The suppressor of cytokine signaling (SOCS) family of proteins has been implicated in the negative regulation of several cytokine pathways, particularly the receptor-associated tyrosine kinase/signal transducer and activator of transcription (Jak/STAT) pathways of transcriptional activation. SOCS-1 (also known as JAB and SSI-1) inhibits signaling by many cytokines. Because of the previously observed hypermethylation-associated inactivation of SOCS-1in hepatocellular carcinoma and the critical role of interleukin-6 (IL-6) as a survival factor in multiple myeloma (MM), we examined CpG island methylation of the SOCS-1 gene in MM cell lines and primary MM samples. Aberrant SOCS-1methylation was found in the IL-6–dependent MM cell lines U266 and XG1, which correlated with transcriptional silencing. Treatment of these cell lines with the demethylating agent 5-aza-2′-deoxycytidine (DAC) up-regulated SOCS-1 expression. Methylation-associated inactivation of SOCS-1 in hematopoietic cell lines correlated with greater sensitivity to the chemical JAK inhibitor AG490. Using methylation-specific polymerase chain reaction (MSP), we found that SOCS-1 is hypermethylated in 62.9% (23/35) of MM patient samples. In contrast, methylation analysis of malignant lymphomas of various histologies revealed SOCS-1 hypermethylation in only 3.2% (2/62), and there was no methylation of SOCS-1 in normal peripheral blood leukocytes or bone marrow cells. We conclude thatSOCS-1 is frequently inactivated by hypermethylation in MM patients. Silencing of the SOCS-1 gene may impair negative regulation of the Jak/STAT pathway and therefore result in greater responsiveness to cytokines, thus supporting survival and expansion of MM cells.
Introduction
Cytokines are known to play important roles in the growth and differentiation of hematopoietic cells. In particular, cytokines, the most well studied of which is interleukin-6 (IL-6), play important roles in B-cell growth and differentiation into plasma cells and are essential growth and survival factors for neoplastic cells in the pathogenesis of multiple myeloma (MM).1-3 Like other cytokines, IL-6 binds to a specific membrane receptor (IL-6R), and the IL-6/IL-6R complex induces the dimerization of the receptor subunit gp130. After receptor binding, members of the Janus kinase (Jak) family become activated, which in turn cross-phosphorylate both each other and the cytoplasmic domains of the receptors on tyrosine residues. This provides docking sites for latent transcription factors called signal transducers and activators of transcription (STATs). These transcription factors become phosphorylated after docking and dimerize before entering the nucleus and initiating transcription of target genes.4 5
Recently, a family of proteins functioning as negative regulators of Jak/STAT signaling has been identified, revealing one mechanism for the regulation of cytokine signal transduction. Proteins belonging to this family are termed suppressors of cytokine signaling (SOCS) and are characterized by the presence of a src homology (SH2) domain and a carboxyl-terminal conserved domain called the SOCS box. So far, 8 members of the SOCS family have been identified, termed SOCS-1 to SOCS-7 and CIS (cytokine-inducible SH2-containing protein).4-7SOCS-1 was discovered in 1997 by 3 independent experimental approaches. In one study, an expression library was used to identify molecules capable of blocking IL-6–mediated differentiation of murine M1 cells8; the second group used a yeast-2–hybrid screen to detect proteins able to interact with Jak29; and the third approach used a screen with antibodies to find proteins with homology with the SH2 domain of STATs.10 The SOCS-1 protein has been shown to interact directly with active Jaks by binding to their activation loop in a phosphorylation-dependent manner. The blocking of Jak activation results in inhibition of the Jak/STAT signal transduction pathway. SOCS-1 suppresses signaling by a wide variety of cytokines including IL-6, IL-4, leukemia inhibitory factor, oncostatin M, growth hormone, prolactin, thrombopoietin, and interferons.11 It has been shown that regulation of cytokine signaling by SOCS-1 is important in normal lymphocyte development and differentiation.12 13
The first report demonstrating involvement of SOCS-1 in carcinogenesis revealed that SOCS-1 is frequently silenced by hypermethylation in hepatocellular carcinoma (HCC).14Restoration of SOCS-1 in HCC cell lines by transient transfection resulted in growth suppression and induction of apoptosis.14 The silencing of gene transcription associated with aberrant DNA methylation of CpG dinucleotides in normally unmethylated gene promoter regions is the most widely studied epigenetic abnormality in tumorigenesis. The binding of protein complexes consisting of methyl-CpG–binding domains, transcriptional corepressors, chromatin remodeling proteins, and histone deacetylases to hypermethylated DNA regions result in a transcriptionally repressive chromatin state.15-17
In hematopoietic malignancies, hypermethylation of genes, includingE-cadherin, DAP-kinase, and the cell-cycle regulators p15INK4B andp16INK4A, has been established to be associated with gene inactivation.18-21 Based on methylation-associated transcriptional silencing of SOCS-1in HCC and the importance of the Jak/STAT pathway in perturbed hematopoiesis,22 we examined CpG island methylation of theSOCS-1 gene in MM cell lines and primary MM samples.
Patients, materials, and methods
MM patient and control samples
After informed consent was given, bone marrow (BM) specimens were aspirated during routine clinical assessment of 35 MM patients (28 at diagnosis, 7 at relapse; 22 men, 13 women), who presented at the University Hospital Aachen, Germany between 1995 and 2000. MM diagnosis and staging classification were made in accordance with standard criteria.23 Median age of the patients was 65 years (range, 40-79 years). Obtained were 2 control bone marrow aspirates from patients with nonmetastatic solid tumors or malignant lymphoma without bone marrow infiltration or hematopoietic dysfunction as part of the routine staging procedure. Peripheral blood (PB) samples were collected from 5 healthy volunteers. Mononuclear cells from BM and PB were separated by density gradient centrifugation prior to further analysis. Details of the lymphoma samples have been described previously.19 24
Cell culture and drug treatment
We obtained the cell lines HL60, U266, and Raji from the American Type Culture Collection (Manassas, VA) and the cell lines KG1a and K562 from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). XG1 cells were kindly provided by P. C. Heinrich (RWTH, Aachen, Germany). Cell lines KG1a and HL60 were routinely cultured in Iscove modified Dulbecco medium (IMDM; Life Technologies) with 20% fetal calf serum (FCS; Gemini Bio-Products, Woodland, CA). U266 cells were routinely cultured in RPMI 1640 (Life Technologies, Rockville, MD) with 15% FCS; K562 and Raji cells, in RPMI 1640 with 10% FCS; and XG1 cells, in IMDM with 20% FCS and 10 U/mL IL-6 (Roche, Indianapolis, IN).
For gene expression studies, U266 and XG1 cells were incubated in complete culture medium with a final concentration of 1.0 μM 5-aza-2′-deoxycytidine (DAC; Sigma, St Louis, MO). After 4 days, drug-treated and untreated cells were harvested and subjected to RNA extraction. In order to assess the sensitivity to AG490, cell lines K562, U266, and XG1 were incubated with or without a final concentration of 50 μM AG490 for 4 days prior to analysis. For STAT3 phosphorylation analysis, cell lines U266 and XG1 were initially starved for 16 hours in culture medium supplemented only with 1% FCS and no cytokine. Cells were then washed in cold phosphate-buffered saline (PBS) and resuspended in their appropriate complete medium with or without 50 μM AG490 and harvested for protein analysis after 1 hour and 24 hours.
