Abstract
Multiple myeloma (MM) is a plasma-cell malignancy characterized by marked epidemiological, biological, and clinical heterogeneity. The goal of this study was to find a genetic basis for this heterogeneity. Using fluorescence in situ hybridization, we analyzed a prospective cohort of 901 patients with various plasma-cell disorders—monoclonal gammopathies of undetermined significance, smoldering MM, MM, and primary plasma-cell leukemia—for genetic abnormalities involving the 13q14 and 14q32 chromosomal regions; the patients were consecutively enrolled in the Intergroupe Francophone du Myélome clinical trials, We performed statistical analyses comparing these chromosomal abnormalities in terms of immunological (ie, immunoglobulin types and light-chain subtypes) and clinical status and, to some exent, prognostic features. It was found that 14q32 translocations and del(13) are the most frequent chromosomal abnormalities, observed in 75% and 45% of the patients, respectively, and are not randomly distributed, but interconnected. Second, correlations between them allowed us to define 4 major genetic categories of patients: (1) patients lacking any 14q32 abnormality (25%) and generally also lacking del(13); (2) patients presenting either t(4;14) or t(14;16), almost always associated with a del(13) (15% of patients); (3) patients with other 14q32 abnormalities and presenting del(13) (25%); and (4) patients with other 14q32 abnormalities but not presenting del(13) (35%). Third, we show that this genetic stratification is highly correlated with immunological status and clinical presentation and with some major prognostic factors. For the first time, this study gives genetic support to the heterogeneity observed in patients with MM and demontrates that the 14q32 and 13q chromosomal abnormalities are not randomly distributed. The strong correlations we found might be the basis for a novel genetic classification of MM, as has been previously demonstrated for leukemias and lymphomas. Furthermore, our study supports different models for MM oncogenesis.
Introduction
Multiple myeloma (MM) is a monoclonal malignant plasma cell (PC) disorder with an apparent homogeneity as opposed to leukemias and lymphomas.1 Actually, prognosis is highly variable, with survival ranging from a few days to more than 10 years. A certain degree of heterogeneity is also associated with some immunological features, mainly immunoglobulin types and light-chain subtypes. Furthermore, epidemiological studies have shown that at least one third of MMs emerge from a pre-existing benign monoclonal PC disorder, ie, monoclonal gammopathy of undetermined significance (MGUS).2 These studies suggest that 2 types of MM could exist: MM secondary to pre-existing MGUS and primary, ie, de novo, MM that bypasses the MGUS stage. Finally, some patients present with a very aggressive form of MM at diagnosis, ie, primary PC leukemia (PCL). The origin of this heterogeneity is unknown. We and others have recently shown that PC disorders are associated with recurrent chromosomal abnormalities, mainly 14q32 translocations,3-5 involving the immunoglobulin H(IGH) locus, and 13q deletions, del(13).6-8 In the current study, we evaluated such chromosomal abnormalities in 901 patients with MGUS, smoldering MM (SMM), overt MM, and primary PCL. For the first time, we show that these chromosomal abnormalities are not randomly distributed, but strongly interconnected and correlated with natural history (ie, preceded or not preceded by a history of MGUS) immunological (in term of immunoglobulin type and light-chain subtype) and clinical status, and prognostic presentation. Thus, our data show that heterogeneity of MM is highly dependent on early genetic events, as in leukemias and lymphomas, and support different models for MM oncogenesis.
Patients, materials, and methods
Patients
Included in this study were 669 consecutive patients (median age, 62 years; range, 29-76 years) with MM, referred to the Intergroupe Francophone du Myélome (IFM) clinical centers. There were 582 patients evaluated at diagnosis before any therapy, and 87 at the time of first relapse. Immunoglobulin types were as follows: immunoglobulin (Ig)–G, 399 patients; IgA, 175 patients; and light chains only, 95 patients. We also analyzed 46 patients with primary PCL and 186 individuals with either MGUS or SMM. The diagnostic criteria used in this study were those commonly used.9-12 MGUS was assessed when individuals had serum monoclonal protein lower than 30 g/L and bone marrow plasmacytosis lower than 10% in the absence of any clinical symptoms, lytic bone lesions, anemia, hypercalcemia, and renal function impairment. Patients with SMM fulfilled the same criteria, except that the monoclonal component was higher than 30 g/L and/or bone marrow plasmacytosis was higher than 10%. Primary PCL was defined by the presence of at least 2000 PCs per microliter in the peripheral blood and/or if PCs represented more than 20% of peripheral white blood cells. Finally, diagnostic criteria for MM were those proposed by the Southwest Oncology Group.12 All the patients analyzed at diagnosis were consecutively enrolled in the IFM clinical trials and were analyzed between June 2000 (time of activation of the new IFM trials) and January 2001. Consequently, no outcome data are available for survival analyses at the present time. Approval was obtained from the Institutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki.
Human myeloma cell lines
In order to compare our data obtained in patients with those found in human myeloma cell lines (HMCLs), we collected the characteristics at diagnosis of the patients who gave birth to 35 HMCLs, for whom these data, as well as chromosomal analyses, were available. Drexler et al13 have recently reviewed 29 of these HMCLs (FLAM-76, Karpas 620, KMS-12-PE, MOLP-5, NOP-2, SKMM-1, SKMM-2, U266, XG1, XG2, XG5, XG6, XG7, KMM-1, JIM-3, KMS-18, LP1, H929, OPM-2, UTMC-2, KMS-11, L363, ANBL6, JJN-3, MM-1, RPMI 8226, EJM, TH, AMO-1); the other 6 HMCLs (NAN-1 to NAN-3, SBN1, BCN, and MDN) were recently established in our laboratory. Most of the cytogenetic data have been published by Bergsagel's group3,14 15 or established in our own laboratory.
PC preparation
Heparinized bone marrow samples were sent from the IFM biological centers to the Central Laboratory of Hematology in Nantes (France). After dilution with RPMI-1640 medium, mononuclear bone marrow cells were separated by density gradient centrifugation over Ficoll-Hypaque. Then, PCs were positively selected by means of anti-CD138–coated magnetic beads (Miltenyi Biotech, Paris, France), as previously reported.16 Purity of selected PCs was evaluated by morphology. Then, 500 000 to 1 million purified PCs (purity, 98%; range in all patients, 92%-100%) were then fixed in Carnoy fixative (methanol/acetic acid 3:1 [vol/vol]) for fluorescence in situ hybridization [FISH] analysis. Cells were then kept frozen in suspension, and slides were prepared immediately before hybridization.
FISH analysis
FISH analyses were performed as previously reported.16 For each probe, 100 PCs were scored. Chromosome 13 deletions were identified by means of a D13S319 probe (Vysis, Voisins-le-Bretonneux, France), mapping at 13q14. Cutoff for a deletion assessment was fixed at 10% of the cells. Rearrangements of the 14q32 region were determined by means of a dual-color FISH assay previously described.16 Briefly, we used the Ig10 cosmid (obtained from T. Rabbitts, MRC, Cambridge, United Kingdom) and the yIgH6-9 cosmid17 (a kind gift from M. Kuehl, National Cancer Institute, Bethesda, MD), mapping at the centromeric and telomeric borders of the IGH locus, respectively. Illegitimate rearrangements were assessed by a separation of the 2 probes. Because physiological rearrangements of the IgH gene occur in all the cells of the B-lineage, the threshold of positivity was evaluated on 2000 nuclei from 5 patients with myeloid malignancies (either chronic myelogenous leukemia or acute myelogenous leukemia), and fixed at 14.6% (mean + 3 SDs). Patients with 14q32 rearrangements were then analyzed with the Ig10 cosmid and probes specific for the 4 most frequent partners: 11q13, 4p16, 16q23, and 8q24. These loci were examined by means of the following probes: the 6.22 cosmid18 (kind gift from E. Schuuring, Department of Pathology, University of Leiden, Leiden, The Netherlands) for 11q13, a PAC probe containing the FGFR3gene14 (kind gift from L. Bergsagel, Department of Pathology, Cornell University, Ithaca, NY) for 4p16, a PAC probe containing the c-maf gene15 (kind gift from L. Bergsagel) for 16q23, and the 934E1 YAC probe (obtained from the Centre d'Etudes du Polymorphisme Humain (CEPH), Paris, France) for 8q24. Translocations were observed as fusions between the Ig10 cosmid and one of the other probes. Translocation t(11;14) was identified by means of the Ig10 cosmid probe (labeled in green) and the 6.22 cosmid probe (labeled in orange), kindly provided by E. Schuuring, mapping at 11q13 and containing the CCND1 gene. To test this probe, 50 patients with MM and a t(11;14) previously diagnosed with a commercial probe (Vysis), were reanalyzed with this set of probes. All 50 patients were also t(11;14)–positive with the use of this set. The analysis of 2000 peripheral blood or bone marrow cells from healthy individuals enabled us to fix the positivity threshold: 10.5% (mean + 3 SDs). Translocation t(4;14) was sought with the use of the Ig10 cosmid (labeled in green) and anFGFR3-specific PAC probe (labeled in orange, kindly provided by L. Bergsagel. In patients with t(4;14), the FGFR3 probe is translocated on the der(14). The cutoff for positivity was calculated under the same conditions as for t(11;14) and was fixed at 10.3% (mean + 3 SDs). Translocation t(14;16) was analyzed by means of the Ig10 cosmid probe (labeled in green) and ac-maf–specific PAC probe (labeled in orange), kindly provided by L. Bergsagel. This probe is localized on the telomeric side of all the 16q23 breakpoints of the t(14;16) reported so far.19 Moreover, to test the ability of this set of probes to detect t(14;16), we blindly analyzed 4 HMCLs (JJN3, ANBL-6, BCN, and NAN-2) and 4 patients with cytogenetically proven t(14;16). In all 8 cases, IgH/c-maf fusions were observed in interphase PCs. The cutoff for positivity was fixed at 9.8% (mean + 3 SDs). Finally, t(8;14) was analyzed by means of Ig10 and a YAC spanning thec-myc locus (934E1) obtained from the CEPH. The cutoff value for this probe set was 9%. Patients were systematically analyzed with the whole panel of probes, even those presenting a 14q32 translocation with the first tested set of probes, in order to detect possible double translocations.
Statistical analysis
Fisher exact test and chi-squared test were used for the statistical evaluation of the results.
Results
14q32 rearrangements
Results are summarized in Tables 1 and2. Rearrangements of the 14q32 region were observed in 73% of MMs at diagnosis or relapse, 84% of primary PCLs, but only 47% and 48% of MGUSs and SMMs, respectively (P < 10−9). This incidence of 14q32 rearrangements is higher than that previously reported by our group, probably because of the more telomeric position of the IgH-5′ probe used in this study, enabling us to pick up translocations involving more telomeric breakpoints. Subsequent analyses did not show any difference between MGUS and SMM. Thus, both entities were analyzed together.
The most frequent 14q32-specific rearrangement was translocation t(11;14)(q13;q32), observed in 16% of MMs, 33% of primary PCLs (P = .006), and 15% of MGUSs/SMMs. Of note, whereas t(11;14) was observed in 14.5% and 10.3% of IgG and IgA MMs, respectively, it occurred in 30.5% of light-chain-only MMs (P < .0005 and P < .0001 for difference with IgG and IgA, respectively). This association with the light-chain-only isotype directly explained the excess of t(11;14) in primary PCL, since 7 of 15 primary PCLs displaying the translocation secreted only light chains.
The second specific rearrangement was t(4;14)(p16;q32), observed in 10% of MMs, in 13% of primary PCLs, but only 2% of MGUSs/SMMs (P < .001). Of note, whereas t(4;14) was observed in 8% of IgG MMs, it was identified in 19% of IgA MMs (P < .0001). In contrast, its incidence was very low in light-chain-only MM (4.2%, P = .001 for difference with IgA MMs).
The incidence of the 2 other specific translocations was significantly lower. Translocation t(14;16)(q32;q24) was observed in only 14 patients with MM, but in 5 patients with primary PCLs (P = .002) and only 1 with MGUS/SMM. Thus, t(14;16) might be associated with specific biological features allowing PCs to disseminate within the peripheral blood. Translocation t(8;14)(q24;q32) was identified in 12 patients with MM, none with primary PCL, and 2 with MGUS/SMM. No specific correlation with immunoglobulin type and light-chain subtype was observed. In 3 patients, this translocation was associated with another 14q32-specific translocation (1 case of each).
Rearrangements of the 14q32 region with other, unknown partners were observed in 44% of MMs, 27% of primary PCLs (P = .03), and 30% of MGUSs/SMMs (P < .001 for difference with MM). Such rearrangements were seen in 44% of IgG MMs, 46% of IgA MMs, and 44% of light-chain-only MMs. In contrast, a germline 14q32 configuration was observed significantly more frequently in IgG MM (32%) than in other subtypes (22% in IgA MM, P = .02, and 17% in light-chain-only MM, P = .007 for difference with IgG MM).
Thus, for the first time, a strong correlation was found in MM and primary PCL between the 14q32 chromosomal abnormalities and the most common genetic feature of the patients, the switch-dependent immunoglobulin types (but not light-chain subtypes). Furthermore, striking differences between MGUS/SMM and MM/primary PCL were confirmed, suggesting that some 14q32 abnormalities may influence the natural history of MM in terms of the pre-existence or absence of MGUS, ie, t(4;14), or of the existence or absence of a presenting leukemic phase, ie, t(14;16).
Chromosome 13 deletion
We identified del(13) in 43% of MMs at diagnosis or relapse, in 70% of primary PCLs (P < .001), and in 23% of MGUSs/SMMs (P < 10−6 for difference with MMs) (Table 1). The deletion was observed in 75% of PCs (range, 16%-95%) in MM, and in 80% of PCs (range 40%-94%) in primary PCL, but in 42% (range, 14%-86%) in MGUSs/SMMs (P < 10−8). The incidence of the deletion was 40%, 49%, and 48%, in IgG, IgA, and light-chain-only MM, respectively (Table 2). In contrast, del(13) was significantly more frequent in patients with a λ subtype (48%) than in those with a κ subtype (38%; P < .03).
Correlations between del(13) and 14q32 rearrangements
We then analyzed the incidence of del(13) in each 14q32 category of MM (Table 3). Patients analyzed at relapse were pooled with newly diagnosed patients for these correlation analyses, since we and others have previously shown that the correlations occurred at diagnosis and not during evolution. This is also confirmed by the current study (see previous sections). Moreover, a separate analysis of only newly diagnosed patients did not modify the results (data not shown). Strong associations, especially with t(4;14) and with t(14;16), were identified: 85% of patients with t(4;14) and 92% of those with t(14;16) displayed del(13), respectively. These percentages are dramatically higher than those observed in the other 14q32 categories. Comparisons of del(13) incidence in t(4;14) MM patients with the incidence observed in the overall population revealed highly significant differences (85% versus 43%;P < 10−9); this was true as well as for comparisons with t(11;14) MM patients (40%;P < 10−9) or with patients with other 14q32 rearrangements (42%; P < 10−9). The difference is even more significant when patients lacking 14q32 abnormality are compared; del(13) was observed in only 22% of these patients (P < 10−9).
Correlations with β-2 microglobulin and creatinine serum levels
To get information on the possible prognostic value of these different subgroups, we analyzed the β-2 microglobulin serum levels in each of them (Table 3) and analyzed the creatinine levels in a subset of 264 newly diagnosed patients for whom the data were available. Whereas no correlation was found between del(13) and β-2 microglobulin serum levels, strong associations were found between β-2 microglobulin and 14q32 abnormalities. In the t(4;14) and t(14;16) subgroups, only 7% and 21% of the patients were in the low β-2 microglobulin group (β-2 microglobulin below 3 mg/L), respectively (P < 10−9 for difference with the other patients), whereas 54% of the patients lacking 14q32 abnormality belong to this group (P < .001 for difference with patients with 14q32 abnormality). Finally, patients with t(11;14) were not different from the whole population, with 49% of them in the low β-2 microglobulin group. Regarding renal function, patients were classified as stage A (creatinine level below 180 μM) or stage B. At diagnosis, 42 patients displayed renal function impairment. The only significant correlation was between stage B and del(13) (P = .05). No correlation with illegitimateIGH rearrangements were observed.
Using our novel and powerful prognostic staging system,8we isolated 3 categories of patients, depending on the number of adverse prognostic parameters presented at diagnosis, ie, low or high β-2 microglobulin level and presence or absence of del(13). We tested this prognostic system in each 14q32 category of patients. Patients lacking any 14q32 abnormality were essentially in groups 0 and 1, ie, good prognosis groups (only 13% of these patients were in group 2; P < .0001), whereas patients with t(4;14) or t(14;16) were mostly in group 2 (64%,P < 10−9, and 71%, P < .001, respectively).
Human myeloma cell lines
We analyzed 35 HMCLs. For all of them, we collected the clinical and immunological data of the corresponding patients at diagnosis and the chromosomal characteristics available in the literature or established in our laboratory. Of note, 11 of these HMCLs (31%) derived from patients with primary PCL, whereas 23 derived from patients with MM. Only 6 of these HMCLs (17%) were established as soon as the diagnosis was made, whereas 28 derived from patients in relapse (and especially from extramedullary sites). Analysis of immunoglobulin types also confirmed a biased distribution, showing predominance of non-IgG patients. Ten patients were IgG (29%); 11 were IgA (32%); 10 secreted only light chains (29%); 2 did not secrete any immunoglobulin chain; and 1 was IgE. This first analysis confirms a striking bias between patients and HMCLs, with an excess of primary PCL, relapse (especially extramedullary relapse), and non-IgG types in patients from whom HMCLs emerged. Analysis of the 14q32 translocations revealed a t(11;14) in 11 cases (involving CCND1), a t(4;14) in 8 cases (involving FGFR3), a t(14;16) in 7 cases (involvingc-maf), a t(14;20) in 4 cases (involving MAF-B), a t(6;14) in 2 cases (involving CCND3 in 1 HMCL andIRF4 in a second one), and a t(12;14) in 1 case. Of note, one HMCL (KMS11) presented 2 rearrangements: 1 with c-mafand 1 with FGFR3. Three cell lines did not display any 14q32 abnormality. Despite the bias of selection for the patients able to generate an HMCL, striking associations with clinical status at diagnosis and immunoglobulin types were observed (Table4), confirming the data obtained in the patients, ie, the following associations: t(11;14) with light-chain-only isotypes, t(4;14) with IgA type, and t(14;16) with primary PCL.
Discussion
Our study demonstrates for the first time that the 2 most frequent cytogenetic abnormalities, 14q32 translocations and 13q deletions, are not randomly distributed in MM. In contrast, striking associations were found among these cytogenetic abnormalities as well as between these cytogenetic abnormalities and immunoglobulin types and light-chain subtypes, natural history, and some clinical and prognostic presenting features. These correlations led us to propose a genetic stratification of patients with PC disorders, stratification that corresponds to a true model of oncogenesis, including different pathways of malignant transformation.
We and others have shown that 14q32 rearrangements involve theIGH locus, which is rearranged with several chromosomal partners.3-5 We found a similar incidence of 14q32 rearrangements at diagnosis or at relapse as well as in at least 50% of MGUSs. Thus, these data suggest that such illegitimateIGH rearrangements (1) occur very early in the clonal development and (2) do not influence response to treatment, as previously shown for del(138). However, about 25% of patients with MM or primary PCL do not display such rearrangements, even when the focus is on patients with available clonal metaphases (data not shown). Thus, the role of these chromosomal changes in malignancy is currently not fully understood. All the translocations cloned to date lead to the upregulation of the partner gene. A hypothesis for understanding the role of the 14q32 translocations could be to consider the deregulation of the partner gene as the principal consequence of the genetic alteration. In this case, oncogenetic pathways would be different from patient to patient, depending on the nature of the partner. So far, 3 major 14q32 translocations have been identified in human myeloma cell lines, each of them being observed in roughly 25% of cases: t(4;14), t(11;14), and t(14;16).20 Thus, these 3 14q32 chromosomal abnormalities cover the majority of the HMCLs. Target genes, belonging to different families of oncogenes, have been cloned in each of them:FGFR3, CCND1, and c-maf, respectively. Our present analysis demonstrates a less documented situation in the patients since only 2 of these translocations, t(11;14) and t(4;14), are really recurrent, in 16% and 10% of the cases, respectively; translocation t(14;16) is encountered in only 2% of MM patients. No systematic analysis of t(14;16) incidence in MM has been reported so far. Regarding t(4;14) and t(14;16), our results confirm our previous analyses5,16,21 and other studies,22 even though some studies analyzing fewer patients found a slightly higher incidence of t(4;14),23 probably reflecting patient selection bias. Thus, 14q32 analysis enables the identification of 4 main chromosomal categories in MM: with t(11;14) (16%); with t(4;14) (10%); with no 14q32 rearrangement (approximately 25%); and with other 14q32 rearrangements (48%).
Although del(13) is observed in all stages, its incidence varies in each condition and appears to be correlated with disease aggressiveness, but not with immunoglobulin type. Whereas del(13) is observed in only 22% of individuals with MGUS, it occurs in 43% of patients with MM (either at diagnosis or at relapse) and in 70% of those with primary PCL. These facts confirm that it is an early event in MM oncogenesis, discriminating 2 types of patients, as 14q32 translocations do, with a more or less aggressive disease as soon as the diagnosis is made. Indeed, we and others have shown that del(13) is associated with a short survival, independently of β-2microglobulin levels.6-8 Thus, a tempting hypothesis would be to suppose a direct correlation between del(13) in MGUS and a subsequent malignant transformation, as we have previously suggested.21 This model would agree with the transformation rates observed in MGUS during 20 years. However, further analyses (including larger cohorts of patients and a longer follow-up) will be needed to definitely validate this hypothesis. A more detailed analysis of the 4 14q32 categories, in relation to del(13) shows strong correlations between 13q and some 14q32 translocations and allows us to describe 4 subsets of patients with probably different oncogenesis.
Translocation t(4;14) identifies a homogeneous group of patients characterized by a very high incidence of del(13q14) (85%). Of note, t(4;14) is the most frequent translocation in IgA MM patients, but has a very low incidence in light-chain-only MM. Although its incidence is not different in MM and in primary PCL, it is almost never encountered in MGUS/SMM, confirming, on a much larger series, our previously reported results.21 The most likely hypothesis to explain these highly significant differences is that t(4;14), throughFGFR3 deregulation, directly precipitates clonal PCs into fully malignant PCs, bypassing a stable MGUS phase. Thus, t(4;14)-positive MM may correspond to true de novo primary MM. These results confirm our previously published data21 (on a much larger number of patients), but are in disagreement with a recent report by Fonseca et al22 finding t(4;14) in 5 of 52 individuals (9.6%) with MGUS/SMM, a similar incidence as in their MM series (10.3%). Further analyses focusing on such individuals are clearly needed to accurately interpret these differences. Finally, it is noteworthy that such MMs have many poor prognostic factors at diagnosis: del(13), high β-2 microglobulin levels, IgA types (the current data), and a higher proliferative activity (unpublished data, September 2001).
Despite its lower incidence, t(14;16) displays particularities similar to those of t(4;14). In this series, it has been identified in fewer than 1% of MGUS cases and in 2% of MMs. On the other hand, it has been observed in 11% of primary PCLs. A similar association with primary PCL has been found in HMCLs. Finally, a highly significant association was observed with del(13) (95%) and with poor prognosis (71%). Thus, this translocation, like t(4;14), could correspond to de novo MM with a poor prognosis.
The situation is dramatically different for patients with t(11;14). In contrast to t(4;14) and t(14;16), t(11;14) is observed in all types of PC disorders, from MGUS to primary PCL, with the same incidence in MGUS/SMM and MM (15%). Because about one quarter of patients with t(11;14) have light-chain-only MM (which cannot be post-MGUS MM), we can expect that at least one quarter of MMs with t(11;14) are de novo MMs. No specific correlation was observed with del(13). Finally, according to our novel prognostic staging system,8 the analysis of patients with t(11;14) reveals a balanced distribution within each subgroup, suggesting that this translocation should not be associated with a specific prognostic subgroup.
The group of patients with 14q32 rearrangements, but with other partners, is probably a highly heterogeneous group. In our model, if the subgroups of patients are defined by the partner identity, this group should be split in an unknown number of different subgroups, with specific characteristics. As expected from such an important heterogeneity, this subgroup does not present any significant correlation (with either del(13) or immunoglobulin isotypes) when compared with the entire population. In this context, the discovery of novel partners represents a major challenge for the future.
Finally, the subgroup of patients lacking any 14q32 abnormality turns out to be a homogeneous entity, as frequent in primary PCL as in MM. These patients rarely display del(13) (22%) and are significantly more frequently of the IgG type. For the majority of these patients, oncogenesis would be independent of IGH rearrangements and of del(13).
As previously emphasized, the relative incidences of the specific 14q32 translocations differ significantly in patients and in HMCLs. In this HMCL series, CCND1, FGFR3, and c-mafrearrangements were found in 31%, 23%, and 20% of the HMCL cases, respectively, versus 16%, 10%, and 2% in the patients. The question was whether these differences were true differences or relative differences due to a bias in HMCL generation. Of interest, a careful analysis of the HMCLs revealed strong similarities with the patients in terms of correlations between translocations, immunoglobulin types, and clinical presentation. Indeed, in HMCLs, as in MM patients, t(11;14) was highly correlated with light-chain-only MM (6 of 11 HMCL cases, 55%). Similarly, t(4;14) was strongly associated with the IgA type (5 of 8 HMCL cases, 63%), again as in MM patients. Finally,c-maf rearrangements were significantly more frequent in HMCLs derived from primary PCL (4 of 7 HMCL cases, 57%), again as in patients. Furthermore, striking differences in immunoglobulin types and clinical presentation were observed between patients and HMCLs, eg, a higher incidence of primary PCL (10 of 35 HMCL cases, 29%, versus less than 5% in patients) or differences in the immunoglobulin types repartition (29% of IgG, 32% of IgA, and of light-chain-only in HMCLs, versus 60%, 26%, and 14%, respectively, in our patients). Thus, the data observed in HMCLs (1) confirm the strong correlations we found in patients, but (2) suggest a bias in the selection of patients able to generate a cell line. A likely hypothesis would be that activation of 1 of these 3 oncogenes is an almost necessary condition for the establishment of an HMCL. In contrast, patients lacking 14q32 abnormality and those presenting a rearrangement with another or an unknown chromosomal partner are not likely to generate a cell line, since only 3 HMCLs (9%) lacked 14q32 abnormality (versus 25% in patients), and 6 are rearranged with other partners (17%), including 4 cases of t(14;20) (versus 44% in patients). These data suggest that this latter group of patients could be more homogeneous than originally supposed, at least in their inability to generate HMCLs. Whether this finding is related to different oncogenetic pathways is a major question for the future.
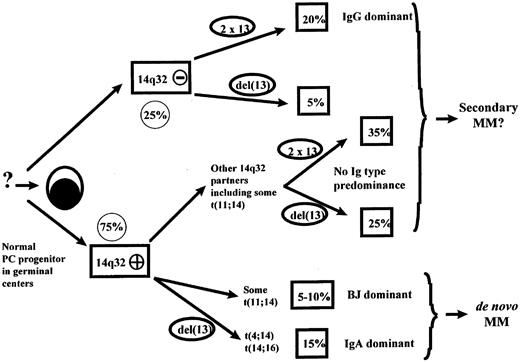
In conclusion, when one considers both 14q32 rearrangements and del(13), our study for the first time makes possible the definition of different categories of MM based on significant correlations between these genetic abnormalities, immunoglobulin types, and light-chain subtypes, natural history, and clinical and prognostic presentation. This leads us to propose a multistep model of PC oncogenesis (Figure1). In this model, 14q32 translocations represent the earliest genetic events in a subset of MMs, probably occurring upstream of PC progenitors in the germinal centers, and defining some subsets of MM according to whether the first “choice” is to illegitimately rearrange the IGH locus or not to do so. This theory is also supported by the correlations found between some recurrent translocations and immunoglobulin isotypes. The immunoglobulin type of the PCs is dependent on the selection of the constant region during the switch process, presumably in the germinal centers. Since most of these recurrent 14q32 translocations, t(4;14), t(11;14), and t(14;16), involve switch regions, they are likely to occur at the same time. These data support the hypothesis that 14q32 translocations make up one of the earliest oncogenetic events, preceding del(13). In this model, del(13) would be a secondary (although early) event, possibly driven by some specific 14q32 translocations, such as t(4;14) or t(14;16).
The first “choice,” ie, no 14q32 illegitimate rearrangement, observed in 25% of the cases, represents a first oncogenetic pathway that is clearly triggered by unknown mechanisms. The group of patients in this subset is characterized by a high incidence of IgG type but a low incidence of del(13), and thus a rather genomic stability and probably an indolent presentation, with the exception of 20% of these patients with del(13). The second oncogenetic pathway is 14q32 dependent and involved 75% of the cases. In this pathway, diversity is introduced by the nature of the 14q32 partner, which can be recurrent or not. Within recurrent translocations, a first group is represented by cases with t(4;14), t(14;16), and probably a subset of t(11;14). These translocations are characterized, for t(4;14), by preponderant IgA; for t(11;14), by light-chain-only types; for t(4;14) and t(14;16), by a frequent association with del(13); and, for t(4;14), t(14;16), and t(11;14) in light-chain-type MM, by their virtual absence in MGUS/SMM stages. Thus, patients in this subgroup are prone to represent true de novo, ie, primary, MM and primary PCL. A second group is represented by MM in which oncogenesis is driven by translocations with other partners. Natural history of the disease in these cases could be dependent on the nature of the partner and the presence or absence of del(13). Patients presenting t(11;14) may be more heterogeneous. Patients presenting this translocation are more likely to secrete light chains only, but are not characterized by any correlation with del(13) or β-2 microglobulin level. Moreover, t(11;14) is encountered as well in MGUS as in primary PCL. Thus, t(11;14)-associated MM may correspond (at least in part) to de novo MM for those of the light-chain-only type (because MGUS cases are almost never of the light-chain-only type), and to secondary post-MGUS MM, again at least in part. The outcome of these patients is probably related to other intrinsic parameters such as del(13) and β-2 microglobulin level.
Recently, another oncogenetic model based on the location of the 14q32 breakpoints has been proposed (P. L. Bergsagel, oral communication, 8th International Myeloma Workshop, Banff, Scotland, May 2001). Translocations with FGFR3, CCND1, andc-maf mostly involve breakpoints within the switch andJH regions and would be considered primary events. In contrast, other translocations would occur at other sites and would be considered secondary events. Our data do not totally support this hypothesis, especially because (1) whatever the partner, the chromosomal change is usually observed in the large majority of PCs, and (2) the high incidence of unknown partners is observed equally in MGUS (ie, in nonmalignant clonal PCs) and in MM, supporting the hypothesis that 14q32 abnormalities are early (perhaps even primary) oncogenetic events. However, our model would agree with P. L. Bergsagel's regarding the location of the breakpoints. HMCLs with (4;14) and most of those with t(11;14) and t(14;16) display breakpoints located within the switch or JH regions (P. L. Bergsagel, oral communication, 8th International Myeloma Workshop, Banff, May 2001). These cases would represent true de novo MM, whose oncogenesis would involve primary IgH rearrangements occurring during the switch or somatic mutation processes. This group of patients would constitute a homogeneous population, prone to generate cell lines. In contrast, other (perhaps nonrecurrent) breakpoints would not involve switch or JH regions, but would occur at random sites within the IgH locus (P. L. Bergsagel, oral communication, 8th International Myeloma Workshop, Banff, May 2001), mediated by other mechanisms such as genomic instability. In these cases, as for the patients lacking any 14q32 abnormality, oncogenesis would depend on other, unknown mechanisms. Because these latter cases are observed with a similar incidence in MGUS, they are likely to represent secondary (post-MGUS) MM. This group of patients can be integrated in our model. Whether the 14q32 abnormalities occur in the germinal centers or later in the evolution is an unresolved issue. The demonstration of this model will require the mapping of the breakpoints in patients with nonrandom or unknown 14q32 partners and represents a major challenge for the future.
In conclusion, this study, which is the largest study focused on the oncogenesis of MM, demonstrates a so far unsuspected genetic heterogeneity in MM. Because we found that genetics correlate with the main factors of heterogeneity previously documented, ie, natural history, clinical and prognostic presentation, immunoglobulin types, and light-chain subtypes, our study provides novel bases for a more accurate classification of PC disorders. Finally, our study shows that myelomas resemble leukemias and lymphomas and that they should benefit from large-scale microarray studies enabling one to perform better stratifications.
We thank Axelle Daviet and Stéphanie Saunier for excellent technical assistance in cell processing and in situ hybridization experiments.
Supported by grants from the Association pour la Recherche contre le Cancer (ARC), the Fondation de France, and Programme Hospitalier de Recherche Clinique.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hervé Avet-Loiseau and Régis Bataille, Laboratoire d'Hématologie, Institut de Biologie, 9 quai Moncousu, 44093 Nantes, France; e-mail: havetloiseau@chu-nantes.fr orfrb@sante.univ-nantes.fr.