Friedreich ataxia (FA) is caused by decreased frataxin expression that results in mitochondrial iron (Fe) overload. However, the role of frataxin in mammalian Fe metabolism remains unclear. In this investigation we examined the function of frataxin in Fe metabolism by implementing a well-characterized model of erythroid differentiation, namely, Friend cells induced using dimethyl sulfoxide (DMSO). We have characterized the changes in frataxin expression compared to molecules that play key roles in Fe metabolism (the transferrin receptor [TfR] and the Fe transporter Nramp2) and hemoglobinization (β-globin). DMSO induction of hemoglobinization results in a marked decrease in frataxin gene (Frda) expression and protein levels. To a lesser extent, Nramp2messenger RNA (mRNA) levels were also decreased on erythroid differentiation, whereas TfR and β-globinmRNA levels increased. Intracellular Fe depletion using desferrioxamine or pyridoxal isonicotinoyl hydrazone, which chelate cytoplasmic or cytoplasmic and mitochondrial Fe pools, respectively, have no effect on frataxin expression. Furthermore, cytoplasmic or mitochondrial Fe loading of induced Friend cells with ferric ammonium citrate, or the heme synthesis inhibitor, succinylacetone, respectively, also had no effect on frataxin expression. Although frataxin has been suggested by others to be a mitochondrial ferritin, the lack of effect of intracellular Fe levels on frataxin expression is not consistent with an Fe storage role. Significantly, protoporphyrin IX down-regulates frataxin protein levels, suggesting a regulatory role of frataxin in Fe or heme metabolism. Because decreased frataxin expression leads to mitochondrial Fe loading in FA, our data suggest that reduced frataxin expression during erythroid differentiation results in mitochondrial Fe sequestration for heme biosynthesis.
Introduction
Friedreich ataxia (FA) is an inherited neurodegenerative condition1,2 with an incidence of 1:30 000 in the European population.3 The gene FRDA that is defective in FA encodes a mitochondrial protein known as frataxin.2,3 In 97% of individuals with FA the defect in this gene is due to a GAA triplet repeat expansion in intron 1 (chromosome 9q13) that results in a marked decrease in gene expression.2,3 Over the last few years evidence has accumulated to suggest that frataxin plays a role in mitochondrial iron (Fe) metabolism.3-24 However, the function of mammalian frataxin remains unclear.24
Experiments using the yeast, Saccharomyces cerevisiae, led to a model that may explain the pathogenesis of FA.10,12The yeast gene YFH1 is homologous to the human geneFRDA,12 and encodes a mitochondrial protein (Yfh1p) involved in Fe homeostasis and respiration.5,9,10,12 When the YFH1 gene was deleted, a marked accumulation of mitochondrial Fe occurred, resulting in loss of mitochondrial DNA and impaired respiration.6,7,12,13 Thus, this led to a hypothesis that the accumulated Fe resulted in the production of free radicals that damage biologic targets. When YFH1 was reintroduced into the yeast, mitochondrial Fe was exported back out into the cytosol as free non–heme-bound Fe.10,12 Therefore, Yfh1p appeared to regulate Fe export from mitochondria in yeast cells.10However, Yfh1p may not be an Fe transporter, because it has no transmembrane sequences necessary for this role.10
Both the human and mouse frataxin genes (FRDA andFrda, respectively), are highly expressed in tissues that are rich in mitochondria, including heart, liver, and skeletal muscle.5,14 Consistent with the yeast knockout model, fibroblasts from FA patients were more sensitive than normal cells to oxidant stress,24 and contained higher levels of mitochondrial Fe.16,21 Further, Fe accumulation and reductions of mitochondrial DNA, complex I/II/III, and aconitase have been noted in the heart4,11 and central nervous system of patients with FA.22,23 Other studies in FA patients showing Fe deposits within heart myofibrils18,19 and defective mitochondrial respiration7,8 also suggest mitochondrial Fe overload. Considering these observations, it is significant that antioxidants have successfully treated the cardiomyopathy of FA.25
If frataxin plays a role in Fe metabolism, an understanding of the pathogenesis of FA will require the examination of mitochondrial Fe processing pathways. The best characterized function of mammalian mitochondria in Fe metabolism comes from studies in erythroid cells, where it is involved in the production of heme for hemoglobin synthesis.24,26 In these cells, the Fe-transport protein transferrin (Tf) binds to the transferrin receptor (TfR) and is endocytosed. Iron is released from Tf by a decrease in endosomal pH and traverses the membrane via the transporter, Nramp2 (also known as the divalent metal ion transporter 1).24 The Fe is then targeted to the mitochondrion where it is incorporated into protoporphyrin IX (PIX) to form heme (for reviews, see Becker and Richardson24 and Ponka26). This molecule is then exported out of the mitochondrion to combine with globin chains to form hemoglobin. The rate-limiting step in the heme synthesis pathway is the acquisition of Fe from Tf.26 When the heme biosynthesis pathway is inhibited using isonicotinic acid hydrazide (an inhibitor of δ-aminolevulinic acid synthase)28 or succinylacetone (SA; a competitive inhibitor of δ-aminolevulinic acid dehydratase),29 Fe accumulates within the mitochondrion and to some degree the cytoplasm.26,27,30-33 Only when PIX is added does Fe become incorporated into this molecule to form heme that is transported out of the mitochondrion.10 31
The role of frataxin in Fe and heme metabolism remains unknown. It is possible to suggest a hypothesis that the decreased levels of frataxin in patients with FA may lead to some defect in either the process of Fe incorporation into PIX, or Fe influx or export, which results in mitochondrial Fe accumulation. Recently, it has been suggested that the yeast frataxin homologue (Yfh1p) may polymerize to store Fe-like ferritin.34 Another study suggested that an anionic surface patch on frataxin could oxidize Fe++ to Fe+++ when this molecule combines with itself or another ligand.35 In this way, frataxin may accumulate Fe similarly to ferritin.35 However, structural and functional studies of mammalian frataxin have shown that this molecule cannot bind Fe directly and does not polymerize to sequester Fe.36 Indeed, frataxin shares little homology with other known molecules involved in Fe or heme metabolism.36 The clustering of conserved residues onto one face of frataxin suggests an interaction with potential ligands and may indicate that frataxin acts indirectly on Fe metabolism.36
In the present study we have assessed the role of frataxin in Fe and heme metabolism by implementing a well-characterized model of erythroid differentiation, namely, Friend cells induced with dimethyl sulfoxide (DMSO).37-41 This model has previously been used to assess the role of other molecules involved in Fe and heme metabolism.37-41 We demonstrate that DMSO induction of erythroid differentiation and hemoglobinization results in a marked decrease in frataxin expression. In addition, incubation of uninduced Friend cells and other cell types with PIX resulted in down-regulation of frataxin protein levels, suggesting a possible regulatory role of frataxin in Fe and heme metabolism. Because decreased frataxin expression leads to mitochondrial Fe loading in FA, our data suggest the decrease in frataxin levels during erythroid differentiation favors mitochondrial Fe sequestration and heme synthesis. These results may explain why there are no marked effects of decreased frataxin expression in the erythroid cells of FA patients; that is, decreased frataxin expression may physiologically facilitate Fe uptake and heme synthesis.
Materials and methods
Cell treatments and reagents
Hemin, phenylmethylsulfonylfluoride (PMSF), PIX, and SA were obtained from Sigma Chemical (St Louis, MO). All experiments with PIX and heme were performed in tubes wrapped in aluminium foil to prevent the photodynamic generation of free radicals. Desferrioxamine (DFO) was obtained from Novartis (Basel, Switzerland). PIH was synthesized as described previously.42
Preparation of 59Fe-transferrin
Apotransferrin (Sigma Chemical) was labeled with59Fe (Dupont NEN, MA) to produce59Fe2-transferrin (59Fe-Tf).43
Cell culture
The mouse Friend erythroleukemia cell lines 707 and 745 as well as the human SK-N-MC neuroepithelioma cell line were obtained from the American Type Culture Collection (ATCC; Rockville, MD). Friend cells were induced to differentiate into hemoglobin-producing cells by incubation with 1.5% DMSO.
The mouse fibroblast cell line, LMTK−, and the mouse HepA1-6 hepatoma cell line were obtained from the European Collection of Cell Cultures (Salisbury, Wiltshire, United Kingdom). The mouse M15 kidney cell line was obtained from Dr Veronica van Heyningen (Medical Research Council of Edinburgh, Edinburgh, United Kingdom). All cell lines above were grown in Eagle modified minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS; Commonwealth Serum Laboratories, Melbourne, Australia), 1% (vol/vol) nonessential amino acids (Gibco BRL, Sydney, Australia), 1 mM sodium pyruvate (Gibco), 2 mM l-glutamine, 100 μg/mL streptomycin, 100 U/mL penicillin (Gibco), and 0.28 μg/mL Fungizone (amphotericin B; Squibb Pharmaceuticals, Montréal, Quebec, Canada). The RAW264.7 and J774 mouse macrophage cell lines were obtained from Dr David Cestor (Centre for Molecular and Cellular Biology, University of Queensland) and the ATCC, respectively. Both RAW 264.7 and J774 cells were grown in RPMI containing 10% Serum Supreme (Biowhittaker, Walkersville, MD), 1% (vol/vol) nonessential amino acids (Gibco BRL), 1 mM sodium pyruvate (Gibco), 2 mM l-glutamine, 100 μg/mL streptomycin (Gibco), 100 U/mL penicillin (Gibco), and 0.28 μg/mL Fungizone (Squibb Pharmaceuticals).
Assays to measure Fe uptake and Fe incorporation into heme
Friend cells were incubated in the presence or absence of DMSO for 24 to 120 hours. After this, aliquots of cells (1 × 107) were placed in tubes and incubated with59Fe-Tf (10 μM) for 1 to 120 minutes at 37°C.37 After appropriate incubation times the tubes were plunged into ice and the cells washed 3 times in a large excess of ice-cold phosphate-buffered saline.27 Incorporation of59Fe into heme was performed by standard procedures.27 Radioactivity was measured using a γ-scintillation counter (LKB Wallace 1282 Compugamma, Turku, Finland).
Northern blot analysis
Northern blot analysis was performed as described previously.44 The Frda probe consisted of an 870-base pair (bp) fragment from the coding region of murineFrda cloned into the pT7T3pac plasmid (from Dr R. Williamson, Murdoch Institute, Melbourne, Australia). The TfRprobe of 800 bp was obtained from the coding region of the murineTfR cloned into the vector pUC18 (from Dr G. Anderson, The Queensland Institute of Medical Research, Brisbane, Queensland). TheNramp2 probe consisted of 1.7 kb of coding sequence from murine Nramp2 cloned into the pMT2 plasmid (from Dr N. C. Andrews, Howard Hughes Medical Institute, Children's Hospital, Boston, MA). The β-globin probe of 612-bp was excised from the pMG5 vector (from Dr P. Ponka). The β-actin probe consisted of a 1.4-kb fragment of β-actin complementary DNA (cDNA) cloned into pBluescript SK− (ATCC catalogue no. 37997).
Western blot analysis
All procedures used for Western blotting were the same as those described previously.45 The monoclonal antibody (MoAb) against frataxin (clone 1G2) was from Chemicon International (Temecula, CA), and the MoAb against β-actin (clone AC-15) was from Sigma. The TfR MoAb (clone H68.4) was purchased from Zymed Laboratories (San Francisco, CA).
Statistics
Data were compared using the Student paired t test. Results were considered statistically significant whenP < .05.
Results
Frda (frataxin) expression is down-regulated during erythroid differentiation
Friend cells exposed to DMSO undergo erythroid differentiation resulting in an increase in Fe uptake from Tf, heme synthesis, and hemoglobinization.26,37-41 Our initial studies examined the effect of DMSO on 59Fe uptake from 59Fe-Tf (10 μM) and the incorporation of 59Fe into heme by the Friend 707 cell line (Figure 1). Cells were incubated with 1.5% DMSO or control media for incubation times of 24 to 120 hours and the effect on 59Fe uptake and incorporation into heme assessed by incubating cells with59Fe-Tf for 1 to 120 minutes (Figure 1A,B). Our studies showed that after a 24-hour incubation with DMSO there was little effect on 59Fe uptake from 59Fe-Tf and incorporation of 59Fe into heme, as shown previously (Figure 1A).40 However, after a 96-hour incubation with DMSO, there was a significant (P < .001) increase in59Fe uptake and incorporation of 59Fe into heme when compared to control cells for time points from 30 to 120 minutes (Figure 1B). Similar results to those found in Figure 1B were also found after incubating cells with DMSO for 48, 72, and 120 hours (data not shown). These data examining Fe uptake are similar to those found by others using Friend cells.40 50
Effect of inducing erythroid differentiation in Friend 707 cells on59Fe uptake and 59Fe incorporation into heme.
Cells were incubated with DMSO for 24 hours (A) or 96 hours (B). Cells were incubated in the presence and absence of 1.5% DMSO for 24 hours or 96 hours at 37°C. After this incubation the cells were then labeled with 59Fe-transferrin (10 μM) for incubations of 1 to 120 minutes at 37°C. The uptake of cellular 59Fe and its incorporation into heme were measured using standard techniques (see “Materials and methods” for details). The results are means of duplicate determinations in a typical experiment from 3 experiments performed.
Effect of inducing erythroid differentiation in Friend 707 cells on59Fe uptake and 59Fe incorporation into heme.
Cells were incubated with DMSO for 24 hours (A) or 96 hours (B). Cells were incubated in the presence and absence of 1.5% DMSO for 24 hours or 96 hours at 37°C. After this incubation the cells were then labeled with 59Fe-transferrin (10 μM) for incubations of 1 to 120 minutes at 37°C. The uptake of cellular 59Fe and its incorporation into heme were measured using standard techniques (see “Materials and methods” for details). The results are means of duplicate determinations in a typical experiment from 3 experiments performed.
To examine the possible role of frataxin in Fe or heme metabolism, experiments were designed to assess the effect on Frdamessenger RNA (mRNA) levels by inducing erythroid differentiation in Friend 707 cells (Figure 2). To further investigate the function of frataxin, we also assessed changes in expression of β-globin mRNA (Figures 2, 6, and 7), andTfR and Nramp2 mRNAs (Figures 6 and 7). The expression of these genes was relevant to examine because β-globin is essential for hemoglobinization,26 whereas TfR and Nramp2 are involved in Fe uptake.24
Effect of inducing erythroid differentiation by DMSO in Friend 707 cells on the mRNA levels of
frataxin (Frda),β-globin, and β-actin(loading control). Northern blot analysis was performed by extracting total mRNA from cells incubated in the presence (+) or absence (−) of 1.5% DMSO for 120 hours at 37°C. The isolated RNA was electrophoresed on a 1.2% formaldehyde gel, transferred to nylon membrane, and sequentially probed under high stringency conditions (see “Materials and methods” for details). (A) Ethidium bromide staining of the agarose gel; (B) Frda; (C) β-globin; and (D) β-actin. Densitometric analysis of the results normalized to the β-actin loading control are shown to the right of each relevant blot. The result illustrated is a typical experiment from 5 experiments performed.
Effect of inducing erythroid differentiation by DMSO in Friend 707 cells on the mRNA levels of
frataxin (Frda),β-globin, and β-actin(loading control). Northern blot analysis was performed by extracting total mRNA from cells incubated in the presence (+) or absence (−) of 1.5% DMSO for 120 hours at 37°C. The isolated RNA was electrophoresed on a 1.2% formaldehyde gel, transferred to nylon membrane, and sequentially probed under high stringency conditions (see “Materials and methods” for details). (A) Ethidium bromide staining of the agarose gel; (B) Frda; (C) β-globin; and (D) β-actin. Densitometric analysis of the results normalized to the β-actin loading control are shown to the right of each relevant blot. The result illustrated is a typical experiment from 5 experiments performed.
A 120-hour incubation with DMSO resulted in a marked decrease inFrda mRNA levels (Figure 2B) compared to cells incubated with medium alone. After normalization to β-actin, densitometric analysis demonstrated that compared to control cells, induction of erythroid differentiation using DMSO resulted in a significant (P < .006) 6 ± 2-fold decrease in Frda mRNA levels in 5 separate experiments (Figure 2B). From this point on, all reported changes in gene or protein expression quantitated below are normalized to the β-actin loading control. The Frdatranscript migrated as a single band at approximately 1.2 kb just below the 18 S ribosomal RNA (rRNA) band (Figure 2B), as reported previously.14 In some experiments, 2 Frda bands were observed, the main 1.2-kb band and a less intense band that may correspond to an alternative Frda transcript (Figure 6B). As a positive control for erythroid differentiation β-globinmRNA levels were assessed, and increased markedly after incubation with DMSO (Figure 2C).37 38
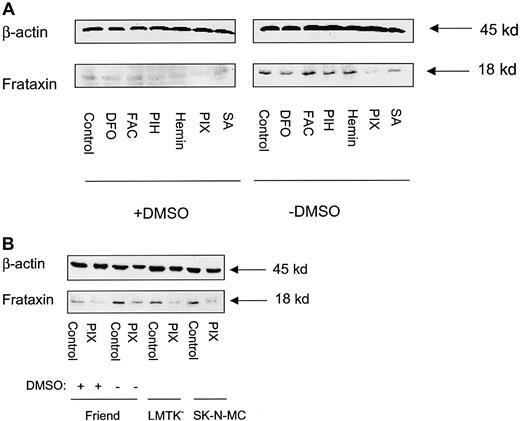
In agreement with the decrease in Frda mRNA levels (Figure2B), Western blot analysis also demonstrated that frataxin protein levels were decreased after induction of erythroid differentiation with DMSO in Friend 707 cells (Figure 3). In all experiments a band at approximately 18 kd was identified that was in good agreement with the relative molecular mass of frataxin reported previously.6 The decrease in frataxin protein levels began after 24 hours of induction with DMSO, with the greatest decrease becoming clear between 72 and 120 hours (Figure 3). Further, the effect of erythroid differentiation on decreasing frataxin protein levels was not specific for the Friend cell clone, with similar results being observed in the 707 and 745 cell lines (Figure4). All subsequent experiments described were performed using the Friend 707 cell line.
Induction of erythroid differentiation in Friend 707 cells by DMSO decreases frataxin protein levels.
Western blot analysis was performed using proteins extracted from cells incubated in the presence (+) or absence (−) of 1.5% DMSO for 24 to 120 hours at 37°C. The membrane was sequentially probed with antifrataxin and anti-β-actin antibodies (see “Materials and methods” for details). Densitometric analysis of the results normalized to the β-actin loading control are shown below the blot. The result illustrated is a typical experiment from 4 experiments performed.
Induction of erythroid differentiation in Friend 707 cells by DMSO decreases frataxin protein levels.
Western blot analysis was performed using proteins extracted from cells incubated in the presence (+) or absence (−) of 1.5% DMSO for 24 to 120 hours at 37°C. The membrane was sequentially probed with antifrataxin and anti-β-actin antibodies (see “Materials and methods” for details). Densitometric analysis of the results normalized to the β-actin loading control are shown below the blot. The result illustrated is a typical experiment from 4 experiments performed.
The effect of DMSO on decreasing frataxin protein levels is not Friend cell clone specific.
Western blot analysis was performed using proteins extracted from the Friend 707 and 745 cell lines incubated in the presence (+) or absence (−) of 1.5% DMSO for 120 hours at 37°C. The membrane was sequentially probed with antifrataxin and anti-β-actin antibodies (see “Materials and methods” for details). Densitometric analysis of the results normalized to the β-actin loading control are shown below the blot. The result illustrated is a typical experiment from 4 experiments performed.
The effect of DMSO on decreasing frataxin protein levels is not Friend cell clone specific.
Western blot analysis was performed using proteins extracted from the Friend 707 and 745 cell lines incubated in the presence (+) or absence (−) of 1.5% DMSO for 120 hours at 37°C. The membrane was sequentially probed with antifrataxin and anti-β-actin antibodies (see “Materials and methods” for details). Densitometric analysis of the results normalized to the β-actin loading control are shown below the blot. The result illustrated is a typical experiment from 4 experiments performed.
Collectively, these data above indicate that the induction of erythroid differentiation results in a decrease in frataxin expression at both the mRNA and protein level. This decrease is inversely related to hemoglobinization (as judged by cell pellet color) and the expression of β-globin mRNA (Figure 2C).
DMSO does not nonspecifically down-regulateFrda gene expression
Control experiments were performed to ensure that the effect of DMSO on Frda expression was not due to nonspecific down-regulation of gene expression (Figure5). In these studies, nonerythroid LMTK− fibroblast or RAW264.7 macrophage cell lines were incubated in the presence or absence of DMSO for 120 hours and thenFrda mRNA levels assessed (Figure 5). Incubation with DMSO had no effect on Frda mRNA expression when compared to cells exposed to control medium (Figure 5). These results indicate thatFrda expression was not nonspecifically down-regulated by DMSO.
The effect of incubation with DMSO on the mRNA levels of
frataxin (Frda) and β-actin(loading control) in mouse LMTK− fibroblasts and the RAW264.7 macrophage cell line. Northern blot analysis was performed as described in Figure 2 using total mRNA extracted from cells after a 120-hour incubation at 37°C in the presence (+) or absence (−) of 1.5% DMSO. (A) Ethidium bromide staining of the agarose gel, (B) Frda, and (C) β-actin. The result illustrated is a typical experiment from 3 experiments performed.
The effect of incubation with DMSO on the mRNA levels of
frataxin (Frda) and β-actin(loading control) in mouse LMTK− fibroblasts and the RAW264.7 macrophage cell line. Northern blot analysis was performed as described in Figure 2 using total mRNA extracted from cells after a 120-hour incubation at 37°C in the presence (+) or absence (−) of 1.5% DMSO. (A) Ethidium bromide staining of the agarose gel, (B) Frda, and (C) β-actin. The result illustrated is a typical experiment from 3 experiments performed.
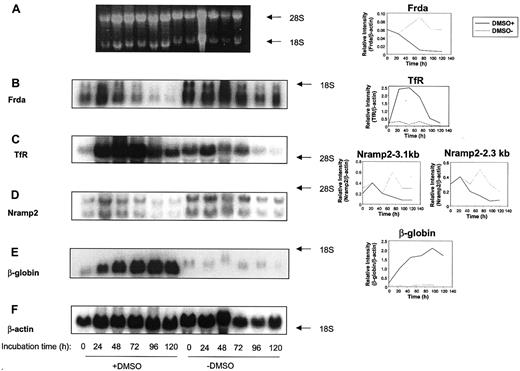
Kinetic analysis of the effects of DMSO on the expression of Frda, Nramp2, TfR, β-globin, and β-actin
To examine the kinetics of the effect of DMSO on gene expression, Friend cells were incubated in the presence or absence of this agent for 24 to 120 hours and the mRNA isolated (Figure6). The levels of Frda mRNA decreased markedly in the presence of DMSO as the incubation time increased to 120 hours (Figure 6B). For example, in the presence of DMSO, densitometric analysis demonstrated that normalizedFrda mRNA expression significantly (P < .001) decreased 14 ± 4-fold (3 experiments) comparing levels at time 0 and 120 hours (Figure 6B). In contrast, in the absence of DMSO, normalized Frda mRNA levels increased and peaked at 72 hours, and then decreased somewhat at later time points (Figure 6B). The normalized levels of Frda mRNA in the absence of DMSO were greater than those observed in the presence of this agent for all time points more than 24 hours (Figure 6B).
The effect of incubation time with DMSO on the mRNA levels of
frataxin (Frda),transferrin receptor (TfR),Nramp2, β-globin, orβ-actin in Friend 707 cells. Total mRNA was extracted from cells incubated in the presence (+) or absence (−) of 1.5% DMSO for 0 to 120 hours at 37°C. Northern blot analysis was performed as described in Figure 2. (A) Ethidium bromide staining of the agarose gel, (B) Frda, (C) TfR, (D) Nramp2, (E) β-globin, and (F) β-actin. Densitometric analysis of the results normalized to the β-actin loading control are shown to the right of each relevant blot. The result illustrated is a typical experiment from 3 experiments performed.
The effect of incubation time with DMSO on the mRNA levels of
frataxin (Frda),transferrin receptor (TfR),Nramp2, β-globin, orβ-actin in Friend 707 cells. Total mRNA was extracted from cells incubated in the presence (+) or absence (−) of 1.5% DMSO for 0 to 120 hours at 37°C. Northern blot analysis was performed as described in Figure 2. (A) Ethidium bromide staining of the agarose gel, (B) Frda, (C) TfR, (D) Nramp2, (E) β-globin, and (F) β-actin. Densitometric analysis of the results normalized to the β-actin loading control are shown to the right of each relevant blot. The result illustrated is a typical experiment from 3 experiments performed.
In contrast to Frda mRNA expression, normalizedTfR mRNA levels significantly (P < .0001) increased up to 25-fold (3 experiments) in the presence of DMSO between 24 and 48 hours compared to initial levels (time 0; Figure 6C). The levels of TfR mRNA then markedly decreased as the incubation was continued up to 120 hours (Figure 6C). In contrast, in the absence of DMSO, normalized TfR mRNA levels significantly (P < .001) decreased more than 6-fold (3 experiments) as the incubation increased from 0 to 120 hours (Figure 6C).
From Figure 6D, it is clear that there are 2 Nramp2transcripts at 2.3 and 3.1 kb in Friend cells, as observed in previous studies for murine tissues and cell lines.46,47 Although the precise identification of these 2 transcripts is uncertain, they may correspond to the Nramp2 splice variants with and without the iron-responsive element (IRE; 2.3 and 3.0 kb, GenBank accession codes AF029758 and L33415, respectively). The change in expression of the Nramp2 transcripts after incubation with DMSO was generally similar (Figure 6D). In the presence or absence of DMSO, densitometric analysis showed there was a slight increase in the normalized expression of both Nramp2 transcripts in the first 24 hours. In the presence of DMSO, this initial increase inNramp2 mRNA expression was then followed by a significant (P < .01) decrease in 3 experiments (Figure 6D). In one typical experiment (representative of 3) in the presence of DMSO, the 2.3-kb and 3.1-kb Nramp2 transcripts decreased 3.7-fold and 2.5-fold, respectively, comparing initial levels and those found after a 120-hour incubation (Figure 6D). In the absence of DMSO, the levels of both the 3.1-kb and 2.3-kb transcripts peaked at 72 hours and then decreased to initial levels or slightly below initial levels, respectively, at 120 hours (Figure 6D). The expression ofNramp2 and TfR mRNA was important to assess because their protein products act as partners involved in Fe uptake from Tf,48,49 which is increased on erythroid differentiation.40 50
In accordance with previous studies,38 we found that the expression of β-globin mRNA increased markedly and significantly (P < .00005) during the incubation with DMSO (3 experiments), whereas it remained very low in the absence of this inducing agent (Figure 6E). In fact, the normalized level ofβ-globin mRNA expression was 85-fold greater in induced compared to uninduced Friend cells at the 120-hour time point (Figure6E). Collectively, the results above clearly indicate that the expression of Frda mRNA is markedly different from that ofTfR mRNA and β-globin mRNA.
The effect of iron chelators, iron donors, and the heme precursor, PIX, on the expression ofFrda, TfR,Nramp2, β-globin, and β-actin
To further assess the possible role of frataxin in Fe or heme metabolism, we examined the effect on gene expression of chelators, which deplete cellular Fe, compounds which donate Fe to cells, and the heme precursor, PIX (Figure 7). Two chelators were used: pyridoxal isonicotinoyl hydrazone (PIH), which depletes cytoplasmic and mitochondrial Fe pools,51,52 and DFO, which chelates cytoplasmic Fe.53-55 Because either or both Fe pools could be involved in regulating Frdaexpression, it was necessary to use both chelators so that Fe levels in each pool could be depleted. Two Fe donors, hemin and ferric ammonium citrate (FAC), were also used. These donate Fe to cells by different mechanisms,56-58 resulting in down-regulation of the TfR59 and up-regulation of ferritin expression.60 The heme precursor PIX was used because it acts as a substrate for heme synthesis.27 31
The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], and the heme precursor, protoporphyrin IX (PIX), on the mRNA levels of frataxin (Frda),transferrin receptor (TfR), Nramp2,β-globin, and β-actin in Friend 707 cells.
Cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence or absence of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), or PIX (50 μM). Northern blot analysis was performed as described in Figure 2. (A) Ethidium bromide staining of the agarose gel, (B) Frda, (C) TfR, (D) Nramp2, (E) β-globin, and (F) β-actin. The result illustrated is a typical experiment from 4 experiments performed.
The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], and the heme precursor, protoporphyrin IX (PIX), on the mRNA levels of frataxin (Frda),transferrin receptor (TfR), Nramp2,β-globin, and β-actin in Friend 707 cells.
Cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence or absence of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), or PIX (50 μM). Northern blot analysis was performed as described in Figure 2. (A) Ethidium bromide staining of the agarose gel, (B) Frda, (C) TfR, (D) Nramp2, (E) β-globin, and (F) β-actin. The result illustrated is a typical experiment from 4 experiments performed.
Friend cells were incubated for 96 hours with control medium or medium containing 1.5% DMSO and then exposed for a further 20 hours to either control medium, DFO (100 μM), PIH (50 μM), FAC (100 μg/mL), hemin (50 μM), or PIX (50 μM) in the presence or absence of 1.5% DMSO. Levels of Frda mRNA (Figure 7B) and Nramp2 mRNA (Figure 7D) were significantly (P < .003) greater under all conditions in cells not induced with DMSO in 4 experiments. Densitometric analysis (not shown) demonstrated that none of the modulatory agents above had any significant effect on normalizedFrda mRNA levels when compared to the control (Figure 7B). In contrast, DFO increased TfR mRNA levels in uninduced cells, whereas it had less effect on TfR mRNA expression in cells incubated with DMSO (Figure 7C). A similar but less marked effect was also seen for PIH. The Fe donors, FAC and hemin, had a greater effect at reducing TfR mRNA in uninduced than induced Friend cells (Figure 7C). The fact that DFO, FAC, and hemin affectedTfR expression provided a positive control that showed these agents acted in accordance with their reported effects to remove or donate Fe to cells.41,44,47,54-56 60
Studies on the effect of DFO and FAC on the expression ofFrda mRNA compared to TfR mRNA were also examined in 5 nonerythroid cell lines available in our laboratory (LMTK− fibroblasts, HepA1-6 hepatoma cells, M15 kidney cells, and J774 and RAW264.7 macrophages). In each case, FrdamRNA levels did not change significantly, in contrast to TfRmRNA that was up-regulated by DFO and down-regulated by FAC (data not shown).
None of the Fe chelators, Fe donors, or PIX, had any significant effect on normalized Nramp2 mRNA levels compared to the control in 4 separate experiments (Figure 7D). Certainly, the level ofNramp2 mRNA was not regulated in concert with TfRmRNA (Figure 7C). This observation is of interest because likeTfR mRNA, Nramp2 mRNA contains an IRE in its 3′ untranslated region.49 These results are similar to those reported in our previous studies examining the effects of intracellular Fe levels on Nramp2 expression in nonerythroid cell types.44 47
Collectively, the results above suggest that Frda mRNA was not regulated by Fe in a similar way to TfR mRNA. To confirm this, we also incubated cells with the heme synthesis inhibitor, SA, which causes accumulation of mitochondrial Fe and, to a lesser extent, cytoplasmic Fe.26,27,30-33 In these studies, Friend cells were incubated for 96 hours with control medium or medium containing 1.5% DMSO and then exposed for a further 20 hours with either control medium or medium containing 2 mM SA. These experiments showed that SA had no significant effect on the expression of Frda mRNA orNramp2 mRNA (data not shown). In contrast, SA down-regulatedTfR mRNA levels of induced cells (data not shown), as also reported by others.41 61 This latter result provided a positive internal control for the inhibitory effect of SA on heme synthesis. Hence, inhibition of heme biosynthesis had no effect onFrda mRNA levels.
PIX down-regulates frataxin protein levels in uninduced Friend cells and other cell types
Western blot analysis was performed to assess whether DFO, FAC, PIH, hemin, PIX, or SA had any regulatory effects on frataxin protein levels. Friend cells were incubated for 96 hours with control medium or medium containing DMSO and then exposed for a further 20 hours with either control medium or medium containing DFO (100 μM), FAC (100 μg/ml), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM) in the presence or absence of DMSO (Figure8A). As shown in Figure 3, induction of erythroid differentiation with DMSO resulted in a marked decrease in frataxin protein levels (Figure 8A). Of all compounds tested, only incubation with PIX consistently decreased the level of frataxin protein in uninduced Friend cells over 8 independent experiments (Figure 8A). After normalization to β-actin, PIX significantly (P < .003) decreased the level of frataxin protein 2.2 ± 0.7-fold compared to the relevant control in 8 experiments (Figure 8A). For induced cells, PIX reduced frataxin expression (Figure 8A) but this was not significant (P > .05) in 8 experiments. All other agents tested in induced cells also had no significant effect on frataxin expression.
Protoporphyrin IX (PIX) decreases frataxin protein levels in uninduced Friend 707 cells, and LMTK− fibroblasts and SK-N-MC neuroepithelioma cells.
(A) Friend cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence (+) or absence (−) of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM). (B) The SK-N-MC and LMTK− cell lines were incubated for 20 hours at 37°C with PIX (50 μM). These results are compared with induced and uninduced Friend cells incubated with (+) or without (−) 1.5% DMSO for 96 hours and then exposed to PIX (50 μM) for 20 hours at 37°C. Western blot analysis was performed as described in “Materials and methods.” The result illustrated in panel A is typical of 8 experiments performed, and that in panel B is typical of 4 experiments.
Protoporphyrin IX (PIX) decreases frataxin protein levels in uninduced Friend 707 cells, and LMTK− fibroblasts and SK-N-MC neuroepithelioma cells.
(A) Friend cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence (+) or absence (−) of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM). (B) The SK-N-MC and LMTK− cell lines were incubated for 20 hours at 37°C with PIX (50 μM). These results are compared with induced and uninduced Friend cells incubated with (+) or without (−) 1.5% DMSO for 96 hours and then exposed to PIX (50 μM) for 20 hours at 37°C. Western blot analysis was performed as described in “Materials and methods.” The result illustrated in panel A is typical of 8 experiments performed, and that in panel B is typical of 4 experiments.
Similar to uninduced Friend cells, PIX also reduced frataxin expression in mouse LMTK− fibroblasts and human SK-N-MC neuroepithelioma cells (Figure 8B). These results further suggest that frataxin plays a regulatory role in Fe or heme metabolism.
To assess if the above treatments had appropriately influenced cellular Fe metabolism, their effects on TfR expression were also assessed by Western blotting (Figure 9). As expected from previous studies,41 DFO increased TfR expression in both uninduced and induced Friend cells, but this was more pronounced in uninduced cells (Figure 9). In contrast, FAC and hemin reduced TfR levels in uninduced cells more than induced cells. In 5 experiments, densitometric analysis demonstrated that PIH, PIX, and SA had no consistent nor significant effect on TfR protein levels in induced or uninduced cells (Figure 9). The failure of PIH to increase TfR levels is of interest and may relate to its different site of action to DFO. Previous studies have also noted that in contrast to DFO, PIH did not increase iron regulatory protein-IRE–binding activity.62Interestingly, SA did not decrease TfR protein levels of induced cells (Figure 9), and this was in contrast to results at the mRNA level (see text above).
The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], the heme precursor, protoporphyrin IX (PIX), and the heme synthesis inhibitor, succinylacetone (SA), on the protein levels of the transferrin receptor (TfR) in Friend 707 cells.
Friend cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence (+) or absence (−) of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM). Western blot analysis was performed as described in “Materials and methods.” The result illustrated is typical of 5 experiments performed.
The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], the heme precursor, protoporphyrin IX (PIX), and the heme synthesis inhibitor, succinylacetone (SA), on the protein levels of the transferrin receptor (TfR) in Friend 707 cells.
Friend cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence (+) or absence (−) of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM). Western blot analysis was performed as described in “Materials and methods.” The result illustrated is typical of 5 experiments performed.
Discussion
The role of frataxin in mitochondrial Fe homeostasis is critical to understand because the decrease in expression of this molecule leads to mitochondrial Fe accumulation that may contribute to FA.1,2,24 Furthermore, understanding the function of frataxin may result in new strategies for the treatment of this disorder.24,62 We examined the role of frataxin in Fe and heme metabolism by using a well-characterized model of erythroid differentiation, namely, Friend cells induced with DMSO.37-41
To our knowledge this is the first report showing down-regulation ofFrda expression on erythroid differentiation. In contrast, and as expected,26,37,38,41,56 expression ofTfR and β-globin mRNA increased markedly after incubation of Friend cells with DMSO (Figure 6). Because there was a marked decrease in frataxin expression on induction of heme and hemoglobin synthesis, it is unlikely that frataxin plays a direct role in these processes. In addition, we demonstrate that modulation of intracellular Fe levels or inhibition of heme synthesis does not regulate the expression of Frda mRNA in a way similar to TfR mRNA in Friend cells. The effect of Fe and heme on frataxin levels was important to investigate because the expression of many molecules involved in Fe and heme synthesis is regulated by the concentration of intracellular Fe or heme.26,60 Furthermore, although frataxin levels decreased during differentiation, there is increased expression of a variety of molecules involved in heme synthesis and other functions in the cytoplasm or mitochondrion.26 61-65 Indeed, there are only a few specific markers that are known to be down-regulated by DMSO treatment of Friend cells. In this respect, the decreases inFrda and Nramp2 gene expression are interesting (Figure 6), but they are not the only molecules that decrease during erythroid differentiation (eg, ribosomal proteins).
On erythroid differentiation there is a marked up-regulation of proteins involved in controlling Fe uptake (eg, TfR),41,61heme synthesis,26,40,56,63 and hemoglobinization (eg, β-globin).37,38 The marked up-regulation of TfRexpression results in a significant increase in Fe uptake (Figure1B) as demonstrated by others.26,40,50,56,64 This Fe is used for heme synthesis within the mitochondrion (Figure1B).26,40,50,56 The decrease in frataxin protein levels began after 24 hours of induction with DMSO, with the greatest decrease becoming obvious between 72 and 120 hours (Figure 3). The decreased frataxin expression is correlated with increased Fe uptake by the mitochondrion and the induction of heme biosynthesis.40Considering this, using Friend cells, the incorporation of59Fe and 14C-aminolevulinic acid into heme as well as the heme concentration becomes pronounced between 72 and 120 hours of incubation with DMSO.40 In comparison, δ-aminolevulinic acid synthetase activity begins to slowly increase after 24 hours of incubation with DMSO, with its activity becoming increasingly marked after 3 days of exposure to this agent.40 For both δ-aminolevulinic acid dehydratase and uroporphyrinogen-I synthetase, their activities increase between 24 and 48 hours of incubation with DMSO and then plateau after 72 hours.40
The finding that erythroid differentiation resulted in down-regulation of frataxin expression provides insight into the function of this protein. Indeed, it is likely that the Fe loading and oxidative damage of the mitochondrion in FA patients is due to a decrease in frataxin levels.6 11 Considering this, and in conjunction with our data, 2 possible roles of frataxin could be suggested that lead to mitochondrial Fe sequestration: (1) down-regulation of frataxin expression may increase mitochondrial Fe import or decrease Fe export by regulating metal transporters, leading to Fe accumulation for heme synthesis, or (2) frataxin could act as a metabolic switch, resulting in the diversion of Fe from one metabolic pathway (eg, Fe incorporation into ferritin or the synthesis of Fe-S clusters) to another, namely, Fe incorporation into heme.
For both of the above hypotheses, increased retention of mitochondrial Fe would occur that could favor heme synthesis.26 In support of the first hypothesis, it has been reported that Yfh1p plays a role in the release of nonheme mitochondrial Fe into the cytoplasm.10 Thus, the decrease in frataxin expression in erythroid cells may prevent mitochondrial Fe release and its use for heme synthesis rather than cytosolic Fe metabolism. Alternatively, or in combination with a decrease in mitochondrial Fe efflux, down-regulation of frataxin expression may increase mitochondrial Fe uptake. In this latter mechanism, frataxin could act as a negative regulator of a mitochondrial Fe import process. Considering this, a yeast gene known as CCC1 has been identified that inhibits mitochondrial Fe uptake.66 We suggest a similar molecule could be regulated by frataxin in mammalian cells. At this point it should be discussed that the regulation of cellular Fe metabolism is complex. The net flux of Fe coming into the cell via Nramp2 or into the mitochondrion as possibly regulated by frataxin, may be dependent on integrating the activity of multiple proteins. Thus, the observation that frataxin expression decreases as Fe uptake into heme increases does not prove direct causality.
In accordance with a role of frataxin in the regulation of Fe and heme metabolism, our studies demonstrated that incubation of uninduced Friend cells with PIX resulted in a significant decrease in frataxin protein levels (Figure 8A). Because PIX is the immediate precursor of heme,26 56 an increase in the intracellular PIX concentration may act as a signal to coordinate heme synthesis with mitochondrial Fe uptake. For instance, an elevation of cellular PIX could signal a decrease in frataxin expression that leads to mitochondrial Fe sequestration for heme biosynthesis. It is of interest to note that although PIX reduced frataxin protein expression (Figure8A), it had no significant effect on the mRNA levels of this molecule (Figure 7B).
Considering the recent discussion of frataxin acting as a mitochondrial ferritin,34-36 it is relevant to note that incubation of Friend cells with the Fe donor, FAC, or the Fe chelator, DFO, did not significantly increase or decrease the level of frataxin, respectively (Figure 8A). This was in marked contrast to the known stimulatory and inhibitory effects of FAC and DFO, respectively, on ferritin protein levels.26,56,60 These data suggest that frataxin is not regulated by Fe in a manner similar to ferritin. It could be expected that a vital Fe storage molecule would respond to increased intracellular Fe levels. Hence, the lack of response of frataxin in our experiments is not consistent with an Fe storage role. In erythroid cells, excess mitochondrial Fe may be stored in the recently identified mitochondrial ferritin.67
Interestingly, depletion of frataxin in FA patients does not result in marked defects in heme synthesis.20 Because a reduction in frataxin expression occurs concurrently with an increase in Fe uptake, heme synthesis, and TfR- and β-globin mRNA expression26,37,38,40,50,56,64 (Figures 1B, 2, and 6), this decrease in frataxin levels may facilitate these latter processes. Indeed, it is well known in erythroid cells that the limiting factor for heme synthesis is Fe acquisition.26 56 Hence, in erythroid cells, the reduction in frataxin expression appears coupled to the marked increase in heme synthesis that is exported from the mitochondrion preventing toxic Fe accumulation. In contrast, in the nonerythroid cells affected in FA, there is a decrease in frataxin expression that results in mitochondrial Fe loading. However, because nonerythroid cells have very low rates of heme production, the increased Fe uptake is not matched by a similar rise in heme synthesis that is necessary for Fe incorporation into PIX and its transport out of the mitochondrion as heme. Under these circumstances, Fe will accumulate and cause toxicity via free radical production.
It is relevant to discuss that mitochondrial Fe must be in the Fe++ state to contribute to heme synthesis.26,56 This is exemplified by sideroblastic anemia, where ferric Fe accumulates and cannot be used for heme synthesis via ferrochelatase. In this study we used a well-characterized model of erythroid differentiation where incubation of Friend cells with DMSO results in initiation of heme biosynthesis, a significant increase in Fe uptake from Tf, and incorporation of Fe++ into PIX to form heme (Figure1B).26,37,40,41,50,56,61 63-65 Hence, our experiments have focused on the role of frataxin during a physiologic response rather than a pathologic condition.
Previous studies by Cossee and associates68 showed that homozygous deletions of frataxin cause lethality in a homozygous mouse knockout model without apparent Fe accumulation. In the case of a developing embryo, an inappropriate increase in mitochondrial Fe levels may not need to be marked to cause apoptosis and death. Moreover, in this latter study,68 there was no attempt to quantitatively examine mitochondrial Fe concentrations by isolating mitochondria, and it is difficult to assess if the methods used (Perl staining and electron microscopic investigation of tissues) were sensitive enough to determine a small but critical increase in Fe. Nonetheless, considering the work of Cossee68 and others,69 it cannot be excluded that frataxin deficiency triggers apoptosis by a mechanism independent of Fe accumulation. It is notable that no significant mitochondrial Fe deposition takes place in sideroblastic anemia at day 11.5 during the embryonic life of the erythroid 5-aminolevulinate synthase knockout mouse.70Relevant to this, Cossee and colleagues68 reported that frataxin knockout mice die soon after implantation. Thus, it is likely that these authors may have missed the Fe accumulation that occurs later in development.
In summary, we demonstrate that erythroid differentiation in Friend cells results in down-regulation of Frda mRNA levels and frataxin protein levels. Further, the down-regulation of frataxin expression during erythroid differentiation may favor the marked mitochondrial Fe uptake, heme synthesis, and hemoglobinization that occur. Indeed, the fact that incubation with PIX decreased frataxin protein levels suggests a role for this protein in the coordination of mitochondrial Fe and heme metabolism. Further studies on cell lines where overexpression or underexpression of frataxin is controlled by inducible promoters may establish the precise role of frataxin in Fe metabolism.
Professor Roger Dean, Dr Anna Baoutina, and members of our laboratory are kindly thanked for their detailed comments on the manuscript prior to submission. We gratefully acknowledge the expert assistance of Mr Ralph Watts in preparing the figures. Dr Jin Gao is also thanked for his help with Western blot analysis.
Supported by a PhD scholarship (to E.M.B.) from Cecily and Neville Cox of Brisbane, Queensland, and by grants from the National Health and Medical Research Council of Australia and an Australian Research Council Large Grant (to D.R.R.). We also kindly acknowledge the Heart Research Institute for financial support.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Des R. Richardson, Heart Research Institute, Iron Metabolism and Chelation Group, 145 Missenden Rd, Camperdown, Sydney, New South Wales, 2050 Australia; e-mail:d.richardson@hri.org.au.







![Fig. 7. The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], and the heme precursor, protoporphyrin IX (PIX), on the mRNA levels of frataxin (Frda),transferrin receptor (TfR), Nramp2,β-globin, and β-actin in Friend 707 cells. / Cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence or absence of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), or PIX (50 μM). Northern blot analysis was performed as described in Figure 2. (A) Ethidium bromide staining of the agarose gel, (B) Frda, (C) TfR, (D) Nramp2, (E) β-globin, and (F) β-actin. The result illustrated is a typical experiment from 4 experiments performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/10/10.1182_blood.v99.10.3813/6/m_h81022541007.jpeg?Expires=1765091072&Signature=R80ItYKRcmQdoFo2HTGzk8D0WXVkOIWJ9d9INAWy3ON1gqgk8v~uGaVLdiZwTJ0yMJPbdOk9ok9pXHV54BgDWlqztByvxPgDcIjWSJRR0QcKdSirUyBM9SV8hP2CQeB-TwBJ6XM9hQq3Kwi2t9jG5tcT2rZ577K994MNml6qSCECcJ3rf9QHKIX7azOFu8a83EHvr5lpk6JrYoNj0G6LG9b~ZsyUBVmdDqWQOjCqKBDOK6dVx8A0~EJrgNAzlrSzWTASQZitV6JZgOCknf7aDQZ0hVgsf-0RrwhMGYB0pIrHq1Svw0-1bSoAL5Yvm8uasGneqC-ig5cwtd5De1fo6A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], the heme precursor, protoporphyrin IX (PIX), and the heme synthesis inhibitor, succinylacetone (SA), on the protein levels of the transferrin receptor (TfR) in Friend 707 cells. / Friend cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence (+) or absence (−) of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM). Western blot analysis was performed as described in “Materials and methods.” The result illustrated is typical of 5 experiments performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/10/10.1182_blood.v99.10.3813/6/m_h81022541009.jpeg?Expires=1765091072&Signature=E2RFb1OtcghMAYaJFYV99tqpOOy5Y6McuKiYNTRb~wWqRCCGFPlP-fPfAZjaQlYUrnfWWfdUIAk3tpbAEq04J~G-z3ZAWqECBEM7-ROcsqOroWm3odCpYL3lckY3k0F50txuF0IhIvVUJJ7AD8LMLiql6fdiEw5gjXo37ShkOPzSz4zl5EBdWTPZO5vdB0Zx-todek0Jtj3-n5Xx-k5OFQB-ffCBiyuQNpaiE1k2ur9F-UfjMD3kcXgRU67WvkazL4vM1XVi6M2KxrqO0pJ3JXf-n5GZRMvfniFvTFn7YHSSZP0LFrHLUwxE3Lfx06oJifhKuoCBZeqwUx5NrjoqGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)






![Fig. 7. The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], and the heme precursor, protoporphyrin IX (PIX), on the mRNA levels of frataxin (Frda),transferrin receptor (TfR), Nramp2,β-globin, and β-actin in Friend 707 cells. / Cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence or absence of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), or PIX (50 μM). Northern blot analysis was performed as described in Figure 2. (A) Ethidium bromide staining of the agarose gel, (B) Frda, (C) TfR, (D) Nramp2, (E) β-globin, and (F) β-actin. The result illustrated is a typical experiment from 4 experiments performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/10/10.1182_blood.v99.10.3813/6/m_h81022541007.jpeg?Expires=1765091073&Signature=err4oemfSLjsuVcpeIFQeeF2A~Za6nUOqW993nj2kxh~H7xgU858RbaDFu7StFzBj5xX1B8-WEPZTFmtTAJXxflCUKRiNo0kyn5Vcu2AMt82Pw98Tj4GysQ43INJG0dDLJ4SGw6gCv9pdTqIok2ilCR4UgKV6Dhw4N9MoQ21FPcFpPFFuxGddErEa17VOFLguRvcAsVbtcnFQ02LPQmTnoQKT9lPfKWwzb0osFDzCQnI5aqL-CoJ-qWZmdYUB-oLltmarFpvFb5xX~uY8QjvUDpRaCtHXcGvgfk0maFbGDO7UOTIUEsPU2JvTyRMnUiYxALhjOfudXo8EtfvRr~~jg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 9. The effect of iron chelators [desferrioxamine (DFO) and pyridoxal isonicotinoyl hydrazone (PIH)], iron donors [ferric ammonium citrate (FAC) and heme], the heme precursor, protoporphyrin IX (PIX), and the heme synthesis inhibitor, succinylacetone (SA), on the protein levels of the transferrin receptor (TfR) in Friend 707 cells. / Friend cells were incubated in the presence (+) or absence (−) of 1.5% DMSO for 96 hours at 37°C. After this, the incubation was continued for a further 20 hours at 37°C in the presence (+) or absence (−) of 1.5% DMSO alone (controls) or in these media containing either DFO (100 μM), FAC (100 μg/mL), PIH (50 μM), hemin (50 μM), PIX (50 μM), or SA (2 mM). Western blot analysis was performed as described in “Materials and methods.” The result illustrated is typical of 5 experiments performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/10/10.1182_blood.v99.10.3813/6/m_h81022541009.jpeg?Expires=1765091073&Signature=0VhWdkfyqlZBMuM1hyEn4H1ZuzBBDIjQJ9NuKUENG-gbZkbK8-k53JLRdjjeqYe550Rez~Dv8uvv9xrKF7f1y6~NpHH4-zjv18iCdHdpqx4cOItIwjtZd0IaofZsi82pwjHiyWCNhgXpeu3JgCJSebhPQxdvL9arDInIgqtrFHu1ZyqtjGGn2rPMkAZtqbD-iUTvnJY6ApcPS0dM9B-Yp4vgK8ZwYYie5jfk7SHIGyBI3AjgWzVGvLpA8dlI8rZIrUYptehNCp6J-Lq-IJy-lNVM2EHXSC5HpCrGYq4HUl9ZPbLAglie2E~3kmA4pjKQzFEaUtZo7VaMn9GZVSZgCw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)