Sodium bisulfite treatment and methylation-specific polymerase chain reaction (MSP)
Genomic DNA was isolated from cell lines and primary tissues by standard methods. The methylation status of SOCS-1 was analyzed using the MSP technique.25 This assay distinguishes unmethylated from methylated alleles in a given gene on the basis of sequence changes induced by sodium bisulfite treatment of DNA, which converts all unmethylated, but not methylated, cytosines to uracil. Subsequently, the DNA region of interest is amplified with primer pairs specific for methylated versus unmethylated DNA. ForSOCS-1 MSP analysis, approximately 1 μg of DNA was sodium bisulfite–modified, and amplified using primers that were described previously.14 Normal DNA from PB was treated in vitro withSssI methyltransferase (New England Biolabs, Beverly, MA) in order to generate a positive control for methylated alleles of SOCS-1.26
RNA isolation and reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA was isolated using a commercially available kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Approximately 3 μg RNA per sample were reverse transcribed with SuperScript reverse transcriptase (Invitrogen, Carlsbad, CA). For PCR, 1 μL cDNA was amplified usingSOCS-1 primers 5′-CCC GGA GCA TGC GCG AGA GC-3′ (sense) and 5′-TGC GGG CTC TGC TGC TGT GG-3′ (antisense) and GAPDHprimers 5′-GAC CAC AGT CCA TGC CAT CAC-3′ (sense) and 5′-GTC CAC CAC CCT GTT GCT GTA-3′ (antisense). Reactions were hot-started at 95°C for 5 minutes and held at 80°C before addition of 1.25 units Taq polymerase (Invitrogen). Temperature conditions were as follows forSOCS-1: 26 cycles of 95°C for 30 seconds, 64°C for 30 seconds, and 72°C for 30 seconds, followed by 1 cycle of 72°C for 5 minutes; and for GAPDH, 23 cycles of 95°C for 1 minute, 63°C for 1 minute, and 72°C for 1 minute, followed by 1 cycle of 72°C for 5 minutes.
Flow cytometric analysis of apoptosis
Apoptosis was assessed by annexin V–binding and counterstaining with propidium iodide (PI) using a commercially available kit (Pharmingen, San Diego, CA) according to the manufacturer's recommendations. Fluorescence analysis was performed on a BD LSR flow cytometer (Becton Dickinson, San Jose, CA).
Western blot analysis
Cell lysis and Western blot analysis for STAT3 phosphorylation were performed as described previously.14 In brief, 60 μg total protein per sample was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotted. For detection of phosphorylation of STAT3 we used anti–phospho-STAT3 antibody (New England Biolabs) according to the manufacturer's recommendations. After antibody removal, the blot was analyzed with anti-STAT3 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Results
SOCS-1 methylation status in hematopoietic cell lines and normal tissues
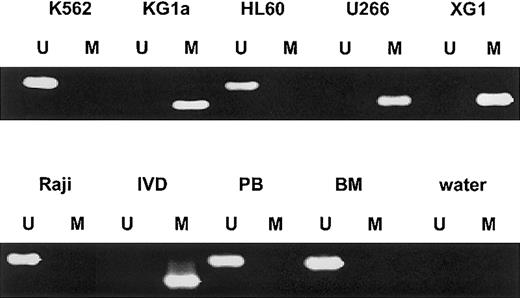
We first analyzed the SOCS-1 methylation status by MSP in various human hematopoietic cell lines (Figure1). SOCS-1hypermethylation was found in both IL-6–dependent MM cell lines U266 and XG1 and in the acute myelogenous leukemia (AML) cell line KG1a. The AML cell line HL60, the chronic myelogenous leukemia cell line K562, and the Burkitt lymphoma cell line Raji did not show aberrant methylation in that region. There was no SOCS-1methylation in normal peripheral blood mononuclear cells (PBMNCs) (n = 5) and nonmalignant bone marrow cells (n = 2).
SOCS-1 MSP analysis of hematopoietic cell lines, normal peripheral blood (PB), and nonmalignant bone marrow (BM).
Lane U indicates amplified product with primers recognizing unmethylated SOCS-1 sequence. Lane M indicates amplified product with primers recognizing methylated SOCS-1 sequence. The cell lines KG1a, U266, and XG1 show aberrant hypermethylation of the SOCS-1 CpG island. In vitro–methylated DNA (IVD) and water served as controls.
SOCS-1 MSP analysis of hematopoietic cell lines, normal peripheral blood (PB), and nonmalignant bone marrow (BM).
Lane U indicates amplified product with primers recognizing unmethylated SOCS-1 sequence. Lane M indicates amplified product with primers recognizing methylated SOCS-1 sequence. The cell lines KG1a, U266, and XG1 show aberrant hypermethylation of the SOCS-1 CpG island. In vitro–methylated DNA (IVD) and water served as controls.
SOCS-1 expression in MM cell lines
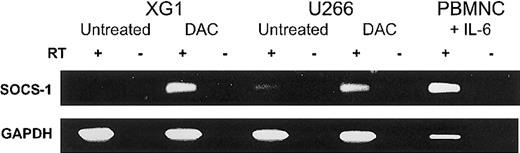
We next examined SOCS-1 expression in both MM cell lines by semiquantitative RT-PCR (Figure2). XG1 cells require the addition of exogenous IL-6,27 while U266 cells depend on an IL-6 autocrine loop.28 Despite the presence of IL-6 in the cell-culture medium, the MM cell lines showed low (U266) or undetectable (XG1) SOCS-1 transcript levels (Figure 2). For comparison, normal PBMNCs incubated in IMDM with 20% FCS and 10 U/mL IL-6 for 2 hours showed strong SOCS-1 expression. Unstimulated PBMNCs expressed SOCS-1 only at low levels (data not shown). In order to determine whether methylation was responsible for the loss of SOCS-1 expression, we incubated U266 and XG1 cells with the demethylating agent DAC (1.0 μM) for 96 hours in complete medium. DAC treatment reactivated SOCS-1expression in both MM cell lines (Figure 2).
Reactivation of SOCS-1 expression by DAC treatment in MM cell lines.
Cell lines XG1 and U266 were incubated with or without 1.0 μM DAC in complete medium for 96 hours. Peripheral blood mononuclear cells (PBMNCs) after stimulation with IL-6 for 2 hours served as positive control. The presence or absence of reverse transcriptase is indicated by RT+ or RT−, respectively. RT-PCR for GAPDH was performed for all samples to ensure the integrity of the reverse transcription reactions.
Reactivation of SOCS-1 expression by DAC treatment in MM cell lines.
Cell lines XG1 and U266 were incubated with or without 1.0 μM DAC in complete medium for 96 hours. Peripheral blood mononuclear cells (PBMNCs) after stimulation with IL-6 for 2 hours served as positive control. The presence or absence of reverse transcriptase is indicated by RT+ or RT−, respectively. RT-PCR for GAPDH was performed for all samples to ensure the integrity of the reverse transcription reactions.
Sensitivity of hematopoietic cell lines to AG490
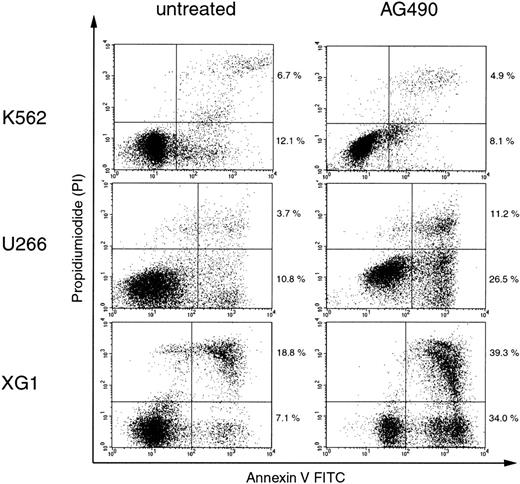
It was previously demonstrated that in HCC cell lines in whichSOCS-1 was silenced by hypermethylation, restoration of SOCS-1 by transient transfection resulted in growth suppression and induction of apoptosis. These effects were also achieved by treatment with the chemical Jak inhibitor AG490.14 In order to investigate the activation of the Jak/STAT pathway in the context of hypermethylation-associated silencing of SOCS-1in hematopoietic cell lines, we treated K562, U266, and XG1 cells with 50 μM AG490 and determined the percentage of apoptotic cells after 96 hours by annexin V–binding and PI counterstaining. Figure3 shows that AG490 treatment induced apoptosis in U266 and XG1 cells, while it had only little effect on K562 cells. Thus, cell lines that carry a hypermethylatedSOCS-1 gene showed greater sensitivity to apoptosis induction by AG490 compared with cell lines without SOCS-1hypermethylation.
AG490 induces apoptosis in cell lines with hypermethylation-associated silencing of SOCS-1.
Cells were incubated with or without 50 μM AG490 for 96 hours before annexin V/PI staining and analysis by flow cytometry. Cells that are annexin V+ and PI− (lower right quadrants) represent early apoptotic cells, and cells that are annexin V+ and PI+ (upper right quadrants) represent late apoptotic and/or necrotic cells. The percentages of cells in the upper and lower right quadrants are indicated.
AG490 induces apoptosis in cell lines with hypermethylation-associated silencing of SOCS-1.
Cells were incubated with or without 50 μM AG490 for 96 hours before annexin V/PI staining and analysis by flow cytometry. Cells that are annexin V+ and PI− (lower right quadrants) represent early apoptotic cells, and cells that are annexin V+ and PI+ (upper right quadrants) represent late apoptotic and/or necrotic cells. The percentages of cells in the upper and lower right quadrants are indicated.
STAT3 phosphorylation
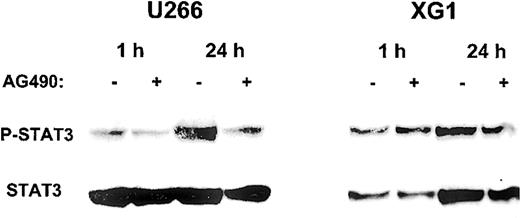
Having demonstrated that AG490 is capable of inducing apoptosis in the MM cell lines U266 and XG1, we then analyzed the impact of AG490 on the phosphorylation status of STAT3, which represents a downstream event in the Jak/STAT pathway. Figure 4shows that AG490 was able to inhibit STAT3 phosphorylation in U266 cells, while there were only minor effects in XG1 cells.
Inhibition of STAT3 phosphorylation by AG490 in MM cell lines.
After starvation for 16 hours, cells were resuspended in complete culture medium with (+) or without (−) 50 μM AG490 and incubated for 1 hour and 24 hours. Cell lysates were analyzed by Western blot with anti–phospho-STAT3 antibody, and subsequently with anti-STAT3 antibody.
Inhibition of STAT3 phosphorylation by AG490 in MM cell lines.
After starvation for 16 hours, cells were resuspended in complete culture medium with (+) or without (−) 50 μM AG490 and incubated for 1 hour and 24 hours. Cell lysates were analyzed by Western blot with anti–phospho-STAT3 antibody, and subsequently with anti-STAT3 antibody.
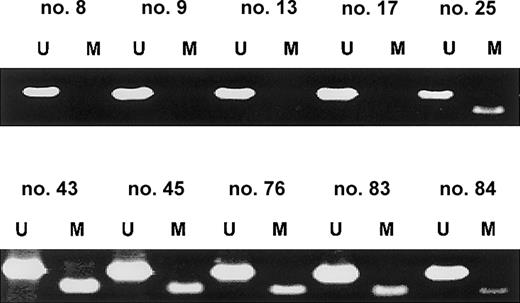
SOCS-1 methylation in primary patient samples
In order to examine the prevalence of SOCS-1methylation in noncultured cells, we analyzed the SOCS-1methylation status in primary patient samples by MSP. Aberrant methylation was present in 62.9% (23/35) of MM cases. Representative MSP results are shown in Figure 5. MSP analysis of malignant lymphomas of various histologies (55 B-cell lymphomas, 4 T-cell lymphomas, and 3 Hodgkin lymphomas) revealedSOCS-1 hypermethylation in only 3.2% (2/62) (data not shown). These 2 cases included one large B-cell lymphoma and one T-lymphoblastic lymphoma.
SOCS-1 is frequently hypermethylated in MM patient samples.
Representative MSP results from primary MM samples are shown. Lane U indicates amplified product with primers recognizing unmethylated SOCS-1 sequence. Lane M indicates amplified product with primers recognizing methylated SOCS-1 sequence. None of these samples were enriched for plasma cells, so the occurrence of both signals in the same sample indicates either the presence of nonmalignant cells in the bone marrow aspirate or heterogeneity of aberrant methylation within the MM cell population.
SOCS-1 is frequently hypermethylated in MM patient samples.
Representative MSP results from primary MM samples are shown. Lane U indicates amplified product with primers recognizing unmethylated SOCS-1 sequence. Lane M indicates amplified product with primers recognizing methylated SOCS-1 sequence. None of these samples were enriched for plasma cells, so the occurrence of both signals in the same sample indicates either the presence of nonmalignant cells in the bone marrow aspirate or heterogeneity of aberrant methylation within the MM cell population.
Discussion
Stimulation of the Jak/STAT pathway by cytokines or growth factors results in transactivation of target genes. Under physiologic conditions, SOCS-1 is simultaneously up-regulated and blocks Jak activation, leading to termination or attenuation of the signal. If hypermethylation-associated silencing of SOCS-1 occurs in tumor cells, this negative feedback loop is disrupted, and consequently those cells may become more responsive to stimulation by cytokines and growth factors that use the Jak/STAT pathway for signal transduction. In MM pathogenesis, IL-6 has been identified as an essential growth and survival factor. Loss of SOCS-1 function may therefore result in increased IL-6 signal transduction, and thus support survival and expansion of MM cells. In this report, we have found that in the MM cell lines U266 and XG1, aberrant SOCS-1 hypermethylation is associated with transcriptional silencing, and treatment of these cell lines with the demethylating agent DAC resulted in SOCS-1re-expression.
The importance of Jak activation in those cell lines that carry a hypermethylated SOCS-1 gene is underscored by their sensitivity to the chemical Jak inhibitor AG490. These results are in accordance with the finding that in HCC cell lines silencing ofSOCS-1 by hypermethylation was associated with constitutive activation of Jak2.14 When we examined STAT3 as a downstream target of the Jak/STAT pathway, we detected that in U266 cells, AG490 is capable of inhibiting the phosphorylation of STAT3. Similar effects of AG490 have been described in the MM cell line XG2.29
In our study, aberrant methylation within the SOCS-1 CpG island was identified in 62.9% of MM patient samples, but in only 3.2% of primary tissues from different malignant lymphomas. This finding is in accordance with the importance of the Jak/STAT pathway in MM pathogenesis compared with other lymphoid neoplasms.3Previous molecular studies have largely focused on genetic aberrations in MM.2,30 Important changes include chromosomal translocations involving the immunoglobulin heavy chain locus on chromosome 14q32 and various partner genes such as cyclin D1, cyclin D3, fibroblast growth factor receptor 3, and c-maf, as well as mutations of N-ras and K-ras.2,30 Additionally, recent reports have identified hypermethylation ofp15INK4B, p16INK4A, andDAP-kinase in MM patients and cell lines,21,31-34 demonstrating that epigenetic silencing of genes that effect cell-cycle regulation and apoptosis pathways is an additional mechanism of gene inactivation in MM. In addition to its physiologic role in antagonizing cytokine signaling, ectopic expression of SOCS-1 was shown to be capable of blocking the transforming activity of oncogenic forms of Jak2, kit, and abl.35,36 Furthermore, the TEL-Jak2 and v-abl–mediated phosphorylation of STAT proteins was inhibited by SOCS-1overexpression.36 Rottapel et al concluded from their data that inactivation of SOCS-1 may contribute to cellular transformation, and that SOCS-1 is a candidate tumor suppressor gene, whose loss of function may favor the development of hematopoietic malignancies.36
Our findings reveal an important novel epigenetic event in the pathogenesis of MM. Silencing of SOCS-1 may result in increased responsiveness of the neoplastic clone to IL-6 signaling, thus supporting survival and expansion of MM cells. These findings provide not only new insights into the pathogenesis of MM but also support novel approaches in MM therapy, since hypermethylation-associated gene silencing is a potentially reversible phenomenon.37 Demethylating agents such as DAC have been shown to exert clinical efficacy in patients with AML and myelodysplastic syndrome.38,39 The high prevalence ofSOCS-1 hypermethylation in MM indicates that this pathway may be a potential target for therapeutic intervention. While reactivation of genes like p15INK4B andp16INK4A could contribute to reconstitution of disrupted cell-cycle regulation, reactivation of SOCS-1might functionally restore the endogenous negative feedback loop of cytokine signaling. Since IL-6 has been shown to contribute to MM resistance to conventional therapy options such as corticosteroids,40 inhibition of IL-6 signaling and other cytokines by SOCS-1 reactivation could potentially overcome such resistance. The combination of demethylating agents with conventional therapeutics or tyrosine kinase inhibitors might be a promising new strategy in MM therapy.
We thank L. Meszler (Johns Hopkins Oncology Cell Imaging Facility) for expert technical assistance.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-06-1735.
Supported by National Institutes of Health (NIH) grant CA84986, and by a grant from the Rheinisch-Westfaelische Technische Hochschule Aachen (to O.G.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James G. Herman, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, 1650 Orleans St, Room 543, Baltimore, MD 21231; e-mail: hermanji@jhmi.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal