CD5+ diffuse large B-cell lymphomas (DLBLs) have recently been described as a particular subgroup of DLBLs. Classical banding and interphase cytogenetic analyses targeting ATM, TP53, and P16INK4a genes and theD13S25 locus from 13 CD5+ DLBLs were compared with 55 CD5− DLBLs. Additionally, analysis of somatic mutations of the immunoglobulin heavy chain variable region (IgVH) genes were performed in CD5+ DLBLs. CD5+DLBLs were somatically mutated (7 of 8 cases) and were negative for t(11;14)(q13;q32) and t(14;18)(q32;q21), whereas t(3;14)(q27;q32) was found in only one tumor. Trisomy 3 and gains on chromosomes 16/16p and 18/18q were significantly overrepresented in CD5+DLBLs. No ATM deletions were detected. The prevalence of deletions at the D13S25 locus was significantly higher in CD5+ DLBLs (4 of 12 [33%]) compared with CD5− DLBLs (4 of 42 [10%]), as were p16INK4a deletions (33% versus 8%). On the basis of these findings, CD5+ DLBLs are likely to arise from the same progenitor cell as the mutated variant of CD5+ lymphocytic lymphoma/B-cell chronic lymphocytic leukemia (B-CLL).
Introduction
Diffuse large B-cell lymphomas (DLBLs) are the most common type of non-Hodgkin lymphoma (NHL).1 They represent a heterogeneous category with respect to morphology, immunophenotype, cytogenetics, and gene expression profiles.2-4 Studies identified de novo DLBL with CD5 expression (CD5+ DLBL) as a particular subgroup, suggesting that these tumors may be different from both CD5+ DLBL developing in the setting of small lymphocytic lymphoma/chronic lymphocytic leukemia (Richter syndrome) and CD5− DLBL.5-9 Their clinical characteristics are elderly onset, female predominance, frequent involvement of extranodal sites, and a poor clinical course.9 No BCL1 rearrangements or CyclinD1 expression were demonstrated in those tumors, excluding a possible association with the blastoid variant of mantle-cell lymphoma (MCL).5,7,10 Moreover, most CD5+ DLBLs have been shown to carry somatic mutations in their immunoglobulin heavy chain variable region (IgVH) genes, further separating them from MCL.11 Interestingly, in one study, the extent of somatic mutations in de novo CD5+ DLBL was determined to be similar to that of CD5+ (mutated) B-cell chronic lymphocytic leukemia (B-CLL), thus allowing for speculations on the derivation of CD5+ DLBL and CD5+ B-CLL from the same cell of origin, the B-1 lymphocyte.6
Until now, only rare genetic data,5 and especially no cytogenetic data on CD5+ DLBL, have been reported. In this work, we describe classical cytogenetic results in 13 CD5+ DLBLs. Because deletions at the D13S25locus and of the ATM gene are frequent findings in CD5+ B-CLL,12 we analyzed these chromosomal regions by interphase cytogenetics (fluorescence in situ hybridization [FISH]) and also performed FISH forTP53 and P16INK4a genes, the inactivation of which has been strongly associated with lymphoma progression.13 These data were compared with respective findings in 55 nodal CD5− DLBLs.
Study design
Thirteen CD5+ DLBLs were compared with 55 CD5− DLBLs according to their morphologic features, classical and interphase cytogenetic data, and clinical characteristics. In addition, the molecular configuration of the IgVH genes in CD5+ DLBL was determined. Classification of the tumors was performed on high-quality Giemsa-stained slides according to the World Health Organization (WHO) classification.2
Cytogenetic investigations were done following established protocols,14 and metaphases were evaluated according to the International System for Cytogenetic Nomenclature (ISCN) guidelines.15
Bicolor interphase FISH for TP53, D13S25, and BCL1 (cases 9 and 10) was carried out according to the manufacturer's advice (VYSIS, Stuttgart, Germany). TheP16INK4a deletion status was analyzed by a cosmid contig of approximately 200 kb that had previously been shown to reliably detect genomic deletions of the INK4a cluster region.16 For deletions affecting ATM,biotin-dUTP (deoxyuracil triphosphate)–labeled P1-derived artificial chromosome (PAC) probes specific for the ATM gene locus17 were applied as previously described.18 19 Signal visualization was accomplished by using a Zeiss Axioskop2 fluorescence microscope (ZEISS, Jena, Germany), and illustrations were made by using the ISIS imaging system (MetaSystems, Altlussheim, Germany).
Genomic DNA of 8 CD5+ DLBLs was extracted from cryopreserved tissue blocks. VH-DH-JH gene rearrangements were amplified by polymerase chain reaction (PCR) by using family-specific VH forward primers and consensus JH reverse primers according to the modified protocol of Küppers et al20 and Campbell et al.21 PCR products were ligated into pPCR-Script Amp SK (+) cloning vector and transformed into Epicurian Coli XL-Gold Kan ultracompetent cells (Stratagene, La Jolla, CA). Usually, 9 clones per case were sequenced bidirectionally and compared with the nucleotide sequence database Entrez Blast (www.ncbi.nlm.nih.gov/blast/). VH-DH-JH rearrangements were analyzed by using DNAPLOT (www.dnaplot.de/).
Results and discussion
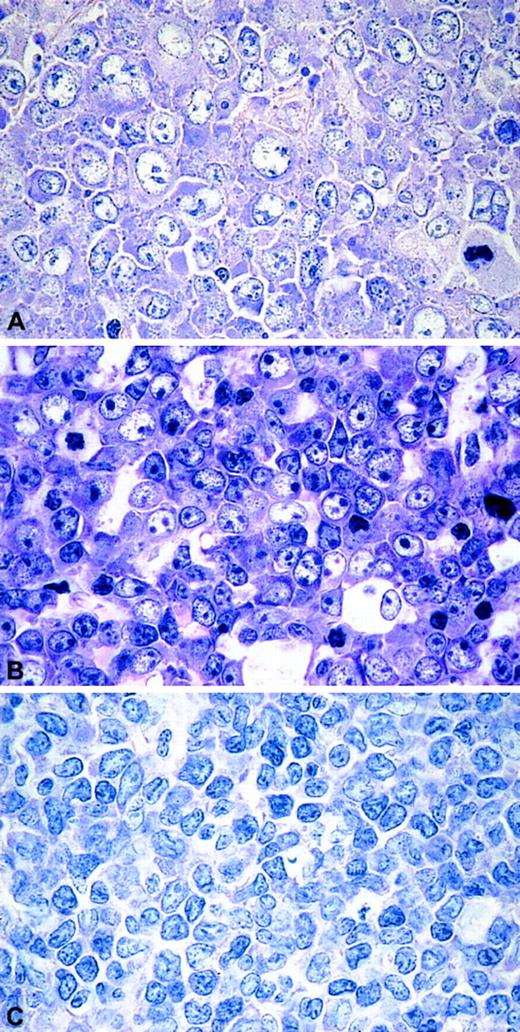
On morphology, the series presented here displayed a remarkable heterogeneity. The CD5+ DLBLs consisted of 8 centroblastic and 3 immunoblastic DLBLs. Two tumors (cases 9 and 10) displayed particular features: their neoplastic cells were medium- to large-sized with scant cytoplasm and slightly irregular nuclei, giving the morphologic impression of blastoid MCL (Figure1). All cases were CD5+ and negative for CD23 and CyclinD1. IgD expression was found in 5 of 13 (39%) CD5+ tumors. In the CD5− DLBLs, 8 of 39 cases tested were IgD+ (difference not statistically significant). Cases 6 and 12, in addition, stained weakly for CD10. Nuclear expression of BCL6 was demonstrated in more than 10% of cells in 12 of 12 CD5+ DLBLs and in 46 of 54 (85%) CD5− DLBLs. There was also no statistically significant difference between the groups, if only high BCL6 expression (> 60% of cells) was taken into account (4 of 12 [33%] CD5+DLBL versus 29 of 54 [54%] CD5− DLBL).
The morphologic spectrum of CD5+DLBLs.
(A) DLBL centroblastic. (B) DLBL immunoblastic. (C) DLBL unclassified. Giemsa stain, original magnifications × 400.
The morphologic spectrum of CD5+DLBLs.
(A) DLBL centroblastic. (B) DLBL immunoblastic. (C) DLBL unclassified. Giemsa stain, original magnifications × 400.
All 68 patients enrolled in this study were treated by a conventional anthracycline-containing regimen (cyclophosphamide, hydroxydaunomycin, vincristine, and prednisone [CHOP]) with or without radiotherapy. In contrast to recently published clinical data,9 there was no statistically significant difference in survival between the CD5+ and CD5− DLBLs.
Analysis of somatic hypermutations of IgVH genes revealed a germ line pattern in one case and somatically mutated or hypermutated IgVH genes in 3 (37%) and 4 (50%) of 8 cases, respectively (Table1). VH4 gene family usage was demonstrated in 7 of 8 (88%) tumors, and VH4-34 gene usage in 4 of 7 cases. Only case 6 showed ongoing somatic mutations. These findings are in accordance with published data, suggesting that the cell of origin of CD5+ DLBL predominantly is of postgerminal origin.6,8 11
Cytogenetic and interphase cytogenetic data of CD5+ DLBL
| Case no. . | Diagnosis . | Karyotype . | IgVH status . | D13S25 . | ATM . | TP53 . | P16INK4a . |
|---|---|---|---|---|---|---|---|
| 1 | CB | 33-50,X,?del(X)(q13)[5] | ND | n | n | n | n |
| 2 | CB | 43-50,XX,t(3;14)(q27;q32),del(6)(q15q21),+11,+18,+19[11] | ND | ND | ND | ND | ND |
| 3 | CB | 86-91P〈4n〉,XXXX,−1[4],del(1)(q11)[4],+3[3],t(3;14)(p21;q32)[3],der(6)t(1;6)(q21;q21)[4], +der(6)t(1;6)(q21;q21)[3],t(8;11)(p23;p12)x2[4],der(16)t(1;16)(q21;q21)[4], +der(16)t(1;16)(q21;q21)[3],−17[4],−17[3][cp4] | Mutated | tet | tet | del | del |
| 4 | IB | 44/48,XY,−6,+21[2] | Hypermutated | del | n | n | n |
| 5 | IB | 43-45,XY,del(2)(q21q31)[15],−6[15],del(9)(p13)[15],del(10)(q22q24)[15],+mar[2][cp15] | ND | del | n | n | del |
| 6 | CB | 46-52,XY,+2[2],+3[8],+5[9],+7[6],+17[5],+18[4],+19[2],+20[3],+22[3][cp13] | Mutated* | n | n | n | n |
| 7 | CB | 48-50,XX,+3[7],del(4)(q21q25)[7],dup(11)(q21q24)[7],+16[7],add(17)(p10)?t(13;17) (q22;p10)[6],+18x2[7][cp7] 50,XX,+3[11],del(4)(q21q25)[11],trp(11)(q21q24)[11], +16[11],iso(17)(p10)[10],+18x2[11][cp11] | Hypermutated | n | tri/tet | del | n |
| 8 | IB | 48-50,XX,t(2;8)(p13or14;q24.1)[14],add(2)(p11)[14],+3[13],del(6)(?q15q23)[14], ?der(9)t(?X;9)(?p11;p11)[14],del(10)(q22q24)[14],t(13;15)(q14;q24)[14],+16[14], iso(18)(q10)[14],+iso(18)(q10)x2[14][cp14] | Hypermutated | del | n | del | del |
| 9 | Unclass | 47-49,X,−Y[15],del(1)(p31or32)[15],+3[15],idic(6)(q16)[15],+idic(6)(q16)[15], del(12)(q11q22)[15],+del(12)(q11q22)[15],+der(12)t(5;12)(q13;p13)[15], −15[15],add(17)(q25)[15],+mar[14][cp15] | Mutated | tri | n | n | n |
| 10 | Unclass | 84-87〈4n〉,XXYY,add(2)(q37)[5],add(2)(q37)[2],add(6)(q11)[3],add(6)(q11)[2],del(9)(q12)x2[5], der(12)t(12;15)(p11;q15)[4],der(12)t(12;15)(p11;q15)[2],add(14)(q32)[4][cp6],inc | Unmutated | del | tet | tet | tet |
| 11 | CB | 39-48,XX,−X[5],add(3)(q21)?t(3;12)(q21;q15)[20],+5[3],−6[5],−8[5],−9[4],−10[5],+12[5], +14[2],add(14)(q32)[20],+15[11],−18[4],+20[7],+20[2],−21[4],−22[5],+mar[7][cp20] | ND | tri | n | n | n |
| 12 | CB | 48-51,X,del(X)(q22orq24)[3],+3[3],+16[3],+der(16)t(X;16)(q22orq24;q13)[2],+18[2], der(19)t(5;19)(q14q31;q13)[3][cp3]/47–49,X,del(X)(q22orq24)[10],+3[10],−8[12], der(9)t(8;9)(q13;p21orp22)[12],+16[12],+der(16)t(X;16)(q22orq24;q13)[5],+18[11], der(19)t(5;19)(q14q31;q13)[12][cp12] | Hypermutated | n | n | n | n |
| 13 | CB | 91-97〈4n〉,XXXX,−X[3],+X[2],−1[6],+1[2],add(1)(p22)[7],−2[3],del(2)(p23)[6],add(2)(q31) [15],+3[2],−4[3],+4[3],−6[5],del(6)(q21)[15],del(6)(q21)[9],del(6)(p22)[15],del(6) (p22)[12],+7[2],der(7)t(7;12)(q21;q13)[6],+8[6],+9[2],−10[6],ins(10;?)(q22;?)[2], +11[4],del(11)(q23or24)[2],add(11)(p15)[6],+13[3],−14[8],−14[3],−15[4],+16[2], −17[4],?add(17)(p13)[3],?add(17)(p13)[2],−18[6],+18[2],−19[3],+19[3],+20[5], +21[5],+mar[7],+mar2[3],+mar3[5],+mar4[5],+mar6[2],+mar7[3],+mar8[2][cp15],inc | ND | tet | tet | del | del |
| Case no. . | Diagnosis . | Karyotype . | IgVH status . | D13S25 . | ATM . | TP53 . | P16INK4a . |
|---|---|---|---|---|---|---|---|
| 1 | CB | 33-50,X,?del(X)(q13)[5] | ND | n | n | n | n |
| 2 | CB | 43-50,XX,t(3;14)(q27;q32),del(6)(q15q21),+11,+18,+19[11] | ND | ND | ND | ND | ND |
| 3 | CB | 86-91P〈4n〉,XXXX,−1[4],del(1)(q11)[4],+3[3],t(3;14)(p21;q32)[3],der(6)t(1;6)(q21;q21)[4], +der(6)t(1;6)(q21;q21)[3],t(8;11)(p23;p12)x2[4],der(16)t(1;16)(q21;q21)[4], +der(16)t(1;16)(q21;q21)[3],−17[4],−17[3][cp4] | Mutated | tet | tet | del | del |
| 4 | IB | 44/48,XY,−6,+21[2] | Hypermutated | del | n | n | n |
| 5 | IB | 43-45,XY,del(2)(q21q31)[15],−6[15],del(9)(p13)[15],del(10)(q22q24)[15],+mar[2][cp15] | ND | del | n | n | del |
| 6 | CB | 46-52,XY,+2[2],+3[8],+5[9],+7[6],+17[5],+18[4],+19[2],+20[3],+22[3][cp13] | Mutated* | n | n | n | n |
| 7 | CB | 48-50,XX,+3[7],del(4)(q21q25)[7],dup(11)(q21q24)[7],+16[7],add(17)(p10)?t(13;17) (q22;p10)[6],+18x2[7][cp7] 50,XX,+3[11],del(4)(q21q25)[11],trp(11)(q21q24)[11], +16[11],iso(17)(p10)[10],+18x2[11][cp11] | Hypermutated | n | tri/tet | del | n |
| 8 | IB | 48-50,XX,t(2;8)(p13or14;q24.1)[14],add(2)(p11)[14],+3[13],del(6)(?q15q23)[14], ?der(9)t(?X;9)(?p11;p11)[14],del(10)(q22q24)[14],t(13;15)(q14;q24)[14],+16[14], iso(18)(q10)[14],+iso(18)(q10)x2[14][cp14] | Hypermutated | del | n | del | del |
| 9 | Unclass | 47-49,X,−Y[15],del(1)(p31or32)[15],+3[15],idic(6)(q16)[15],+idic(6)(q16)[15], del(12)(q11q22)[15],+del(12)(q11q22)[15],+der(12)t(5;12)(q13;p13)[15], −15[15],add(17)(q25)[15],+mar[14][cp15] | Mutated | tri | n | n | n |
| 10 | Unclass | 84-87〈4n〉,XXYY,add(2)(q37)[5],add(2)(q37)[2],add(6)(q11)[3],add(6)(q11)[2],del(9)(q12)x2[5], der(12)t(12;15)(p11;q15)[4],der(12)t(12;15)(p11;q15)[2],add(14)(q32)[4][cp6],inc | Unmutated | del | tet | tet | tet |
| 11 | CB | 39-48,XX,−X[5],add(3)(q21)?t(3;12)(q21;q15)[20],+5[3],−6[5],−8[5],−9[4],−10[5],+12[5], +14[2],add(14)(q32)[20],+15[11],−18[4],+20[7],+20[2],−21[4],−22[5],+mar[7][cp20] | ND | tri | n | n | n |
| 12 | CB | 48-51,X,del(X)(q22orq24)[3],+3[3],+16[3],+der(16)t(X;16)(q22orq24;q13)[2],+18[2], der(19)t(5;19)(q14q31;q13)[3][cp3]/47–49,X,del(X)(q22orq24)[10],+3[10],−8[12], der(9)t(8;9)(q13;p21orp22)[12],+16[12],+der(16)t(X;16)(q22orq24;q13)[5],+18[11], der(19)t(5;19)(q14q31;q13)[12][cp12] | Hypermutated | n | n | n | n |
| 13 | CB | 91-97〈4n〉,XXXX,−X[3],+X[2],−1[6],+1[2],add(1)(p22)[7],−2[3],del(2)(p23)[6],add(2)(q31) [15],+3[2],−4[3],+4[3],−6[5],del(6)(q21)[15],del(6)(q21)[9],del(6)(p22)[15],del(6) (p22)[12],+7[2],der(7)t(7;12)(q21;q13)[6],+8[6],+9[2],−10[6],ins(10;?)(q22;?)[2], +11[4],del(11)(q23or24)[2],add(11)(p15)[6],+13[3],−14[8],−14[3],−15[4],+16[2], −17[4],?add(17)(p13)[3],?add(17)(p13)[2],−18[6],+18[2],−19[3],+19[3],+20[5], +21[5],+mar[7],+mar2[3],+mar3[5],+mar4[5],+mar6[2],+mar7[3],+mar8[2][cp15],inc | ND | tet | tet | del | del |
CB indicates centroblastic; ND, not done; n, normal; Mutated, VH homology in 95% to 98%; tet, tetrasomy; del, deleted; IB, immunoblastic; Hypermutated, VH homology in < 95%; tri, trisomy; Unclass, unclassified; and Unmutated, VH homology in > 98%.
Ongoing mutations.
On banding analysis, the majority (11 of 13 [85%]) of CD5+ DLBLs exhibited complex karyotypic alterations with 12 structural or numerical aberrations on average. All cases were negative for the t(11;14)(q13;q32) and t(14;18)(q32;q21) chromosome translocations, and t(3;14)(q27;q32) targeting the BCL6 gene was found in only 1 tumor (case 2). In contrast, CD5−DLBLs harbored BCL2 and BCL6 rearrangements in 9 of 55 (16%) and 4 of 55 cases (7%), respectively. The sole recurrent structural aberration was deletion del(10)(q22q24) in cases 5 and 8. Trisomies for chromosomes 3 (7 of 13 [54%] versus 7 of 55 [13%]), 16/16p (5 of 13 [38%] versus 8 of 55 [15%]), and 18/18q (6 of 13 [46%] versus 7 of 55 [13%]) were significantly more frequent in CD5+ DLBLs than in CD5− DLBLs (P < .05).
FISH analysis failed to reveal ATM deletions, being strongly associated with B-CLL12 and MCL,17 in both CD5+ and CD5− DLBLs. In contrast, the prevalence of deletions at the D13S25 locus was significantly higher in CD5+ DLBLs (4 of 12 [33%]) compared with CD5− DLBLs (4 of 42 [10%],P < .05). Deletions of TP53 were equally distributed in both groups, whereas p16INK4ahemizygous deletions were more frequent in CD5+ tumors (4 of 12 [33%]) than in the CD5− group (3 of 38 [8%],P < .05).
Deletion of D13S25 is a frequent finding in B-CLL and MCL, but it is rarely found in other malignant lymphomas of B-cell type.12,22,23 The strong association of D13S25deletions with CD5+ DLBL may, therefore, indicate that this aberration hits a CD5+ progenitor B cell at the same differentiation level as in B-CLL. Interestingly, deletions ofATM commonly found in B-CLL12 were not detectable in CD5+ DLBLs. It recently turned out, however, that ATM deletions in B-CLL occur almost exclusively in the unmutated variant of the disease,24 whereasD13S25 deletions cluster in mutated B-CLL.25Because most of our CD5+ cases displayed mutated IgVH genes, their cell of origin is likely to be different from the pregerminal progenitor cell of the unmutated B-CLL cases that are frequently characterized by ATM inactivations. CD5+ DLBLs may, therefore, be viewed as an early transformed aggressive DLBL variant related to CD5+ lymphocytic lymphoma with additional transforming events having occurred before the clonal expansion of the low-grade neoplasm. According to the results of this first study comparing cytogenetic alterations in CD5+ and CD5− DLBLs, one likely candidate gene for this early transforming event is the loss of one p16INK4aallele. The frequent association of p16INK4ainactivations with transformed low-grade lymphomas13 would be in excellent agreement with this hypothesis.
We thank Mrs H. Brückner, Mrs A. Trumpfheller, and Mrs N. Hemmrich for technical assistance and Mr E. Schmitt for artful photographic work. The Resource Center/Reference Library of the German Human Genome Project (Berlin, Germany) generously provided the P1-derived artificial chromosome (PAC) probes specific forATM (PAC ATM-1, LLNLP704G18220Q19 and PAC ATM-2, LLNLP704O01298Q19). These clones were part of the libraries RPCI 1, RPCI 3-5, originating from Roswell Park Cancer Institute, created by Pieter de Jong and Panayiotis A. Ioannou.
Prepublished online as Blood First Edition Paper, August 29, 2002; DOI 10.1182/blood-2002-06-1726.
Supported by the European Commission “Growth program, Research project Molecular and biological risk factors in mantle cell lymphoma” (Contract no. QLG1-CT-2000-00687).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tiemo Katzenberger, Institute of Pathology, University of Würzburg, Josef-Schneider-Strasse 2, 97080 Würzburg, Germany; e-mail:path054@mail.uni-wuerzburg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal