 200 μL media plus drug were pulsed with 40 μL of 5 mg/mL MTS to each well for the last 4 hours of the cultures. Absorbance was measured at 490 nm using a spectrophotometer (Molecular Devices, Sunnyvale, CA).
Cell cycle analysis
MM cell lines and patient MM cells cultured for 0, 8, 12, 18, 36, and 42 hours in NVP-LAQ824 (1 μM), NVP-LAQ824 plus pancaspase inhibitor ZVAD-FMK (Z-Val-Ala-Asp(OMe)-CH2F; 20 μM) (Calbiochem, San Diego, CA), or control media were harvested, washed with phosphate-buffered saline (PBS), fixed with 70% ethanol, and treated with 10 mg/mL RNase (Roche Diagnostics, Indianapolis, IN). Cells were then stained with 5 mg/mL propidium iodide (PI; Sigma, St Louis, MO), and cell cycle profile was determined using the program M software on an Epics flow cytometer (Coulter Immunology, Hialeah, FL). Data were analyzed using MultiCycle for Windows software (Phoenix FlowSystems, San Diego, CA).
Detection of apoptosis
In addition to identifying sub-G1 cells using cell cycle analysis as described in the previous paragraph, apoptosis was also confirmed using annexin V staining. MM cells were cultured in media alone, or with media plus 1 μM NVP-LAQ824 at 37°C for 24 hours. Cells were then washed twice with ice-cold PBS and resuspended (1 × 106 cells/mL) in binding buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4], 140 mM NaCl, 2.5 mM CaCl2). MM cells (1 × 105) were incubated with annexin V-fluorescein isothiocyanate (5 μL; Pharmingen, San Diego, CA) and PI (5 mg/mL) for 15 minutes at room temperature. Annexin V+PI- apoptotic cells were enumerated using the Epics cell sorter (Coulter Immunology).
Immunoblotting
Patient MM cells and MM.1S cells were cultured with 0.01, 0.1, or 1 μM NVP-LAQ824; harvested; washed; and lysed using lysis buffer: radioimmunoprecipitation assay buffer, 2 mM Na3VO4, 5 mM NaF, 1 mM phenylmethyl sulfonyl fluoride, 5 mg/mL leupeptin, and 5 mg/mL aprotinin. Cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride (PVDF) membrane, and immunoblotted with antiacetylated histone (Upstate Biotechnology, Lake Placid, NY), anti-p21 antibody (Ab; Santa Cruz Biotech, Santa Cruz, CA), anti-Poly (adenosine diphosphate [ADP]) ribose (PARP); anti-caspase-8, anti-caspase-9, and anti-caspase-3 (Cell Signaling, Beverly, MA); as well as antipolyubiquitin conjugates, anti-large multifunctional protease 7 (LMP7), and anti-β5 (Affiniti, Mamhead, United Kingdom). Membranes were stripped and reprobed with anti-tubulin or anti-β-actin (Sigma) to ensure equivalent protein loading.
Proteasome activity assay
Proteasome activity in cytosolic extracts was quantified using the fluorogenic proteasome substrate Suc-LLVY-amino-4-methylcoumarin (AMC) (Calbiochem). Cytosolic extract (100 μg of protein in 5 μL) was incubated in a 200-μL reaction containing 20 mM Tris (tris(hydroxymethyl)aminomethane)-HCl (pH 7.8), 0.5 mM EDTA (ethylenediaminetetraacetic acid), and 100 μM Suc-LLVY-AMC at room temperature for 90 minutes. Fluorescence was measured in a microtiter plate fluorometer (excitation, 360 nm; emission, 460 nm). The mean and standard deviations of duplicate well experiments are shown, and the data are representative of 3 experiments.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts for EMSAs were carried out using double-stranded nuclear factor-κB (NF-κB) consensus oligonucleotide probe (5′-GGGGACTTTCCC-3′; Santa Cruz Biotechnology) end-labeled with [γ-32P] adenosine triphosphate (50 μCi [1.85 MBq] at 222 TBq/mM; NEN Products). Binding reactions containing 1 ng of oligonucleotide and 5 μg of nuclear protein were conducted at room temperature for 20 minutes in a total volume of 10 mL of binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 4% glycerol [vol/vol], and 0.5 mg of poly [dI-dC] [Pharmacia, Peapack, NJ]).
Xenograft murine model
Beige-nude-xid (BNX) mice (5- to 6-weeks old) were inoculated subcutaneously into the right flank with 3 × 107 RPMI 8226 MM cells in 100 μL RPMI 1640, together with 100 μL Matrigel basement membrane matrix (Becton Dickinson, Bedford, MA). When tumors were measurable, mice were assigned into one group receiving 25 mg/kg NVP-LAQ824 intraperitoneally daily or into a control group receiving the vehicle alone (0.9% sodium chloride) at the same schedule. Treatment with NVP-LAQ824 was given at 25 mg/kg. Caliper measurements of the longest perpendicular tumor diameters were performed every alternate day to estimate the tumor volume, using the following formula: 4π/3 × (width/2)2 × (length/2), representing the 3-dimensional volume of an ellipse. Animals were killed when their tumors reached 2 cm to prevent unnecessary morbidity to the mice. Survival was evaluated from the first day of treatment until death. Tumor growth was evaluated using caliper measurements from first day of treatment until day of first killing (day 9). Serum paraprotein was not detectable by enzyme-linked immunosorbent assay in this xenograft model.
Statistical analysis
Statistical significance of differences observed in drug-treated versus control cultures was determined using Student t test. The minimal level of significance was a P value less than .05. The survival curves for mice were computed using the Kaplan-Meier method. The survival time differences between the control and treated group were compared using a log-rank test. The median survival times were compared using the Fisher exact test. To compare the rate of tumor growth in the 2 arms, a linear mixed-effect model (random coefficient model) was fitted.
Results
NVP-LAQ824 inhibits DNA synthesis in patient MM cells
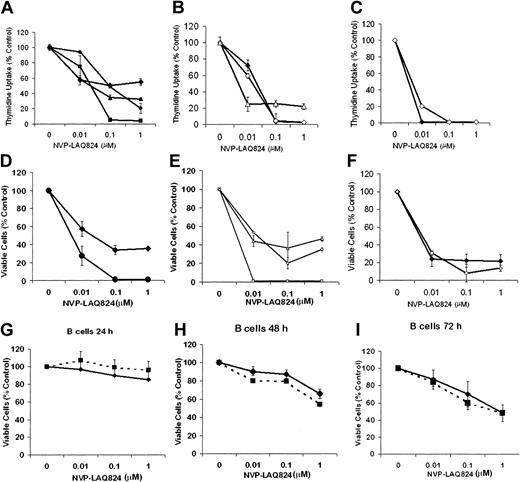
The effect of NVP-LAQ824 on DNA synthesis of freshly isolated primary patient MM cells was first examined at 24, 48, and 72 hours (Figure 1A-C). 3H-thymidine uptake of patients' MM cells was inhibited by NVP-LAQ824 (0.01-1.0 μM). IC50 was observed at 10 to 100 nM by 24 to 48 hours, and at less than 10 nM by 72 hours. Almost complete inhibition was seen with 100 nM by 48 hours, and with 10 nM at 72 hours. The effects of these drugs on proliferation were confirmed using MTS assays (Figure 1D-F). By contrast, an IC50 in B cells was observed only after 72 hours (Figure 1G-I). At 72 hours the IC50 was approximately 1 μM, 100-fold higher than in malignant plasma cells (IC50 < 0.01 μM). Similar results were observed for isolated T cells (data not shown). These results demonstrate potency of NVP-LAQ824 in submicromolar concentrations, and suggest a therapeutic window between the effects on malignant plasma cells versus normal B cells and T cells. NVP-LAQ824 had a modest effect on patient-derived bone marrow stromal cells (BMSCs), reducing viability at 1 μM to 73%, 69%, and 70% at 24, 48, and 72 hours, respectively (P = .02, .05, and .08, respectively; data not shown). These results are consistent with the differential effects of NVP-LAQ824 on myeloma cells versus normal cells.
Cytotoxicity of NVP-LAQ824 against patient MM cells and normal B cells. (A-C) The effect of NVP-LAQ824 on DNA synthesis of freshly isolated primary patient MM cells was examined at 24, 48, and 72 hours. 3H-thymidine uptake of patients' MM cells was inhibited by NVP-LAQ824 (0.01-1 μM). IC50 was observed at 10 to 100 nM by 24 to 48 hours, and less than 10 nM by 72 hours. Almost complete inhibition was seen with 100 nM by 48 hours, and with 10 nM at 72 hours. (D-F) The effects were confirmed using MTS assay. Patient 1 (♦), patient 2 (▪), patient 3 (▴), patient 4 (•), patient 5 (⋄), patient 6 (□), and patient 7 (▵). Thymidine uptake: (A) 24 hours, (B) 48 hours, and (C) 72 hours. MTS assay: (D) 24 hours, (E) 48 hours, and (F) 72 hours. The effect of NVP-LAQ824 was also assessed on normal B cells unstimulated (♦) and stimulated (▪) at 24 hours (G), 48 hours (H), and 72 hours (I). Error bars represent ± 1 SD of a triplicate experiment.
Cytotoxicity of NVP-LAQ824 against patient MM cells and normal B cells. (A-C) The effect of NVP-LAQ824 on DNA synthesis of freshly isolated primary patient MM cells was examined at 24, 48, and 72 hours. 3H-thymidine uptake of patients' MM cells was inhibited by NVP-LAQ824 (0.01-1 μM). IC50 was observed at 10 to 100 nM by 24 to 48 hours, and less than 10 nM by 72 hours. Almost complete inhibition was seen with 100 nM by 48 hours, and with 10 nM at 72 hours. (D-F) The effects were confirmed using MTS assay. Patient 1 (♦), patient 2 (▪), patient 3 (▴), patient 4 (•), patient 5 (⋄), patient 6 (□), and patient 7 (▵). Thymidine uptake: (A) 24 hours, (B) 48 hours, and (C) 72 hours. MTS assay: (D) 24 hours, (E) 48 hours, and (F) 72 hours. The effect of NVP-LAQ824 was also assessed on normal B cells unstimulated (♦) and stimulated (▪) at 24 hours (G), 48 hours (H), and 72 hours (I). Error bars represent ± 1 SD of a triplicate experiment.
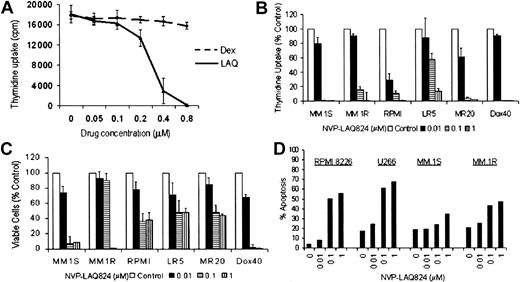
The effect of NVP-LAQ824 on DNA synthesis in MM cell lines was determined by measuring 3H-thymidine uptake during the last 8 hours of 48-hour cultures, in the presence or absence of drug at various concentrations. To examine whether there was cross-resistance between NVP-LAQ824 and conventional therapies, we evaluated RPMI 8226 MM cells resistant to Dox (Dox40 cells), Mit (MR20 cells), or Mel (LR5 cells), as well as MM.1R cells resistant to Dex. Proliferation of MM.1R cells was unaffected by culture with 1 μM Dex, but was completely inhibited by less than 1.0 μM NVP-LAQ824 (Figure 2A). NVP-LAQ824 inhibited 3H-thymidine uptake in MM.1S and RPMI 8226 cells in a dose-dependent fashion. IC50 of proliferation of MM.1S cells was noted between 0.01 to 0.1 μM NVP-LAQ824 and of RPMI 8226 at less than 0.01 μM (Figure 2B) (P = .02). Although MM.1R cells appeared to be less sensitive than MM.1S, both cell lines were completely inhibited well within physiologic concentrations. Similarly, the proliferation of Dox40, MR20, or LR5 cells was unaffected by culture with 400 nM Dox, 20 nM Mit, 5 mM Mel, respectively (data not shown), but was inhibited in cultures with NVP-LAQ824 in a dose-dependent manner (< 0.1 μM for MR20 and Dox40; < 1 μM for LR5) (Figure 2B). The effects of these drugs on proliferation were confirmed using the MTS assay (Figure 2C). Apoptosis was confirmed in human myeloma cell lines by annexin V-PI staining of RPMI 8226, U266, MM.1S, and MM.1R cells cultured with NVP-LAQ824 (Figure 2D). These results indicate that NVP-LAQ824 is toxic to cells resistant to Dox, Mel, Mit, and Dex.
NVP-LAQ824 causes apoptosis in human myeloma cell lines (HMCLs) resistant to conventional chemotherapy. (A-C) The effect of NVP-LAQ824 on DNA synthesis in MM cell lines was determined by measuring 3H-thymidine uptake during the last 8 hours of 48-hour cultures, in the presence or absence of drug at various concentrations. (A) Proliferation of MM.1R cells resistant to Dex was unaffected by culture with 1 μM Dex (dotted line), but was completely inhibited by less than 1.0 μM NVP-LAQ824 (solid line). (B-C) RPMI 8226 MM cells resistant to Dox (Dox40 cells), Mit (MR20 cells), or Mel (LR5 cells) were inhibited in a dose-dependent manner by NVP-LAQ824 at concentrations of 0 (□), 10 nM (▪), 100 nM (▤), and 1 μM ( ), as demonstrated by thymidine uptake and MTS assay. Error bars represent ± 1 SD of a triplicate experiment. (D) Percent of annexin-positive HMCLs (▪), RPMI 8226, U266, MM.1S and dexamethasone-resistant MM.1R cells were quantitated after 24-hour incubation with 0.01 to 1 μM NVP-LAQ824.
), as demonstrated by thymidine uptake and MTS assay. Error bars represent ± 1 SD of a triplicate experiment. (D) Percent of annexin-positive HMCLs (▪), RPMI 8226, U266, MM.1S and dexamethasone-resistant MM.1R cells were quantitated after 24-hour incubation with 0.01 to 1 μM NVP-LAQ824.
NVP-LAQ824 causes apoptosis in human myeloma cell lines (HMCLs) resistant to conventional chemotherapy. (A-C) The effect of NVP-LAQ824 on DNA synthesis in MM cell lines was determined by measuring 3H-thymidine uptake during the last 8 hours of 48-hour cultures, in the presence or absence of drug at various concentrations. (A) Proliferation of MM.1R cells resistant to Dex was unaffected by culture with 1 μM Dex (dotted line), but was completely inhibited by less than 1.0 μM NVP-LAQ824 (solid line). (B-C) RPMI 8226 MM cells resistant to Dox (Dox40 cells), Mit (MR20 cells), or Mel (LR5 cells) were inhibited in a dose-dependent manner by NVP-LAQ824 at concentrations of 0 (□), 10 nM (▪), 100 nM (▤), and 1 μM ( ), as demonstrated by thymidine uptake and MTS assay. Error bars represent ± 1 SD of a triplicate experiment. (D) Percent of annexin-positive HMCLs (▪), RPMI 8226, U266, MM.1S and dexamethasone-resistant MM.1R cells were quantitated after 24-hour incubation with 0.01 to 1 μM NVP-LAQ824.
), as demonstrated by thymidine uptake and MTS assay. Error bars represent ± 1 SD of a triplicate experiment. (D) Percent of annexin-positive HMCLs (▪), RPMI 8226, U266, MM.1S and dexamethasone-resistant MM.1R cells were quantitated after 24-hour incubation with 0.01 to 1 μM NVP-LAQ824.
Effect of Dex and IL-6 on response of MM cells to NVP-LAQ824
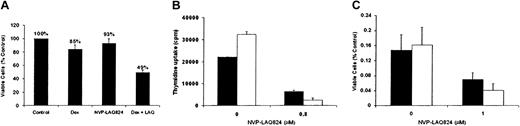
To determine whether the effects of NVP-LAQ824 are additive with conventional therapies, we next examined the effect of Dex (0.025 μM) together with NVP-LAQ824 (0.025 μM) on viability of Dex-sensitive MM.1S cells. As shown in Figure 3A, Dex or NVP-LAQ824 alone produced only 15% and 7% growth inhibition, respectively, but produced 51% inhibition when used in combination. These results have been confirmed 3 times using both thymidine uptake and MTS colorimetric proliferation assays, indicating that the effects of both drugs together are greater than the effects of either alone.
NVP-LAQ824 displays synergy with dexamethasone against MM cells, and NVP-LAQ824 induced cytotoxicity is not overcome by interleukin-6. The inhibitory effects of Dex and NVP-LAQ824 at low concentrations (25 nM), alone and in combination, are shown by MTS assay (A). The sum of the inhibition of each drug alone is 22% (15% inhibition by Dex and 7% inhibition by NVP-LAQ824), significantly less than both drugs (51% viability); (B-C) exogenous IL-6, 100 ng/mL (□) does not protect MM.1S cells (B) or freshly isolated patient cells (C) from the inhibitory effects of NVP-LAQ824, compared with media alone (▪). Error bars represent 1 SD of a triplicate experiment.
NVP-LAQ824 displays synergy with dexamethasone against MM cells, and NVP-LAQ824 induced cytotoxicity is not overcome by interleukin-6. The inhibitory effects of Dex and NVP-LAQ824 at low concentrations (25 nM), alone and in combination, are shown by MTS assay (A). The sum of the inhibition of each drug alone is 22% (15% inhibition by Dex and 7% inhibition by NVP-LAQ824), significantly less than both drugs (51% viability); (B-C) exogenous IL-6, 100 ng/mL (□) does not protect MM.1S cells (B) or freshly isolated patient cells (C) from the inhibitory effects of NVP-LAQ824, compared with media alone (▪). Error bars represent 1 SD of a triplicate experiment.
Given the known role of IL-6 as a growth factor and inhibitor of Dex-induced MM cell apoptosis, we next examined whether NVP-LAQ824 could inhibit DNA synthesis and myeloma cell proliferation in the presence of exogenous IL-6. Figure 3B-C demonstrates that NVP-LAQ824 inhibits DNA synthesis and cell growth, even in the presence of IL-6 (100 ng/mL).
In addition, NVP-LAQ824 completely inhibited the proliferation of MM.1S cells when cocultured with BMSCs, as measured by thymidine uptake, as well as the adhesion-induced up-regulation of IL-6 secretion in BMSCs triggered by coculture with MM.1S (data not shown). Thus, NVP-LAQ824 may further overcome drug resistance by inhibiting the protective effects of the bone marrow microenvironment.
Effect of NVP-LAQ824 on cell cycle profile of MM cell lines and patient MM cells
To further analyze the mechanism of NVP-LAQ824-induced apoptosis of MM cells, we first examined the cell cycle profile of plasma cells from a patient presenting with de novo plasma cell leukemia cultured for 36 hours with media alone or NVP-LAQ824 (1 μM). As shown in Figure 4A, NVP-LAQ824 increased the fraction of sub-G1 apoptotic plasma cells from 24% at baseline to 52% at 10 nM, 63% at 100 nM, and 82% at 1 μM NVP-LAQ824. Thus, induction of apoptosis in patient cells occurred at the same dose-response range noted for inhibition of proliferation. To further investigate the cell cycle effects in MM cell lines and determine reversibility of effects, MM.1S cells were cultured with 1 μM NVP-LAQ824 for 8 hours and 12 hours, washed twice, and then incubated for 24 hours prior to cell cycle examination by propidium iodide staining. As shown in Figure 4B-C, there was a significant increase in G1 phase of the cell cycle at 8 hours, followed by an irreversible increase in apoptotic sub-G1 cells at 12 hours.
NVP-LAQ824 causes G1arrest and apoptosis (increased sub-G1) in HMCLs and patient myeloma cells. (A) Freshly isolated patient cells were incubated with NVP-LAQ824 (0.01-1 μM) and examined for cell cycle profile using propidium iodide. There was a concentration-related increase in the sub-G1 fraction of patient cells, representing apoptosis. (B-C) MM.1S cells were cultured with 1 μM NVP-LAQ824 for 8 hours and 12 hours, washed twice, and then incubated for 24 hours prior to cell cycle examination by propidium iodide staining. The cell cycle changes shown in panel B are graphically represented in panel C. G1 arrest was observed at 8 hours, followed by increased sub-G1 (apoptosis) at 12 hours.
NVP-LAQ824 causes G1arrest and apoptosis (increased sub-G1) in HMCLs and patient myeloma cells. (A) Freshly isolated patient cells were incubated with NVP-LAQ824 (0.01-1 μM) and examined for cell cycle profile using propidium iodide. There was a concentration-related increase in the sub-G1 fraction of patient cells, representing apoptosis. (B-C) MM.1S cells were cultured with 1 μM NVP-LAQ824 for 8 hours and 12 hours, washed twice, and then incubated for 24 hours prior to cell cycle examination by propidium iodide staining. The cell cycle changes shown in panel B are graphically represented in panel C. G1 arrest was observed at 8 hours, followed by increased sub-G1 (apoptosis) at 12 hours.
NVP-LAQ824-induced histone hyperacetylation, up-regulation of p21, and PARP cleavage in MM cells
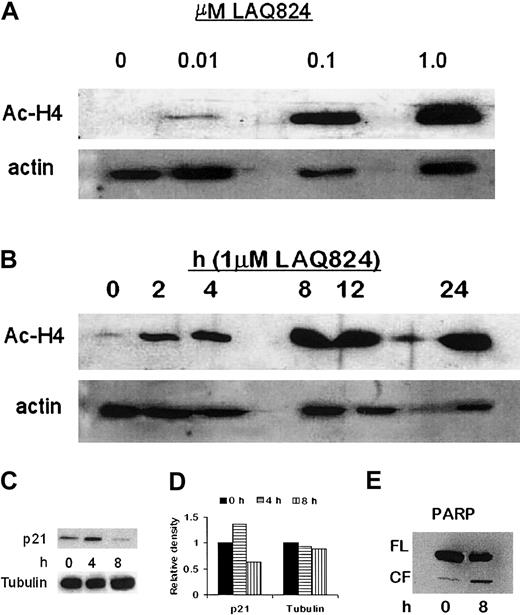
To confirm the mechanism of action of NVP-LAQ824, histone acetylation status was determined in both time- and dose-dependent fashions. CD138+ plasma cells from a patient with plasma cell leukemia were incubated with 1 μM NVP-LAQ824 for 2, 4, 8, 12, and 24 hours, or for 24 hours with 0.01, 0.1, or 1 μM NVP-LAQ824. Whole-cell extracts were then analyzed by Western blot, as described in “Materials and methods.” As shown in Figure 5A-B, NVP-LAQ824 resulted in both a time- and dose-dependent increase in histone acetylation at concentrations as low as 0.01 μM after 24 hours, and as early as 2 hours with 1 μM NVP-LAQ824.
NVP-LAQ824 induces time- and dose-dependent histone hyperacetylation, p21 up-regulation, and PARP cleavage. (A-B) NVP-LAQ824 induced both a time- and dose-dependent increase in histone acetylation at concentrations as low as 0.01 μM after 24 hours, and as early as 2 hours with 1 μM NVP-LAQ824. Immunoblotting with antiactin Ab confirmed equivalent protein loading. Ac-H4 indicates acetylated histone 4. (C) p21 up-regulation was evident as early as 4 hours in treated cells, but returned to below baseline by 8 hours. (D) Densitometric analysis demonstrated a 36% increase in p21 expression at 4 hours. (E) PARP cleavage of an 85-kDa product was observed at 8 hours, consistent with apoptosis. FL indicates full-length fragment; CF, cleavage fragment.
NVP-LAQ824 induces time- and dose-dependent histone hyperacetylation, p21 up-regulation, and PARP cleavage. (A-B) NVP-LAQ824 induced both a time- and dose-dependent increase in histone acetylation at concentrations as low as 0.01 μM after 24 hours, and as early as 2 hours with 1 μM NVP-LAQ824. Immunoblotting with antiactin Ab confirmed equivalent protein loading. Ac-H4 indicates acetylated histone 4. (C) p21 up-regulation was evident as early as 4 hours in treated cells, but returned to below baseline by 8 hours. (D) Densitometric analysis demonstrated a 36% increase in p21 expression at 4 hours. (E) PARP cleavage of an 85-kDa product was observed at 8 hours, consistent with apoptosis. FL indicates full-length fragment; CF, cleavage fragment.
The promoter for p21 transcription is regulated by histone acetylation status,22 and up-regulation of p21 has been widely reported with HDAC inhibitors.23,24 Transcriptional activation of p21 through the specificity protein 1 (Sp1) sites of the WAF1/Cip1 promoter by TSA and trapoxin has been shown to coincide with induced hyperacetylation of histone H4,25,26 and Sp1 can be a target for histone deacetylase 1-mediated transcriptional repression.27 We therefore correlated p21 expression in MM.1S cells with 1 μM NVP-LAQ824 treatment. As can be seen in Figure 5C, p21 up-regulation was evident at 4 hours in treated cells, but returned to below baseline by 8 hours. Densitometric analysis demonstrated a 36% increase in p21 expression at 4 hours (Figure 5D). Increased PARP cleavage was observed at 8 hours (Figure 5E). Immunoblotting with anti-actin Ab confirmed equivalent protein loading.
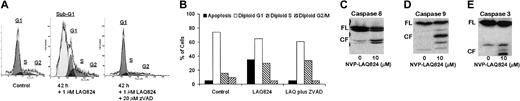
We next investigated whether the apoptotic effects of NVPLAQ824 were caspase dependent (Figure 6A-B). MM.1S cells were incubated with 1 μM NVP-LAQ824 for 18 and 42 hours, with or without the pancaspase inhibitor ZVAD-FMK. There was 35% apoptosis at 18 hours (not shown) and 63% apoptosis at 42 hours, which was in both cases completely inhibited by ZVAD-FMK. The apoptotic signaling triggered by NVP-LAQ824 is therefore caspase dependent. To confirm caspase activation and further characterize the signaling pathways involved, MM.1S cells were incubated with 1 μM NVP-LAQ824 for 24 hours, and examined for caspase cleavage by Western blot as described in “Materials and methods.” As shown in Figure 6C-E, NVP-LAQ824 increased cleavage of caspases-8, -9, and -3.
NVP-LAQ824-induced apoptosis is caspase dependent. (A-B) MM.1S cells were incubated with 1 μM NVP-LAQ824 for 42 hours, with or without the pancaspase inhibitor ZVAD. Apoptosis at 42 hours was completely inhibited by ZVAD. Sub-G1 (▪), G1 (□), S (▧), and G2 (▨). (C-E) A 24-hour incubation with NVP-LAQ824 resulted in cleavage of pro-caspases-8, -9, and -3. FL indicates full-length fragment; CF, cleavage fragment.
NVP-LAQ824-induced apoptosis is caspase dependent. (A-B) MM.1S cells were incubated with 1 μM NVP-LAQ824 for 42 hours, with or without the pancaspase inhibitor ZVAD. Apoptosis at 42 hours was completely inhibited by ZVAD. Sub-G1 (▪), G1 (□), S (▧), and G2 (▨). (C-E) A 24-hour incubation with NVP-LAQ824 resulted in cleavage of pro-caspases-8, -9, and -3. FL indicates full-length fragment; CF, cleavage fragment.
NVP-LAQ824 inhibits proteasome activity and NF-κB activation
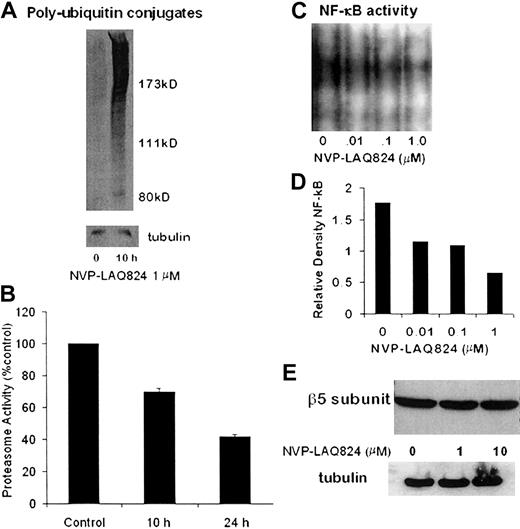
To determine the effect of NVP-LAQ824 treatment on the activity of the 26S proteasome, MM.1S cells were processed after incubation for 10 hours, and whole cell extracts were analyzed for levels of polyubiquitin conjugates by Western blot. As shown in Figure 7A, there was a significant increase in polyubiquitin conjugates at 10 hours. To further characterize the effects on the 20S proteasome, MM.1S cells were processed after incubation for 10 and 24 hours with NVP-LAQ824, and chymotrypsin-like proteasome activity was measured as described in “Materials and methods.” NVPLAQ824 induced a time-dependent decrease in 20S proteasome activity, as shown in Figure 7B. Chymotrypsin-like proteasome activity was 70% at 10 hours (P = .001) and 42% at 24 hours (P = .0003).
NVP-LAQ824 inhibits proteasome activity. (A) Whole-cell extracts were analyzed for levels of polyubiquitin conjugates by Western blot. Polyubiquitin conjugates were significantly increased at 10 hours, indicating inhibition of the 26S proteasome. (B) NVP-LAQ824 induced a time-dependent decrease in 20S proteasome activity, as determined by chymotrypsin-like proteasome activity at 10 hours (P = .001) and 24 hours (P = .0003). (C-D) NVP-LAQ824 inhibits NF-κB activation in a dose-dependent fashion, determined by EMSA and measured by densitometry. (E) There was no change in the levels of β5 proteasome subunits, even after 24-hour incubation with NVP-LAQ824, as determined by Western blot.
NVP-LAQ824 inhibits proteasome activity. (A) Whole-cell extracts were analyzed for levels of polyubiquitin conjugates by Western blot. Polyubiquitin conjugates were significantly increased at 10 hours, indicating inhibition of the 26S proteasome. (B) NVP-LAQ824 induced a time-dependent decrease in 20S proteasome activity, as determined by chymotrypsin-like proteasome activity at 10 hours (P = .001) and 24 hours (P = .0003). (C-D) NVP-LAQ824 inhibits NF-κB activation in a dose-dependent fashion, determined by EMSA and measured by densitometry. (E) There was no change in the levels of β5 proteasome subunits, even after 24-hour incubation with NVP-LAQ824, as determined by Western blot.
The HDAC inhibitor sodium butyrate has been shown to inhibit NF-κB activity,28 and we therefore next investigated the potential effects of NVP-LAQ824 on NF-κB activity. Using the EMSA assay (Figure 7C) analyzed with densitometry (Figure 7D), we show that NVP-LAQ824 inhibited NF-κB activation in a dose-dependent fashion at 24 hours.
Possible mechanisms of proteasome inhibition include transcriptional dysregulation of proteasome subunits and/or ubiquitin-ligase pathways, or direct enzymatic inhibition by NVP-LAQ824. The β5 and LMP7 proteasome subunits mediate the chymotryptic activity of the 20S proteasome, and levels of β5 and LMP7 proteasome subunits were therefore examined by Western blot. Figure 7E shows there was no change in the levels of β5 proteasome subunits, even after 24-hour incubation with NVP-LAQ824. Levels of LMP7 were also unchanged (data not shown). The chymotryptic chromogenic assay substrate does not require ubiquitination for proteasome-mediated degradation, and it is therefore very unlikely the inhibition we observed was due to interference with the ubiquitin-ligase pathways.
NVP-LAQ824 suppresses in vivo tumor growth in an immunodeficient mouse model
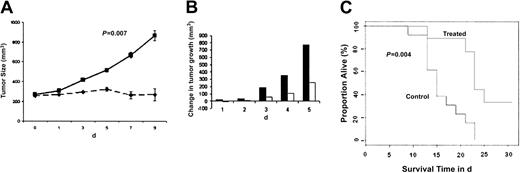
Having shown the signaling mechanisms mediating the anti-MM effects of NVP-LAQ824 in vitro, we next determined whether these in vitro effects correlate with the in vivo activity of NVP-LAQ824 using an immunodeficient mouse model. Immunodeficient BNX mice were inoculated subcutaneously in the flank with 3 × 107 RPMI 8226 MM cells in 100 μL RPMI 1640 medium, together with 100 μL Matrigel. Subcutaneous tumors became palpable in 90% of mice within 3 days and in all mice within 8 days, allowing randomization of mice to either treatment with NVP-LAQ824 or normal saline control cohorts. The data were log-transformed and modeled as a simple linear growth curve. There was no significant difference in the baseline intercept between control and treated groups (P = .83). Daily intraperitoneal administration of NVP-LAQ824 (25 mg/kg) significantly reduced MM tumor growth (Figure 8A-B) and increased survival (Figure 8C), compared with the control group treated with normal saline vehicle only.
NVP-LAQ824 inhibits tumor growth in a BNX myeloma model. Immunodeficient BNX mice were inoculated subcutaneously in the flank with 3 × 107 RPMI 8226 MM cells in 100 μL RPMI 1640 medium, together with 100 μL Matrigel. Daily intraperitoneal administration of NVP-LAQ824 (25 mg/kg) starting after the development of measurable tumor significantly reduced MM tumor growth (A-B) (P = .007), and increased survival (C) (P = .004), compared with the control group treated with vehicle (normal saline) only. (A) Treated (--); control (—). (B) Treated (□); control (▪). Error bars in panel A represent ± 1 SD of 9 mice in the treated group and 13 mice in the control group.
NVP-LAQ824 inhibits tumor growth in a BNX myeloma model. Immunodeficient BNX mice were inoculated subcutaneously in the flank with 3 × 107 RPMI 8226 MM cells in 100 μL RPMI 1640 medium, together with 100 μL Matrigel. Daily intraperitoneal administration of NVP-LAQ824 (25 mg/kg) starting after the development of measurable tumor significantly reduced MM tumor growth (A-B) (P = .007), and increased survival (C) (P = .004), compared with the control group treated with vehicle (normal saline) only. (A) Treated (--); control (—). (B) Treated (□); control (▪). Error bars in panel A represent ± 1 SD of 9 mice in the treated group and 13 mice in the control group.
There is a significantly different rate of growth between control and treated groups (P = .007). The log-rank test P value comparing the 2 curves is .004. The median survival time for the animals in the control group was 15 days. Among the animals in the treated group, one mouse died before 15 days (1/9). The P value from the Fisher exact test comparing these 2 proportions (1/9 vs 8/13) was .03, indicating a statistically significant difference in the proportion of mice alive at 15 days between the 2 groups. No significant toxicity, as evidenced by lack of weight changes, was observed in any treatment groups. Evaluation of terminal bleeds did not reveal any differences between hematologic parameters of treated mice compared with control mice. Histologic examination of the liver, spleen, bone marrow, kidney, lungs, and gut from 6 treated mice did not reveal any toxic changes to these vital organs (data not shown).
Discussion
There is growing evidence that HDAC inhibitors will be useful in the treatment of MM.10,29 Previous experiments in our laboratory have shown that SAHA induces apoptosis in MM and Waldenstrom macroglobulinemia at low micromolar concentrations.30 In this study, we have shown that NVP-LAQ824 inhibits proliferation and induces apoptosis of myeloma cell lines and freshly isolated patient myeloma cells at nanomolar concentrations. Apoptosis has been observed in cell lines resistant to Dex, Dox, Mit, and Mel. Importantly, neither exogenous IL-6 nor adhesion to BMSCs confers cellular resistance to NVP-LAQ824 despite minimal toxicity against BMSCs.
Induction of CDKN1A has previously been shown to be necessary for HDAC inhibitor-induced G1 arrest.31,32 Similarly, in this current study we observed transient up-regulation of p21 coinciding with G1 arrest after treatment with NVP-LAQ824. Cell death signaling pathways subsequent to p21 up-regulation involve cleavage of pro-caspase-8, pro-caspase-9, pro-caspase-3, and PARP cleavage of an 85-kDa product. Caspase-dependent cytotoxicity has previously been seen with sodium butyrate, CBHA, and trichostatin A,33-35 in contrast to SAHA-induced cell death, which is not caspase dependent.30,36 Butyrate and trichostatin A have been shown to induce apoptosis in Jurkat lymphoid and LIM 1215 colorectal cancer cell lines in a manner that is strictly dependent on new protein synthesis (within 10 hours). This led to conversion of the proenzyme form of caspase-3 to the catalytically active effector protease (within 16 hours) and apoptotic death within 24 hours.37 However, up-regulation of procaspases and uncleaved PARP was not seen, nor was it seen in our current study, suggesting that these events are not directly a result of increased procaspase transcriptional activity.
Synergy was observed between Dex and NVP-LAQ824. This synergy can be at least partly explained by apoptotic signaling. Dex-induced apoptosis involves activation of RAFTK (related adhesion focal tyrosine kinase), as well as release of mitochondrial Smac (second mitochondrial activator of caspase) and activation of caspase-9, but not caspase-8.38,39 Release of mitochondrial Smac into the cytosol results in neutralization of the inhibitory effects of IAPs (inhibitors of apoptosis proteins) on caspase-9, providing a mechanism for Dex to augment caspase-9-mediated NVP-LAQ824-induced cytotoxicity. Caspase-8 activation by NVP-LAQ824 provides an additional apoptotic signaling pathway to those already induced by Dex.
The acetyl transferase activities of p300 and calcium-binding protein have been associated with p53-mediated transcription.40 However, we found that MM cell lines with mutated p53 such as RPMI 822641,42 and U26643 were sensitive to NVP-LAQ824, indicating that increased p53 activity is not essential to the cytotoxicity of NVP-LAQ824.
NVP-LAQ824 induced inhibition of 20S and 26S proteasome function, as demonstrated by reduced chymotrypsin-like activity and increased levels of cellular polyubiquitin conjugates. At 24 hours, 20S activity in NVP-LAQ824-treated cells was approximately 40% of control cells, similar to the inhibition previously observed by Yin et al28 using 4 mM of sodium butyrate. The fluorescent substrate assay measures the enzymatic activity of the core 20S proteasome subunits β5/LMP7, but there was no alteration in the levels of these subunits by Western blot, as expected given that the cellular half-lives are 7 to 10 days. In the phase 1 and 2 clinical trials for the proteasome inhibitor PS-341, the maximal effect on proteasome proteolytic activity in peripheral blood mononuclear cells was 65% inhibition, which was achieved at the maximum recommended phase 2 dose.44-46 Therefore, although NVP-LAQ824 resulted in only 60% 20S proteasome inhibition at 24 hours, this may contribute significantly to the cytotoxic effects of NVP-LAQ824 against MM cells in vivo. As reported previously with sodium butyrate, inhibition of NF-κB was also seen after treatment with NVP-LAQ824. After 24 hours of treatment of MM cells with NVP-LAQ824, there was a dose-related inhibition of NF-κB activity. The cytotoxicity of NVP-LAQ824 may therefore be mediated, at least in part, by NF-κB inhibition as a result of proteasome inhibition.
The role of histone deacetylase in oncogenesis may be a consequence of abnormal transcription factors usurping the normal mechanisms regulating gene transcription. For example, in t(15; 17)- and t(8;21)-positive acute myeloid leukemia, the abnormal transcription factor protein products form a corepressor complex with HDAC to block cellular differentiation.19,47-49 In MM, overexpression of c-myc is more common in aggressive disease and myeloma cell lines.50,51 HDAC inhibition represses c-myc expression, and therefore HDAC inhibitors could target a specific oncogene pathogenic to MM. Other HDAC-related myelomaspecific oncogenes may emerge with time to explain the selective efficacy of NVP-LAQ824 against MM. However, it is possible that many less specific HDAC functions serve to promote oncogenesis. For example, histone deacetylation is required to form new chromatin and stabilize chromosomes prior to mitosis.52,53 Proliferating cells may therefore be more vulnerable to the effect of HDAC inhibitors.
In conclusion, NVP-LAQ824 represents a novel hydroxamic acid-derived HDAC inhibitor with activity against MM in nanomolar concentrations, which should be readily achievable in patients. Importantly, NVP-LAQ824 inhibits MM cell growth and prolongs survival in a murine myeloma model, without toxicity to normal marrow or peripheral blood. These results support the use of NVP-LAQ824 to overcome drug resistance in multiple myeloma patients. A phase 1 clinical trial of NVP-LAQ824 in hematologic malignancies is now ongoing at the Dana-Farber Cancer Institute.
Prepublished online as Blood First Edition Paper, June 19, 2003; DOI 10.1182/blood-2003-01-0233.
P.A. and S.R. are employees of Novartis Pharmaceuticals, which hold a patent on NVP-LAQ824.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to acknowledge the help of Dr Roderick T. Bronson and Emily Wu at the Rodent Histopathology Core, Dana-Farber/Harvard Cancer Center, for mouse experiments and histopathologic studies, and Sigui Li and Edie Weller at the Department of Biostatistical Science for their help with statistical analysis.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal