Rac GTPases regulate a wide variety of cellular processes including actin cytoskeleton organization, gene expression, cell-cycle progression, and apoptosis. Here we report that the TRQQKRP motif of Rac2 located near the C-terminus, a region of sequence disparity among Rac proteins, is essential for complementation of Rac2 function in Rac2-deficient cells. Deletion of this sequence can also intragenically suppress the dominant-negative Rac2D57Nmutation in a variety of functional assays. In Rac2-deficient cells, expression of TRQQKRP-deleted Rac2 protein is unable to completely rescue migration and nicotinamide adenine dinucleotide phosphate oxidase deficiencies previously described in these cells. In fibroblasts, the Rac2D57N mutant phenotypes of abnormal proliferation, cell morphology, and membrane ruffling are suppressed by the TRQQKRP motif deletion. In myeloid hematopoietic cells, the deletion of the TRQQKRP motif eliminates a Rac2D57N-induced block in in vitro differentiation of neutrophils not previously described with this mutant. Mechanistically, deletion of the TRQQKRP motif results in diminished geranylgeranylation and delocalization of intracellular Rac2 protein. Taken together, these results indicate that the TRQQKRP motif in Rac2 protein is required for efficient prenylation and correct intracellular localization of Rac2 protein and is essential for Rac2 to mediate a variety of its biologic functions. These data suggest that precise localization of Rac2 protein in intracellular compartments and/or with other proteins/lipids is a prerequisite for its diverse functions.
Introduction
The Rho GTPases belong to the Ras superfamily of small GTP-binding proteins that have a molecular mass of 20 to 30 kDa. Rho GTPases have been shown to play essential roles in a wide variety of cellular functions, such as the regulation of the actin cytoskeleton, membrane trafficking, transcriptional regulation, oxidant generation, cell growth control, chemotaxis, and cell adhesion.1 To date, there are at least 16 members identified in the mammalian Rho subfamily, including Rho, Rac, and Cdc42. By cycling between inactive GDP- and active GTP-bound conformations, the small GTPases function as critical relays in the transduction of signals originating from cell-surface receptors. In fibroblasts, Rac regulates actin reorganization to produce lamellipodia and membrane ruffles, transcriptional activation, cell proliferation, transformation, and apoptosis.2-7
Members of the Rho GTPases are subject to posttranslational modifications of prenylation, proteolysis, and carboxylmethylation. Mammalian Rac and most other Rho GTPases are geranylgeranylated, and prenylation occurs at a conserved cysteine in the carboxy terminal motif of the sequence CAAL, where C is cysteine, A is an aliphatic amino acid, and L is leucine. Subsequent to prenylation, the carboxy terminal tripeptide (AAL) is removed by proteolysis, and the newly generated carboxy terminal amino acid is methylated.8-10Protein prenylation is thought to be crucial for targeting GTPases to cellular membranes, protein-protein interactions, and membrane-associated protein trafficking.11-13
In mammals, 3 isoforms of Rac proteins have been identified: Rac1, Rac2, and Rac3. The 3 Rac isoforms share a high degree of amino acid identity (> 89% overall) but differ substantially in tissue distribution and levels of expression. Rac1 and Rac3 proteins are widely expressed in different tissues, including cells of hematopoietic origin, while the expression of rac2 gene is restricted to cells of hematopoietic origin.14-17 In addition, Rac2 differs from Rac1 and Rac3 in the primary sequence located upstream of the C-terminal prenylation site. Rac2-deficient mice display defects in neutrophil, stem cell, and mast cell functions, including superoxide production, chemotaxis, adhesion, degranulation, and F-actin generation, as well as abnormalities in host defense in spite of continued expression of Rac1 in these cells.18,19,43 These data suggest that the area of sequence divergence may play a key role in the functions of Rac2. A dominant-negative mutation (D57N) of Rac2 is associated with human phagocytic immunodeficiency, suggesting that Rac2 also plays a critical role in human phagocyte cells.20 21
The structure and most of the functional domains of Rho GTPases, including GTP binding and hydrolysis domains, effector domains, an insert domain, GEF interaction domains, and a prenylation site, are well studied and characterized.22-24 However, the motif located just upstream of the C-terminal prenylation site, an area of sequence diversity among Rac proteins, has been investigated less thoroughly. The domain near the C-terminal prenylation site found in Rac1, the related Rho GTPase, Cdc42, and also in K-Ras proteins is termed the polybasic domain, since it usually consists of 4 to 6 basic residues. The polybasic domain in Rac1 is composed of 6 consecutive basic residues, whereas 3 of the 6 residues in the corresponding motif of Rac2, RQQKRP, are replaced with neutral amino acids. Rac1, through its polybasic domain, has been shown to bind to and stimulate the kinase activity of p21-activated kinase (PAK) more efficiently than Rac2.25 Moreover, it has been shown that the polybasic domain of Rac1 but not the TRQQKRP motif of Rac2 is important for nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activation in cell-free systems.26,27 The polybasic domain of Rac1 also has been shown to be required for the oligomerization of Rac1.28 However, it is still not clear to what extent the TRQQKRP motif in Rac2 is critical to the intracellular localization and the functions of this protein.
In this study, we have shown that a TRQQKRP motif deletion mutant of Rac2 cannot rescue phenotypic abnormalities present in Rac2-deficient primary myeloid cells. The same deletion, placed in the dominant-negative mutant D57N Rac2, can intragenically suppress the phenotypic abnormalities of this mutant in a variety of functional assays. Furthermore, deletion of the TRQQKRP motif results in reduced geranylgeranylation and the delocalization of intracellular Rac2 protein. Taken together, these results indicate that the TRQQKRP motif is required for correct intracellular localization of Rac2 protein and is essential for the biologic functions of Rac2, suggesting strongly that precise localization of Rac2 protein in the intracellular compartments and/or interactions of Rac2 with other proteins are prerequisites for its diverse biologic functions.
Materials and methods
Construction of retroviral vectors, transfections, and infection
The bicistronic retroviral vectors expressing the N-terminal Flag epitope–tagged murine wild-type Rac2 and enhanced green fluorescent protein (EGFP) or the dominant-negative mutant (D57N) of human Rac2 or Flag-tagged murine Rac2 and EGFP were constructed as described.21 A polymerase chain reaction–based technique was used to generate C-terminal truncation (Rac2ΔCT; Rac2Δ [182-192]) and the TRQQKRP motif deletion (Rac2ΔM; Rac2Δ[182-188]) mutants of murine Rac2. The dominant-negative mutant of murine Rac2, D57N, was generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The entire sequences of wild-type rac2 and all the rac2 mutants created were confirmed by sequencing multiple times and then subcloned into the murine stem cell virus–based bicistronic retroviral vector, MIEG3.21
Phoenix-Eco and Phoenix-Ampho packaging cell lines (ATCC, Manassas, VA) were transfected with various retroviral constructs using LipofectAMINE reagent (GIBCO BRL Life Technologies, Rockville, MD). The titers of the retroviral supernatants collected from the transfected Phoenix cells were approximately 5 × 105 colony-forming units per mL (CFU/mL). Bone marrow (BM) mononuclear cells were transduced twice on fibronectin fragment CH-296 with the retroviral supernatants as described previously.21 29 NIH3T3 cells were infected in the presence of polybrene (5.3 μg/mL). The transduced EGFP+ cells were isolated by fluorescence-activated cell-sorter (FACS; FACStar Plus; Becton Dickinson, Mountain View, CA) 48 hours after the second infection. NIH3T3 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA). The cells were cultured in Dulbecco modified Eagle medium (DMEM; GIBCO BRL Life Technologies) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 4 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (P/S; GIBCO BRL Life Technologies).
Isolation, culture, and in vitro differentiation of bone marrow cells
Female 10- to 12-week-old C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) or rac2−/− mice back crossed to C57BL/6 (N12) were used for BM isolation. All animal procedures were approved by the Animal Care Committee, Indiana University School of Medicine. BM cells were obtained as previously described.29 After washing, low-density cells were cultured, transduced, and differentiated for 14 days in α-MEM containing 20% FBS (HyClone), 4mM l-glutamine, P/S (GIBCO BRL Life Technologies), recombinant rat stem cell factor (rSCF, 100 ng/mL; Amgen, Thousands Oaks, CA), recombinant murine interleukin-3 (mIL-3, 100 U/mL), and recombinant human interleukin-6 (hIL-6, 100 U/mL; both Pepro Tech, Rocky Hill, NJ). To assay neutrophil differentiation, the cultured BM cells were cytocentrifuged onto a slide, stained with Wright-Giemsa, and examined and photographed using a light microscope. The percentage of abnormal neutrophils was determined based on microscopic enumeration of more than 200 cells in at least 5 independent fields per sample.
Quantitation of NADPH oxidase activity
Reduction of nitroblue tetrazolium (NBT) as a measure of superoxide production was assayed as described with minor modifications.30 Briefly, the transduced and differentiated normal BM cells were added to 800 μL of 0.1% NBT (Sigma, St Louis, MO) in the presence of 1.6 × 10−6 M phorbol myristate acetate (PMA). The cells were then incubated for 20 minutes at 37°C, pelleted, resuspended in 150 μL PBS, and cytocentrifuged onto a slide. After Safranin-O (Sigma) counterstain, the cells were examined by light microscopy to assay for dark purple deposits. The number of NBT+ cells and the total number of cells per high-power field were counted in at least 5 independent fields per sample, and the percentage of NBT+ cells was calculated from > 300 cells. For complementation study, a different protocol was used. Briefly, transduced rac2−/− BM cells (2∼5 × 104) were seeded onto chamber slides in 750 μL of Iscove modified Dulbecco medium (IMDM) containing P/S, and incubated for 1 hour at 37°C to allow cells to adhere to the slide. Subsequently, 200 μL of saturated NBT solution containing 10 μM N-formyl-methionyl-leucyl-phenylalanine (fMLP; Sigma) were added to the cells, and the cells were incubated for 20 minutes at 37°C. After the cells were washed with cold PBS, they were fixed with methanol and stained with Safranin-O. The percentage of NBT+ cells was determined, in duplicate, by evaluating a total of 200 cells.
Chemotaxis
Low-density BM cells isolated fromrac2−/− mice were transduced with vector, wt Rac2, Rac2ΔCT, or Rac2ΔM, and GFP+ cells were sorted by FACS. The sorted cells were then cultured for 7 to 10 days in IMDM supplemented with 10% FCS, 2% P/S, 100 ng/mL stem cell factor (SCF), 100 ng/mL megakaryocyte growth and development factor (MGDF), and 100 ng/mL granulocyte colony-stimulating factor (G-CSF). Chemotaxis of the in vitro differentiated neutrophils was assayed using a 48-well microchemotaxis chamber (Neuro Probe, Cabin John, MD) as described previously18 by counting 4 randomly chosen fields per well using 1 μM chemoattractant fMLP.
Hematopoietic progenitor assays
Transduced and sorted low-density BM cells were plated at 2 × 104 cells/mL in MethoCult M3231 methycellulose medium (StemCell Technologies, Vancouver, BC, Canada) supplemented with 100 ng/mL rSCF, 100 U/mL mIL-3, 100 U/mL hIL-6, 10 ng/mL human granulocyte colony-stimulating factor (hG-CSF, Amgen), 100 ng/mL human megakaryocyte growth and differentiation factor (MGDF, Amgen), and 4 U/mL human erythropoietin (Epo, Amgen). Cultures were incubated at 5% CO2 and 37°C for 11-12 days and then scored using an inverted microscope. For each experiment, triplicate plates were scored per data point. The colony sizes were visually estimated under the microscope.
Scanning electron microscopy
Transduced and in vitro differentiated BM cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for about 1 hour at room temperature followed by one week at 4°C. The cells were then allowed to settle by gravity overnight onto polylysine-coated coverslips at 4°C, cross-linked to polylysine, and postfixed for 30 minutes with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4). Subsequently, the samples were dehydrated in a graded series of ethanol, critical point dried, coated with 40 nm of Au/Pd with a SPUTTER coater (Hummer V, Anatech, Springfield, VA), and examined in an AMR 1000A scanning electron microscope (SEM) (Amray, Burlington, MA).
Immunofluorescence
Transduced NIH3T3 cells were cultured, serum starved for 20 hours, treated with platelet-derived growth factor (PDGF) (Pepro Tech, Rocky Hill, NJ) (5 ng/mL) for 8 minutes, and stained for actin filaments with rhodamine (TRITC) phalloidin as described.21 Images were visualized and recorded using a Bio-Rad confocal microscope. For localization of Flag-tagged Rac2 proteins, cells were treated with PDGF and stained with the purified anti-Flag M2 monoclonal antibody (Sigma) (10 μg/mL) for 90 minutes at room temperature followed by 40 minutes of incubation with affinity purified TRITC-conjugated goat anti–mouse IgG (Jackson ImmunoResearch, West Grove, PA). The cells were then visualized on a Zeiss epifluorescent microscope and recorded with a Spot RT digital imaging system (Diagnostic Instrument, Sterling Heights, MI) as well as Bio-Rad confocal microscope (Hercules, CA).
Western blotting and flow cytometric analysis
The Western blot analysis of the transduced and FACS-sorted cells was performed as previously described using anti-Flag M2 monoclonal antibody (Sigma) and an anti-Rac monoclonal antibody (BD Transduction Laboratories, San Diego, CA).31 FACS analysis was used to determine the expression of specific cell-surface markers on the transduced, sorted, and in vitro differentiated BM cells. Briefly, the cells were first incubated with 10% normal rat serum to block the nonspecific binding sites then incubated with phycoerythrin-labeled isotype control antibody or monoclonal antibodies against c-Kit, Sca-1, Mac-1, or Gr-1 (Pharmingen, San Diego, CA) (∼8-38 μg/mL). The labeled cells were analyzed with a FACScan flow cytometer (Becton Dickinson). In addition, FACS analysis was performed to estimate the percentage of neutrophils present in the EGFP+ and differentiated BM cells based on size (forward scatter).
Subcellular fractionation
Transduced and sorted NIH3T3 cells were grown to 90% confluence and scraped in hypotonic homogenization buffer (15 mM Tris, pH 7.5, 1.5 mM MgCl2, 5 mM KCl, 0.25M sucrose) supplemented with protease inhibitor mixture. Cells were homogenized using tight-fitting Dounce homogenizers (Wheaton Science Products, Millville, NJ) (50 strokes), and the homogenates were clarified by centrifugation at × 1000g for 10 minutes at 4°C to remove unbroken cells and nuclei. The resulting supernatants were centrifuged at × 100 000g for 90 minutes to yield cytosol (S100) and total membrane (P100) fractions. Equivalent proportions of S100 and P100 fractions (cell equivalents) were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with anti-Flag M2 antibody as described above.
Prenylation of Rac2 proteins
Prenylation of wild-type and mutant Rac2 proteins was assessed by determining the amount of label, derived from [3H] mevalonolactone, incorporated into the Rac2 proteins.32,33Briefly, at 60 hours after seeding, the transduced and sorted NIH3T3 cells were incubated in Dulbecco modified Eagle medium containing 40 μM mevastatin (Calbiochem, San Diego, CA) for 4 hours. This was followed by a 12-hour incubation at 37°C with (R, S)-[3H] mevalonolactone (78 μCi/mL [2.886 MBq/mL], 24 Ci/mmol [8.88 × 1011 Bq/mmol]; NEN Life Science Products, Boston, MA) in the presence of 40 μM mevastatin. The cells were solubilized in 1% Triton X-100 lysis buffer containing protease inhibitor mixture and immunoprecipitated with anti-Flag M2 affinity gel (Sigma). The immunoprecipitates were analyzed by SDS-PAGE, and the gels were fluorographed with EN3HANCE (NEN Life Sciences Products).
Results
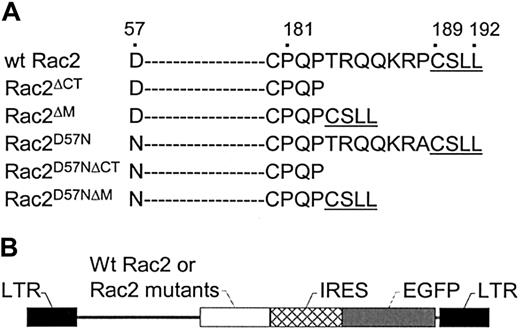
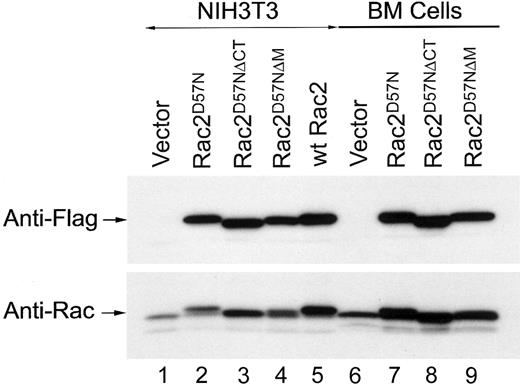
To examine whether the TRQQKRP motif located near the C-terminus of Rac2 is crucial for biologic function, we first generated a Rac2 mutant in which the TRQQKRP motif is deleted (Rac2ΔM) as well as a Rac2 mutant in which dominant-negative mutation (D57N) is combined with the deletion of the TRQQKRP motif (Rac2D57NΔM) (Figure 1A). As controls and for comparison purposes, the wild-type Rac2 (wt Rac2), a dominant-negative mutant (Rac2D57N), a C-terminus deletion mutant (Rac2ΔCT), and a mutant with both the D57N mutation and C-terminus deletion (Rac2D57NΔCT) were also generated (Figure 1A). Wt Rac2 and the Rac2 mutants were cloned into the bicistronic retroviral vector, MIEG3 (Figure 1B). Normal NIH3T3 and low-density mononuclear BM cells were transduced with the various retroviral constructs and then sorted for EGFP+cells. As shown in Figure 2, anti-Flag monoclonal antibody detected a single band of ∼ 20 kDa in cell extracts of Rac2D57N, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 transduced cells (top panel). Rac2D57N, Rac2D57NΔCT, and Rac2D57NΔM proteins were expressed at a level similar to that of wt Rac2. The size of the Rac2D57NΔCT bands, as expected, are slightly smaller than wt Rac2. A duplicate blot was probed with an anti-Rac antibody that recognizes both Rac1 and Rac2 proteins (Figure 2, lower panel). Although freshly transduced Rac2D57N cells express this mutant protein at a level comparable to that of wt Rac2, we have consistently observed that the expression level of Rac2D57N proteins becomes lower in NIH3T3 cells over time. This is also evident when these cells were analyzed by FACS analysis and confocal microscopy for EGFP expression (data not shown).
Rac2 amino acid sequence and vector constructs.
(A) Schematic representation of wild-type and mutant ΔCT,ΔM, D57N, D57NΔCT, and D57NΔMRac2 carboxy-terminal amino acid sequence. The dashes denote amino acid sequences not shown. The amino acid sequences are numbered on top. (B) Schematic representation of recombinant retroviral vectors.
Rac2 amino acid sequence and vector constructs.
(A) Schematic representation of wild-type and mutant ΔCT,ΔM, D57N, D57NΔCT, and D57NΔMRac2 carboxy-terminal amino acid sequence. The dashes denote amino acid sequences not shown. The amino acid sequences are numbered on top. (B) Schematic representation of recombinant retroviral vectors.
Expression of mutant Rac2D57N, Rac2D57NΔCT, Rac2D57NΔM, and wild-type Rac2 proteins in transduced NIH3T3 and BM-derived myeloid cells.
Equal amounts of total protein from cell extracts from each sample was subjected to SDS-PAGE, blotted onto polyvinylidenefluoride membrane, stained with Ponceau-S (Sigma), and probed with either anti-Flag (M2) or anti-Rac antibodies. Lanes 1-5, NIH3T3 cells; lanes 6-9, bone marrow cells.
Expression of mutant Rac2D57N, Rac2D57NΔCT, Rac2D57NΔM, and wild-type Rac2 proteins in transduced NIH3T3 and BM-derived myeloid cells.
Equal amounts of total protein from cell extracts from each sample was subjected to SDS-PAGE, blotted onto polyvinylidenefluoride membrane, stained with Ponceau-S (Sigma), and probed with either anti-Flag (M2) or anti-Rac antibodies. Lanes 1-5, NIH3T3 cells; lanes 6-9, bone marrow cells.
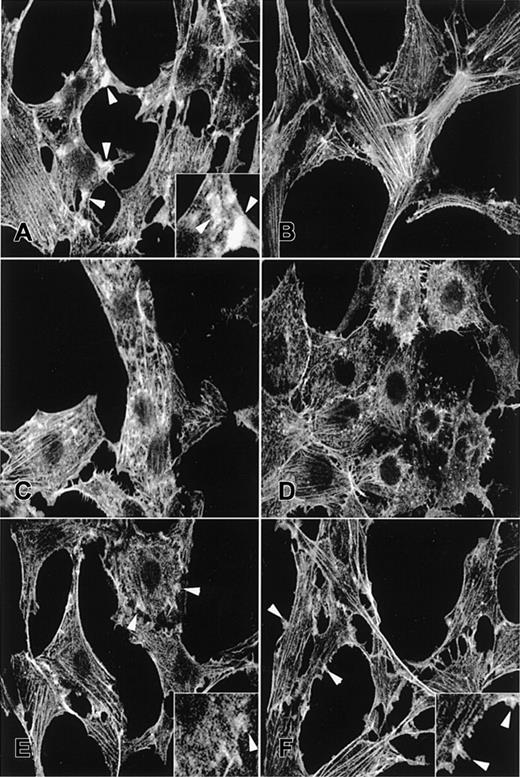
Rac stimulates growth factor–induced membrane ruffling in fibroblast cells, whereas dominant-negative mutants of Rac proteins block this response.1,34 To assess whether the TRQQKRP motif is required for Rac2 to stimulate membrane ruffling, we examined the actin cytoskeleton reorganization by TRITC-phalloidin staining in NIH3T3 cells transduced with each respective retrovirus. As noted previously, PDGF clearly induced actin reorganization and membrane ruffles at the cell periphery of a cell transduced with a control vector (Figure3A, at arrowheads and the insert) compared to unstimulated cells (Figure 3B), whereas expression of Rac2D57N severely reduced membrane ruffling at the cell periphery (Figure 3C). The majority of Rac2D57N-expressing cells exhibited irregular actin cytoskeleton organization in their cytoplasm, characterized by punctate phalloidin staining over the perinuclear region and irregular stress fibers, which existed in the presence or absence of PDGF stimulation (Figure 3C-D). Importantly, the membrane ruffling at the cell periphery was largely restored by expression of either Rac2D57NΔCT or Rac2D57NΔM, and most of these cells displayed regular actin cytoskeleton organization (Figure 3E-F at arrowheads and inserts). Expression of Rac2D57N in NIH3T3 cells also altered cell morphology, as previously demonstrated,21 and reduced growth rate and cell density. All of these characteristics were suppressed by expression of either Rac2D57NΔCT or Rac2D57NΔM (data not shown).
Fluorescence photomicrographs of TRITC-phalloidin–stained NIH3T3 fibroblasts.
(A-B) EGFP alone, (C-D) Rac2D57N, (E) Rac2D57NΔCT, or (F) Rac2D57NΔM. Panels A, C, E, and F were treated with PDGF; panels B and D, no treatment. Arrowheads point to membrane ruffles. Magnification × 400. Inserts show the membrane ruffles at higher magnification (×800).
Fluorescence photomicrographs of TRITC-phalloidin–stained NIH3T3 fibroblasts.
(A-B) EGFP alone, (C-D) Rac2D57N, (E) Rac2D57NΔCT, or (F) Rac2D57NΔM. Panels A, C, E, and F were treated with PDGF; panels B and D, no treatment. Arrowheads point to membrane ruffles. Magnification × 400. Inserts show the membrane ruffles at higher magnification (×800).
Rac proteins have been shown to be essential for activation of the NADPH oxidase enzyme complex that produces superoxide,49,50 and we have previously demonstrated that expression of Rac2D57N inhibits superoxide production induced by some agonists in normal human neutrophils.21 To assess NADPH oxidase function in neutrophils derived from transduced and differentiated wild-type BM cells expressing Rac mutants, an NBT test was performed. The results shown in Table1 indicated that the number of NBT+ cells in Rac2D57N cultures was significantly reduced as compared to that in vector control. In contrast, expression of either Rac2D57NΔCT or Rac2D57NΔM restored the number of NBT+ cells to the level similar to vector control. These data demonstrate that the minimal TRQQKRP motif is required for the inhibition of neutrophil oxidase formation by Rac2D57N. This impaired oxidase function may be due to the effects of Rac2D57N on neutrophil differentiation (see below) as well as the inability of this mutant to support the active oxidase.
NADPH oxidase expression of the myeloid cells derived from the transduced and differentiated BM cells
| . | % NADPH oxidase-positive cells . | |||
|---|---|---|---|---|
| Vector . | Rac2D57N . | Rac2D57NΔCT . | Rac2D57NΔM . | |
| Experiment 1 | 46.9 (318) | 11.5 (356) | 49.0 (382) | 49.1 (387) |
| Experiment 2 | 25.2 (349) | 8.8 (377) | 28.6 (402) | 26.7 (453) |
| . | % NADPH oxidase-positive cells . | |||
|---|---|---|---|---|
| Vector . | Rac2D57N . | Rac2D57NΔCT . | Rac2D57NΔM . | |
| Experiment 1 | 46.9 (318) | 11.5 (356) | 49.0 (382) | 49.1 (387) |
| Experiment 2 | 25.2 (349) | 8.8 (377) | 28.6 (402) | 26.7 (453) |
The numbers in parentheses indicate the total number of cells counted.
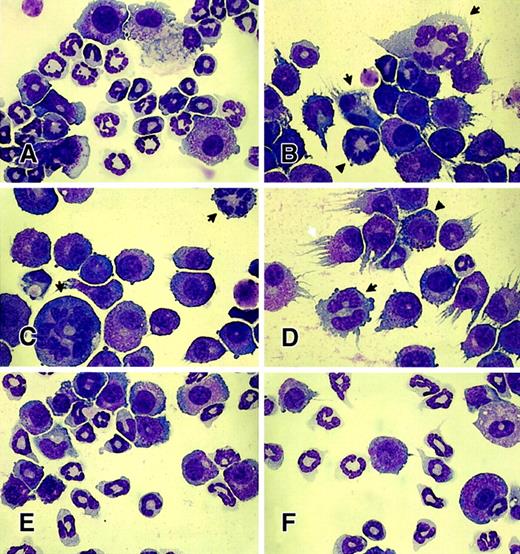
Previous studies from our laboratory have demonstrated significant negative effects of D57N Rac2 on hematopoietic cell growth in vitro and reconstitution in vivo.35 To determine whether the TRQQKRP motif is required for this effect of Rac2D57N, we expressed Rac2D57NΔM in mononuclear BM cells to test whether the deletion of the TRQQKRP motif can suppress the Rac2D57Nphenotypes. Normal mononuclear BM cells were infected with the respective retrovirus, and the EGFP+ cells were sorted by FACS. The sorted cells were differentiated in the presence of SCF, IL-3, and IL-6. In this cytokine mixture, all cultures infected by retrovirus encoding EGFP alone, Rac2D57N, Rac2D57NΔCT, or Rac2D57NΔM yielded similar growth kinetics (data not shown). After a total of 14 days in culture, a significant portion of the vector-transduced (control) cells exhibited mature neutrophil morphology (Figure4A). By contrast, cultures infected with a retrovirus expressing Rac2D57N yielded a significantly reduced number of mature neutrophils (Figure 4B). Moreover, the remaining neutrophils exhibited markedly abnormal morphology, including much larger cell size, multiple (arrows in Figure 4B-D) or hypersegmented nuclei (arrowhead in 4B), intense cytoplasmic granules and irregular granulation (arrowhead in 4D), and asynchronous nuclear and cytoplasmic maturation (Figure 4B-D). Consistent with functional data presented above, expression of either Rac2D57NΔCT or Rac2D57NΔM completely abrogated the Rac2D57Nphenotypes (Figure 4E-F). We found no significant differences among cultures infected by retrovirus encoding EGFP alone, Rac2D57NΔCT, or Rac2D57NΔM. The percentages of morphologically abnormal neutrophils from all cultures were determined (Table 2).
Effect of expression of mutant Rac proteins on neutrophil differentiation.
Wright-Giemsa staining of retrovirus-transduced mononuclear BM cells after in vitro differentiation: (A) EGFP alone, (B-D) Rac2D57N, (E) Rac2D57NΔCT, or (F) Rac2D57NΔM. Wright-Giemsa stained, magnification × 1000. Note the abnormal cell size, multiple nuclei (arrows in B-D), hypersegmented nuclei (arrowhead in B), intense granules and irregular granulation (black arrowhead in D), and asynchronous nuclear and cytoplasmic maturation (B-D) in Rac2D57N-expressing neutrophils. Also note the marked reduction in the number of mature neutrophils present in Rac2D57N-expressing cells. The white arrow in panel D indicates the cell processes extending in a “streamer” morphology predominantly found in Rac2D57N-expressing cells under the experimental conditions. These photomicrographs are representative of the results obtained from 3 independent experiments.
Effect of expression of mutant Rac proteins on neutrophil differentiation.
Wright-Giemsa staining of retrovirus-transduced mononuclear BM cells after in vitro differentiation: (A) EGFP alone, (B-D) Rac2D57N, (E) Rac2D57NΔCT, or (F) Rac2D57NΔM. Wright-Giemsa stained, magnification × 1000. Note the abnormal cell size, multiple nuclei (arrows in B-D), hypersegmented nuclei (arrowhead in B), intense granules and irregular granulation (black arrowhead in D), and asynchronous nuclear and cytoplasmic maturation (B-D) in Rac2D57N-expressing neutrophils. Also note the marked reduction in the number of mature neutrophils present in Rac2D57N-expressing cells. The white arrow in panel D indicates the cell processes extending in a “streamer” morphology predominantly found in Rac2D57N-expressing cells under the experimental conditions. These photomicrographs are representative of the results obtained from 3 independent experiments.
Quantification of the morphological abnormality of neutrophils differentiated in vitro from the transduced bone marrow cells
| . | % Abnormal neutrophils . | |||
|---|---|---|---|---|
| Vector* . | Rac2D57N† . | Rac2D57NΔCT* . | Rac2D57NΔM* . | |
| Experiment 1 | 16.6 (223) | 67.7 (204) | 10.2 (235) | 12.6 (222) |
| Experiment 2 | 10.1 (228) | 73.0 (222) | 9.7 (237) | 11.0 (228) |
| Experiment 3 | 11.8 (220) | 62.4 (218) | 10.3 (234) | 12.3 (245) |
| . | % Abnormal neutrophils . | |||
|---|---|---|---|---|
| Vector* . | Rac2D57N† . | Rac2D57NΔCT* . | Rac2D57NΔM* . | |
| Experiment 1 | 16.6 (223) | 67.7 (204) | 10.2 (235) | 12.6 (222) |
| Experiment 2 | 10.1 (228) | 73.0 (222) | 9.7 (237) | 11.0 (228) |
| Experiment 3 | 11.8 (220) | 62.4 (218) | 10.3 (234) | 12.3 (245) |
The numbers in parentheses indicate the total number of cells counted.
The defects in these neutrophils are relatively mild, consisting only of hypersegmented nucleus (bi-nucleated cells were occasionally found).
The defects in these neutrophils are much more severe. They include multiple nuclei, hypersegmented nuclei, much larger cell sizes, intense granules and abnormal granulation, and asynchronous nuclear and cytoplasmic maturation.
One of the major functions of Rac proteins is the organization of the actin cytoskeleton, which contributes to cell morphology and shape. To test whether expression of Rac2D57N causes abnormal surface morphology in myeloid cells and whether the deletion of the TRQQKRP motif can suppress the Rac2D57N morphological phenotypes, we examined transduced myeloid cells expressing the control vector, Rac2D57N, and Rac2D57NΔM by scanning electron microscopy (SEM). Myeloid cells derived from vector control culture were covered with numerous small microvilli or with surface folds of varying thickness (Figure 5A-B), which resembles the typical polymorphonuclear neutrophil (PMN) surface morphology.36 In contrast, the myeloid cells derived from Rac2D57N culture exhibited abnormally short microvilli, and numerous blebs were distributed over the entire surface of these cells (Figure 5C-D). Importantly, the abnormal surface morphology caused by Rac2D57N mutation was completely reversed by deletion of the TRQQKRP motif (Figure 5E-F). The surface morphology of the myeloid cells from the vector control and Rac2D57NΔM cultures was very similar (Figure 5A-B versus Figure 5E-F). Furthermore, when the myeloid cells from all cultures were cytocentrifuged onto slides, long extrusions from the cell surface, “streamers” (Figure 4D, white arrow), were found in Rac2D57N cells, further suggesting that the cytoskeleton in these cells was more fragile and more susceptible to environmental manipulations as compared to vector control, Rac2D57NΔCT, and Rac2D57NΔM cells.
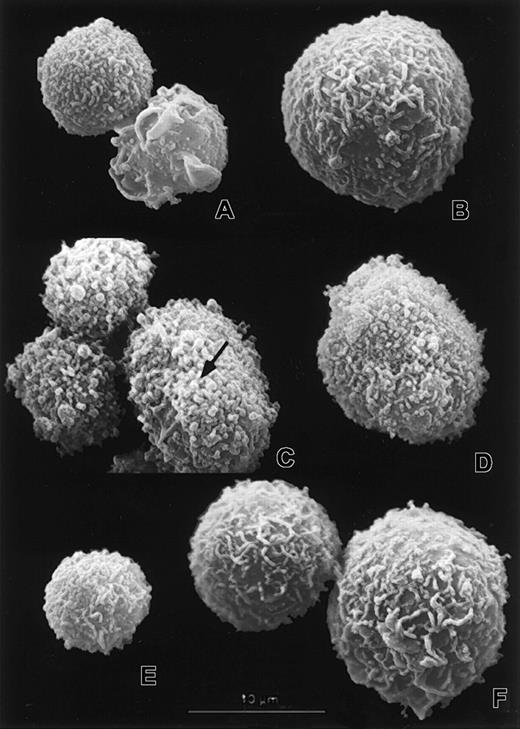
Scanning electron micrographs showing surface morphology of transduced myeloid cells.
The neutrophils (A,C,E) and large myeloid cells (B,D,F) from vector control (A-B), Rac2D57N (C-D), and Rac2D57NΔM(E-F) cultures were examined by SEM. Note the microvilli and surface folds of varying thickness (A,B,E,F) as well as the abnormal stumpy microvilli and blebs (C-D). The arrow in panel C points to a bleb. Scale bar represents 10 μm in all micrographs.
Scanning electron micrographs showing surface morphology of transduced myeloid cells.
The neutrophils (A,C,E) and large myeloid cells (B,D,F) from vector control (A-B), Rac2D57N (C-D), and Rac2D57NΔM(E-F) cultures were examined by SEM. Note the microvilli and surface folds of varying thickness (A,B,E,F) as well as the abnormal stumpy microvilli and blebs (C-D). The arrow in panel C points to a bleb. Scale bar represents 10 μm in all micrographs.
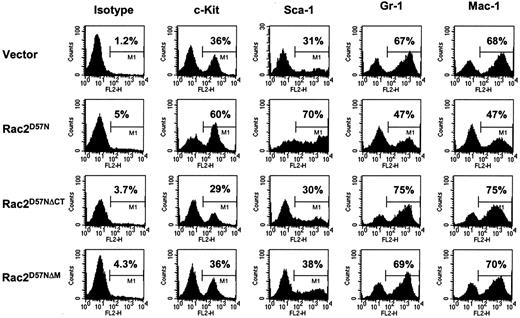
In addition to morphological characterization, expression of specific cell-surface antigens of hematopoietic progenitor cells, including c-Kit (CD117) and Sca-1 (Ly-6A/E), and of the lineage-specific and differentiation-associated surface antigens, such as Gr-1 (Ly-6G) and Mac-1 (CD11b), was examined in the transduced BM cells cultured for 15 days in SCF, IL-3, and IL-6. FACS analyses demonstrated that the majority of the cells in vector control cultures expressed Gr-1 (67%) or Mac-1 (68%), whereas fewer cells expressed c-Kit (36%) or Sca-1 (31%), in agreement with the histological analysis. In contrast, significantly fewer cells in Rac2D57N culture expressed Gr-1 (47%) or Mac-1 (47%), whereas the majority were positive for c-Kit (60%) or Sca-1 (70%), suggesting an increase in the number of immature myeloid cells. Moreover, the level of Sca-1 expression in Rac2D57N cells was increased as compared to that in vector control, Rac2D57NΔCT, and Rac2D57NΔM cells. Importantly, we found that percentages of Gr-1–, Mac-1–, c-Kit–, or Sca-1–expressing cells in either Rac2D57NΔCT or Rac2D57NΔM culture were similar to that in vector control (Figure 6). Thus, abnormal differentiation of myeloid cells due to Rac2D57N mutation is also suppressed by the deletion of TRQQKRP motif within the Rac2 proteins.
Expression of lineage markers on transduced and differentiated BM cells.
After in vitro culture and differentiation for 14 days, BM cells from the respective cultures were stained with PE-conjugated isotype control, antimouse c-Kit, Sca-1, Gr-1, or Mac-1 antibodies. Live cells were gated on EGFP, and expression of the indicated antigens on the EGFP+ cells was analyzed. Percentages of cells in M1 gate are indicated. Similar results were obtained in 3 independent experiments.
Expression of lineage markers on transduced and differentiated BM cells.
After in vitro culture and differentiation for 14 days, BM cells from the respective cultures were stained with PE-conjugated isotype control, antimouse c-Kit, Sca-1, Gr-1, or Mac-1 antibodies. Live cells were gated on EGFP, and expression of the indicated antigens on the EGFP+ cells was analyzed. Percentages of cells in M1 gate are indicated. Similar results were obtained in 3 independent experiments.
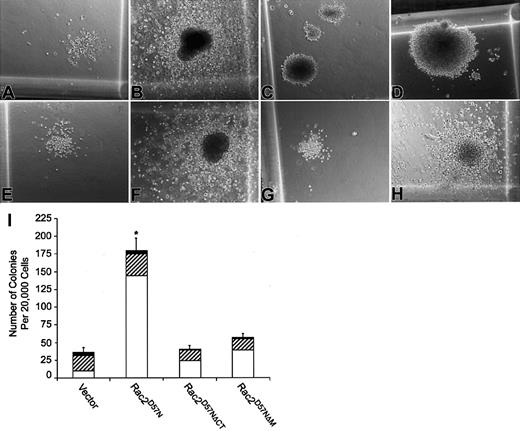
To better assess the effect of Rac2D57N on myeloid differentiation as well as the impact of deletion of TRQQKRP motif on the effect of Rac2D57N in this aspect, the transduced and sorted BM cells were assayed for colony forming capacity in standard methylcellulose assays. As expected, bone marrow transduced with the empty vector exhibited both granulocyte-macrophage (GM) and mast cell colony morphologies (Figure 7A-B). In contrast, the morphology of the colonies generated from the Rac2D57N-expressing progenitors was severely compacted regardless of the colony sizes, suggesting reduced cell migration within the colony (Figure 7C-D). Moreover, the number of myeloid progenitor colonies in Rac2D57N culture was significantly increased as compared to that in control cell cultures (Figure 7I). In contrast, the colony morphology and number of colonies generated from either the Rac2D57NΔCT- or Rac2D57NΔM-expressing progenitors were comparable to those found in the vector control (Figure 7E-H). Thus, expression of Rac2D57N significantly impairs myeloid cell differentiation, and the presence of the TRQQKRP motif is essential for this phenotype.
Colony morphology and colony formation of progenitors from transduced and sorted BM cells.
(A-H) Photomicrographs of small and large GM colonies produced by vector control (A-B), Rac2D57N (C-D), Rac2D57NΔCT (E-F), or Rac2D57NΔM (G-H) progenitors. Magnification × 100. (I) Quantitation of CFU derived from transduced BM cells. Values represent the mean of colony numbers in triplicate cultures. The open, hatched, and black bars denote small, medium, and large size colonies, respectively. The total number of colonies in each genotype was the sum of small, medium, and large colonies. The error bars indicate standard deviations (*P < .004 for comparisons between Rac2D57Nand any other genotype). Data shown are representative of 3 independent experiments.
Colony morphology and colony formation of progenitors from transduced and sorted BM cells.
(A-H) Photomicrographs of small and large GM colonies produced by vector control (A-B), Rac2D57N (C-D), Rac2D57NΔCT (E-F), or Rac2D57NΔM (G-H) progenitors. Magnification × 100. (I) Quantitation of CFU derived from transduced BM cells. Values represent the mean of colony numbers in triplicate cultures. The open, hatched, and black bars denote small, medium, and large size colonies, respectively. The total number of colonies in each genotype was the sum of small, medium, and large colonies. The error bars indicate standard deviations (*P < .004 for comparisons between Rac2D57Nand any other genotype). Data shown are representative of 3 independent experiments.
To further confirm the results obtained using Rac2D57NΔCTand Rac2D57NΔM mutants, we studied the ability of Rac2ΔCT or Rac2ΔM mutants to rescue the abnormal cell phenotypes seen in Rac2-deficient blood cells,18 which include defective chemotaxis and NADPH oxidase activation. After transduction ofrac2−/− BM cells with retroviral vectors expressing EGFP alone, wt Rac2, Rac2ΔCT, or Rac2ΔM, sorted GFP+ cells were studied in a chemotaxis assay. As shown in Table 3, introduction of wt Rac2 into rac2−/−neutrophils restored chemotaxis in response to fMLP to normal levels; however, migration by rac2−/− neutrophils expressing Rac2ΔM was reduced compared with wt Rac2-transduced rac2−/− neutrophils. Migration was impaired in cells expressing Rac2ΔCT. Cells transduced with Rac2ΔCT or Rac2ΔM exhibited increased migration compared with rac2−/−neutrophils, suggesting preservation of some function, which may be related to overexpression of the mutant protein. Similarly, expression of wt Rac2 in rac2−/− neutrophils completely restored the superoxide production to levels similar to wild-type neutrophils. By contrast, the number of NBT+ cells inrac2−/− neutrophils expressing Rac2ΔCT or Rac2ΔM is significantly reduced as compared to that in wt Rac2 transducedrac2−/− neutrophils.rac2−/− neutrophils expressing Rac2ΔM also exhibited an increased number of NBT+ cells as compared withrac2−/− neutrophils, indicating that this mutant can partially rescue the NADPH oxidase deficiencies in Rac2−/− cells (Table 4).
Chemotaxis of the neutrophils derived fromrac2−/− BM cells transduced with indicated retrovirus
| . | Migrated cells per × 400 field . | |||
|---|---|---|---|---|
| Vector . | wt Rac2 . | Rac2ΔCT . | Rac2ΔM . | |
| Experiment 1 | 11 | 200 | 49 | 91 |
| Experiment 2 | 18 | 103 | 38 | 59 |
| Experiment 3 | 7 | 188 | 34 | 44 |
| . | Migrated cells per × 400 field . | |||
|---|---|---|---|---|
| Vector . | wt Rac2 . | Rac2ΔCT . | Rac2ΔM . | |
| Experiment 1 | 11 | 200 | 49 | 91 |
| Experiment 2 | 18 | 103 | 38 | 59 |
| Experiment 3 | 7 | 188 | 34 | 44 |
NADPH oxidase activity of the neutrophils derived fromrac2−/− BM cells transduced with indicated retrovirus
| % NADPH+ cells . | ||||
|---|---|---|---|---|
| rac2+/+ vector . | rac2−/− vector . | rac2−/− wt Rac2 . | rac2−/− Rac2ΔCT . | rac2−/− Rac2ΔM . |
| 88 ± 9 | 49 ± 7 | 87 ± 64-150 | 57 ± 5 | 71 ± 84-151 |
| % NADPH+ cells . | ||||
|---|---|---|---|---|
| rac2+/+ vector . | rac2−/− vector . | rac2−/− wt Rac2 . | rac2−/− Rac2ΔCT . | rac2−/− Rac2ΔM . |
| 88 ± 9 | 49 ± 7 | 87 ± 64-150 | 57 ± 5 | 71 ± 84-151 |
Significantly different fromrac2−/−.
Significantly different from wt Rac2 as well asrac2−/−.
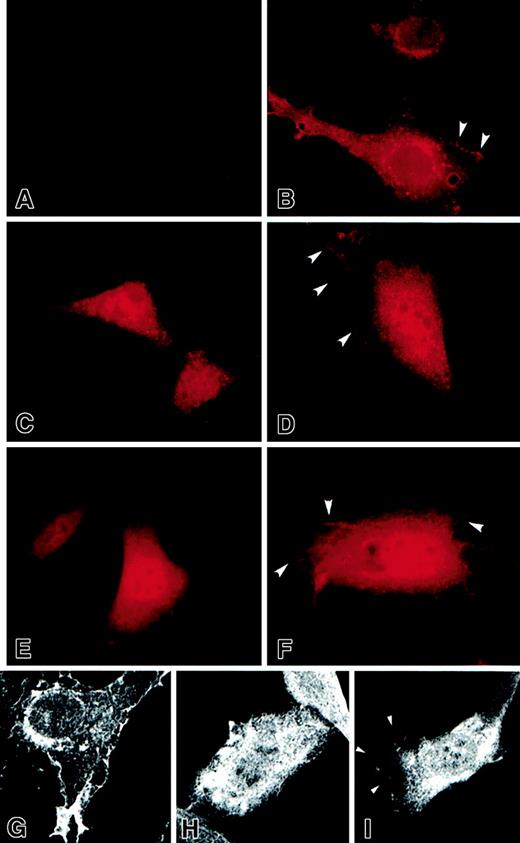
Data presented thus far demonstrate that deletion of TRQQKRP motif can intragenically suppress a variety of phenotypes associated with expression of Rac2D57N and that the TRQQKRP deletion mutant cannot fully complement the rac2−/−phenotypes. One plausible explanation of these findings is that the motif is essential for correct interactions of Rac2 with membranes and/or target proteins. Therefore, we next examined the subcellular localizations of Flag-tagged Rac2 proteins in PDGF-stimulated NIH3T3 cells transduced with each respective retrovirus by immunofluorescence microscopy. To confirm that this localization was related to the tail mutations and not the D57N substitution, we also determined the subcellular localizations of Rac2ΔCT and Rac2ΔM proteins by epifluorescence as well as confocal microscopy. The localization of Rac2ΔCT and Rac2ΔM proteins was identical to D57N mutants. As seen in Figure 8B,G, wt Rac2 proteins were primarily localized around the nucleus and at the cell periphery, consistent with the result of Michaelson et al.37 By contrast, the Rac2D57NΔCT and Rac2ΔCTproteins were diffusely distributed throughout the nucleus and cytoplasm and were barely detectable at the cell periphery (Figure8C,E,H). The intracellular distribution of Rac2D57NΔM and Rac2ΔM proteins was very similar to that of Rac2D57NΔCT, but some of these mutant proteins were also detected at the cell periphery, although to a lesser extent than wt Rac2 (Figure 8D,F,I). Thus, the TRQQKRP motif is required for the correct intracellular localization of Rac2 protein.
Immunofluorescence photomicrographs show the localization of Rac proteins in transduced NIH3T3 cells by epifluorescence and confocal microscopy.
Panel A shows vector control; panels B and G, wt Rac2; panel C, Rac2D57NΔCT; panel D, Rac2D57NΔM; panels E and H, Rac2ΔCT; and panels F and I, Rac2ΔMproteins, which were tagged with a Flag epitope at the N-terminus, using the mouse anti-Flag M2 antibody followed by TRITC-conjugated anti–mouse IgG. (A-F) Epifluorescence images, magnification × 1000; (G-I) immunofluorescence images recorded with a confocal microscope, magnification × 600. Arrowheads indicate the proteins localized to the plasma membrane.
Immunofluorescence photomicrographs show the localization of Rac proteins in transduced NIH3T3 cells by epifluorescence and confocal microscopy.
Panel A shows vector control; panels B and G, wt Rac2; panel C, Rac2D57NΔCT; panel D, Rac2D57NΔM; panels E and H, Rac2ΔCT; and panels F and I, Rac2ΔMproteins, which were tagged with a Flag epitope at the N-terminus, using the mouse anti-Flag M2 antibody followed by TRITC-conjugated anti–mouse IgG. (A-F) Epifluorescence images, magnification × 1000; (G-I) immunofluorescence images recorded with a confocal microscope, magnification × 600. Arrowheads indicate the proteins localized to the plasma membrane.
To further investigate whether the membrane association of Rac2 proteins was affected by deletion of TRQQKRP motif and to validate the localization results, we performed subcellular fractionations of NIH3T3 cells expressing EGFP alone, Rac2ΔCT, Rac2ΔM, and wt Rac2 and determined the amounts of mutant or wt Rac2 proteins in the cytosol (S100) and crude membrane (P100) fractions. Figure 9A shows that a significant portion of wild-type Rac2 protein (44% ± 7%) was detected in the membrane fraction (P) (lane 8, top panel). By contrast, Rac2ΔCT protein in cells expressing the C-terminal deletion mutant was present almost exclusively in the cytosol fraction (S), with only 7% ± 1% detected in the P100 fraction (lanes 3 and 4, top panel). More importantly, cells expressing the TRQQKRP motif deletion mutant have 28% ± 6% of the Rac2ΔM proteins associated with the membrane fraction, considerably fewer compared with wt Rac2, but more compared with Rac2ΔCT. Thus, deletion of the TRQQKRP motif reduces, but does not abolish, Rac2 protein membrane association. These results also further support subcellular localization of Rac proteins determined by immunofluorescence microscopy.
Subcellular distribution and prenylation of mutant and wild-type Rac2 proteins.
(A) Transduced and sorted NIH3T3 cells expressing EGFP alone, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 were fractionated into cytosol (S100) and crude membrane (P100) fractions. Equivalent proportions of each fraction were resolved by SDS-PAGE followed by Western blotting using anti-Flag (M2) antibody to detect various Rac2 proteins. S = S100; P = P100. (B) Transduced and sorted NIH3T3 cells expressing EGFP alone, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 were labeled with [3H] mevalonolactone, and mutant and wt Rac2 proteins contained in these cells were immunoprecipitated with anti-Flag (M2) antibody and analyzed by SDS-PAGE. The gel was treated with EN3HANCE and exposed to Kodak XOMAT film.
Subcellular distribution and prenylation of mutant and wild-type Rac2 proteins.
(A) Transduced and sorted NIH3T3 cells expressing EGFP alone, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 were fractionated into cytosol (S100) and crude membrane (P100) fractions. Equivalent proportions of each fraction were resolved by SDS-PAGE followed by Western blotting using anti-Flag (M2) antibody to detect various Rac2 proteins. S = S100; P = P100. (B) Transduced and sorted NIH3T3 cells expressing EGFP alone, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 were labeled with [3H] mevalonolactone, and mutant and wt Rac2 proteins contained in these cells were immunoprecipitated with anti-Flag (M2) antibody and analyzed by SDS-PAGE. The gel was treated with EN3HANCE and exposed to Kodak XOMAT film.
To determine whether this change in membrane localization is the result of defective prenylation of Rac2 proteins due to the TRQQKRP motif deletion, the transduced cells expressing EGFP alone, Rac2ΔCT, Rac2ΔM, and wt Rac2 proteins were labeled with [3H] mevalonolactone, and Rac2 proteins in these cells were immunoprecipitated and analyzed by SDS-PAGE and fluorography. Figure 9B shows that a considerable amount of [3H] mevalonolactone–derived label is detected in wt Rac2 proteins, whereas little label is detected in Rac2ΔCT proteins. Surprisingly, although the [3H] mevalonolactone–derived label is apparent in Rac2ΔM proteins, the amount of labeling is dramatically reduced as compared to that of wt Rac2 (Figure 9B, compare lanes 3 and 4), indicating that the geranylgeranylation of Rac2ΔMproteins is impaired. Thus, these data strongly suggest that function of Rac2 is critically dependent on correct subcellular localization and/or protein-protein interaction, which is controlled at least in large part by prenylation.
Discussion
Rho GTPase members of the Ras superfamily have been shown to control actin cytoskeleton organization as well as signaling pathways and gene transcription in eukaryotic cells.1,38 Similar to Ras, by cycling between inactive GDP-bound and active GTP-bound states, Rho GTPases are key regulators of a wide spectrum of cellular functions.39
The mammalian Rho-like GTPases consist of several distinct proteins, including the Rac subfamily.40 Among 3 identified Rac proteins, all of which share very high sequence homology, Rac2 is expressed only in hematopoietic cells,41 whereas Rac1 and Rac3 are widely expressed. We have previously generated Rac2-deficient mice by homologous recombination18 and demonstrated phenotypic abnormalities in multiple blood lineages derived from these mice, including neutrophils, mast cells, lymphocytes, and stem/progenitor cells.18,19,42 43 These phenotypic abnormalities occur in spite of continued expression of Rac1 (and presumably Rac3), suggesting that Rac2 subserves distinct functions in blood cells in spite of the high degree of sequence homology between the Rac proteins.
Previous studies have suggested that Rac1 and Rac2 vary in efficiency of interactions with proteins of the NADPH oxidase complex26,27 and differ in binding affinities with certain effector proteins.25 In all of these studies, the region spanning the polybasic domain of Rac1 has been implicated. More recently, Zhang et al have demonstrated that oligomerization of Rac1 is mediated by this domain.28 This region is also critical for function of other GTPases, such as Cdc42, and represents the region of greatest sequence divergence among the Rac proteins. In Rac2, the sequence analogous to the polybasic domain of Rac1 is interrupted by several neutral amino acids. This TRQQKRP motif located near the C-terminus of Rac2 has not been previously studied in detail.
Here we have used primary cells deficient in Rac2 to demonstrate that the TRQQKRP motif is essential for normal functioning on Rac2 in chemotaxis and superoxide generation. In addition, deleting this sequence can intragenically suppress a variety of dominant-negative phenotypes of Rac2D57N including abnormal superoxide production, abnormal cytoskeletal organization and function, and cell growth. Finally, the TRQQKRP motif of Rac2 is required for efficient prenylation and for correct intracellular localization of Rac2 protein. These data strongly suggest that precise localization of Rac2 proteins in various intracellular compartments and/or proper interactions with other proteins are prerequisites for the diverse functions of Rac2. In addition, data presented here suggest that Rac proteins are involved in normal myeloid cell differentiation, although the specific Rac protein involved is not yet clear.
It is surprising that deletion of the TRQQKRP sequence with retention of the CSLL sequence (representing the CAAL motif) completely abrogates the functions of Rac2D57N proteins and results in impaired prenylation and delocalization of Rac2 proteins. These data differ from studies of Ras, where deletion of the hypervariable sequence with retention of the CaaX motif does not affect prenylation. Ras proteins deleted of the hypervariable sequence possess transforming ability.44-47 The implication of these data are that with regard to the function of sequences near the conserved CaaX box, each member of the Ras superfamily is unique and that sequence-dependent functions established for one member may not be readily applicable to other proteins within the same superfamily.
At present it remains unclear how deletion of the TRQQKRP sequence impairs the prenylation of Rac2 proteins. It is possible that for Rac2, both the CXXL and TRQQKRP motifs constitute the biologic substrate for type I geranylgeranyl protein transferases (GGTase I). Thus, deletion of TRQQKRP motif structurally or spatially changes that region of the Rac2 proteins so that it becomes a poor substrate for GGTase I. It is also possible that deletion of TRQQKRP alters the location of Rac2 proteins within the cell so that the mutants are physically unavailable to GGTase I.
Although the TRQQKRP motif of Rac2, similar to the polybasic domain contained in Rac1, Cdc42, and K-ras, is critical for normal functioning of the protein, this motif appears to function in a different fashion than these other polybasic domains. First, the polybasic domain of these other proteins has been thought to facilitate plasma membrane binding and enable cell transformation via ionic interactions with a negatively charged domain of a target protein and/or negatively charged membrane phospholipids. The polybasic domain in Rac1 or K-ras consists of 6 consecutive basic residues, whereas 3 of the 6 residues in the TRQQKRP motif of Rac2 are replaced with neutral amino acids, which reduces the ionic charges significantly. Second, all 6 lysines of the K-ras polybasic domain may be required for efficient transforming activity, since K-ras mutants containing even 2 Lys to Gln substitutions (2Q) have been shown to drastically reduce transforming activity. Moreover, 3Q and 4Q K-ras mutants had a significant decrease in membrane association.32,48 Third, superoxide anion production in Rac-dependent cell-free systems has been shown to be inhibited by the Rac1(178-188) peptide that spans the polybasic domain, but not by the corresponding Rac2(178-188) peptide.26,27Finally, the TRQQKRP motif of Rac2 and the polybasic domain of Rac1 have been shown to possess different binding affinities and kinase-stimulating activities toward one of the effector proteins, PAK.25 More recently, Michaelson et al37 have demonstrated that the sequences upstream of the CAAL motive in Rac1 are sufficient to direct subcellular localization to the plasma membrane and endomembranes in live cells.
Data presented in this paper demonstrate that the sequences contained within this motif play important roles in the function of Rac2 in hematopoietic cells and are required for efficient prenylation and for correct localization of Rac2 proteins. These results may explain, in part, the specificity of Rac2 functions in these cells. Such specificity would imply that the TRQQKRP motif is critical in specifying protein-lipid and/or protein-protein interactions and subcellular localization. Therefore, some specificity of Rac2 protein function may be determined by localization of this protein in specific intracellular microenvironments and by interactions with other proteins at the right place.
We are indebted to Merv Yoder for his invaluable advice and helpful comments on the manuscript and to Gillian B. Bradford for her excellent advice and help on flow cytometry. We are also grateful to Philip Blomgren for his help with composing the SEM images and to Susan Rice and Jeffrey Lay for their technical assistance on the FACStar sorting. We thank Sharon Smoot, Eva Meunier, and Kerry Holleran for administrative assistance.
Supported by National Institutes of Health grant ROI DK59955.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David A. Williams, Division of Experimental Hematology, Children's Hospital Research Foundation, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: david.williams@chmcc.org.

![Fig. 9. Subcellular distribution and prenylation of mutant and wild-type Rac2 proteins. / (A) Transduced and sorted NIH3T3 cells expressing EGFP alone, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 were fractionated into cytosol (S100) and crude membrane (P100) fractions. Equivalent proportions of each fraction were resolved by SDS-PAGE followed by Western blotting using anti-Flag (M2) antibody to detect various Rac2 proteins. S = S100; P = P100. (B) Transduced and sorted NIH3T3 cells expressing EGFP alone, Rac2D57NΔCT, Rac2D57NΔM, and wt Rac2 were labeled with [3H] mevalonolactone, and mutant and wt Rac2 proteins contained in these cells were immunoprecipitated with anti-Flag (M2) antibody and analyzed by SDS-PAGE. The gel was treated with EN3HANCE and exposed to Kodak XOMAT film.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/5/10.1182_blood.v100.5.1679.h81702001679_1679_1688/3/m_h81723074009.jpeg?Expires=1767841550&Signature=evcxDNM~n630W4GW5ffjsdj9zL~EkbxeMvU0YMxZG73ZxMDYDg~yPTh~W4efVGpsxyzKd8LzXWfu7X49yWzLOPs-uJpazPMM2UGR9GvirtM9yJtQQY2wjtCWw0bcFl5pNyHj1~DlxsRxZdFLqhEyhOmVVFvMbwKYHguzlJYYOiWV~gBzwPvvDdkERk5Hy4nOIcdVcVOKywksYtiMeCu28b-6AytJmlW7IkpJTJH0LhitolAFCxIiiyYx8VJ-rYanihx41tGE8nRR7BGqlS4JflvoKOh3B~cpvLWXsXJpICzZFYhrwg74Kmo1c-zVzV75G04mXibTd4MQd7vArK2JwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal