Mutations in the granulocyte colony-stimulating factor (G-CSF) receptor gene are found in a number of patients with severe chronic neutropenia predisposed to acute myeloid leukemia. These mutations result in the absence of the C-terminal domain of the G-CSF-R, a region which has been implicated in differentiation signaling. We generated mice with an equivalent mutation (gcsfr-▵715) by homologous and Cre-mediated recombination in embryonic stem cells. Both wt/▵715 and▵715/▵715 mice have significantly reduced numbers of blood neutrophils compared with their wt/wt littermates. However, under continuous G-CSF administration mutant mice develop peripheral neutrophil counts that significantly exceed those of wild-type littermates. These findings indicate that depending on G-CSF levels in mice, the ▵715 mutation can contribute both to neutropenia and to neutrophilia.
THE PRODUCTION of neutrophilic granulocytes is regulated by a range of cytokines, including stem cell factor (SCF), interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-6 and G-CSF. Studies in mice deficient for these cytokines have established that G-CSF is the major regulator of neutrophil production.1-3 G-CSF mediates its effects through activation of its cognate receptor (G-CSF-R), a single-transmembrane protein of the hematopoietin or cytokine receptor class I superfamily.4-6 Whereas the membrane-proximal cytoplasmic region of G-CSF-R is essential for transduction of mitogenic signals in murine cell lines,6,7 induction of neutrophilic differentiation requires the integrity of the membrane-distal C-terminal region.7-9
Like all members of the hematopoietin receptor superfamily, G-CSF-R lacks intrinsic tyrosine kinase activity but activates tyrosine kinases of the JAK family that associate to the membrane proximal cytoplasmic region of the receptor.10,11 The JAKs then tyrosine phosphorylate STAT (signal transducer and activator of transcription) proteins, which form dimeric or oligomeric complexes and translocate to the nucleus, where they induce gene transcription.12
Severe chronic neutropenia (SCN) is a disease with a variable inheritance, characterized by a profound shortage of circulating neutrophils (<0.2 × 109 neutrophils/L compared with 4 × 109/L in normal individuals).13Consequently, SCN patients suffer from frequent episodes of opportunistic bacterial infections. In the majority of SCN patients, treatment with G-CSF results in increased peripheral neutrophil counts and a reduction of infection-related events.13 The molecular defects underlying SCN are largely unknown but are thought to affect the G-CSF responsiveness of neutrophilic progenitor cells rather than G-CSF production.14 Importantly, approximately 10% to 15% of SCN patients develop acute myeloid leukemia (AML).15 Recently, cases of SCN have been identified with acquired mutations in the GCSFR gene.16,17 These mutations introduce stop codons in a critical region between codons 714 and 732, resulting in the truncation of 82 to 98 C-terminal amino acids. Upon enforced expression in murine myeloid cell lines, these truncated G-CSF-R consistently fail to transduce differentiation signals and interfere with differentiation induced via the wild-type G-CSF-R in a dominant negative manner.16,17 In contrast, proliferation signaling by G-CSF-R is enhanced as a result of the truncation. To date, of 59 SCN patients investigated, 16 harbored GCSFR point mutations.18 Importantly, all eight patients from this series who developed AML had GCSFR mutations, which were invariably present in the leukemic cells.
To study the contribution of GCSFR mutations to the pathogenesis of SCN and AML, and to gain further insight in the role of the G-CSF-R C-terminus on neutrophil development, we have generated an in vivo model by introducing a nonsense mutation at codon 715 (gcsfr-Δ715) in mice. We show thatwt/Δ715 and Δ715/Δ715 mice have reduced numbers of circulating neutrophils as compared with theirwt/wt littermates. However, after daily administration of pharmacological doses of G-CSF, the numbers of circulating neutrophils in mutant mice increase and significantly exceed those of the wild-type littermates. Thus, in mice, the gcsfr-Δ715mutation contributes to a neutropenic state under basal G-CSF levels and to neutrophilia at pharmacological dosages of G-CSF.
MATERIALS AND METHODS
Cells and culture.
Embryonic stem (ES) cells (ES-E14), a gift from M. Hooper (Edinburgh, UK), were cultured as described.19 In short, cells were kept in culture medium consisting of Dulbecco's Modified Eagle's Medium (DMEM; GIBCO-BRL, Breda, The Netherlands), 50% Buffalo rat liver–conditioned medium, 10% fetal calf serum (FCS) (ES-qualified; GIBCO-BRL) supplemented with 1% nonessential amino acids (GIBCO-BRL), 0.1 mmol/L 2-mercaptoethanol (Sigma Chemical Co, St Louis, MO), 100 U/mL penicillin, 100 μg/mL streptomycin (GIBCO-BRL), and 1,000 U/mL leukemia inhibitory factor (LIF; GIBCO-BRL) in dishes coated with 0.1% gelatin (Sigma G-1890). The cells were passaged every 2 to 3 days.
Targeting constructs and probes.
Isolation, cloning, and sequencing of genomic DNA were done according to standard procedures.20 Genomic DNA was isolated from a mouse 129SV/Cosmid library (Cosmid SC1-6 SuperCos 1; Stratagene Cloning Systems, La Jolla, CA). Using a mouse exon 17 cDNA probe, a clone spanning a genomic fragment from exon 5 to 17 of gcsfr was isolated.21 To introduce a stop codon at Gln715 and anEcoRV site in exon 17, a 300-bp EcoNI-Xba I fragment of exon 17 was synthesized by polymerase chain reaction (PCR) using standard site-directed mutagenesis (CCA AGA GAA ATT TCC AAC CAG TCC CAG = amino acids PREISNQSQ to CCA AGA GAG ATA TCC AAC TAG TCC CAG = amino acids PREISNstopSQ). This mutated fragment was cloned into pBluescript (pBs), confirmed by sequence analysis, and enlarged to 6.2 kb by insertion of a 1.4-kb Sma I-EcoNI fragment at the 5′ end and a 4.5-kb Xba I fragment at the 3′ end to produce pEUR9. A cassette, containing a thymidine kinase (TK) gene driven by a herpes simplex virus promoter (HSV-TK) and a neomycin (PGK-Neo) gene driven by a phosphoglycerate kinase promoter (gifts from B. Malissen, Marseille, France), was flanked by LoxP sites and introduced into the Spe I site of pEUR9, generating pEUR10 (Fig 1A). A uniqueNot I site in the vector backbone was used for linearization before transfection. The Cre-expressing vector (pMC-Cre) was a gift from K. Rajewsky (Institute for Genetics, University of Cologne, Germany). Probes to screen for homologous recombination were obtained by PCR from genomic DNA using the primers CTCCTCCTCATGCTCCAGCGCTG and CTAGGGTCCTCAGGGTAAGGCCTG (Fig 1, probe a), and CCAGACCCAGCCCACAGTAGC and GGCAGGGTCTTCAAGATACAAGG (Fig 1, probe b).
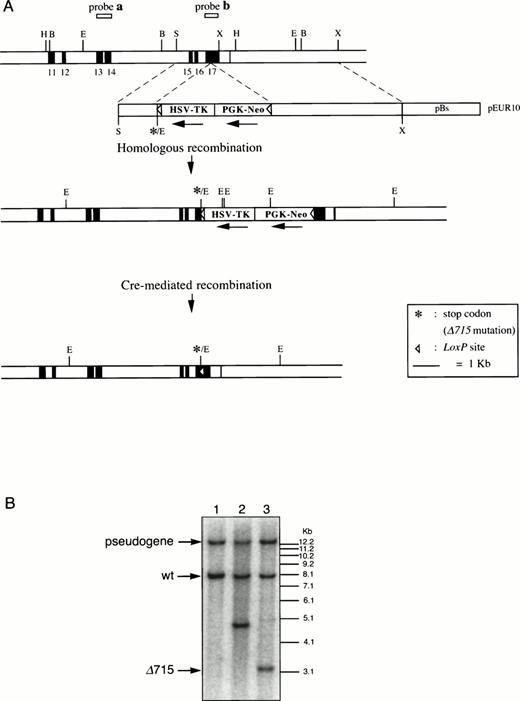
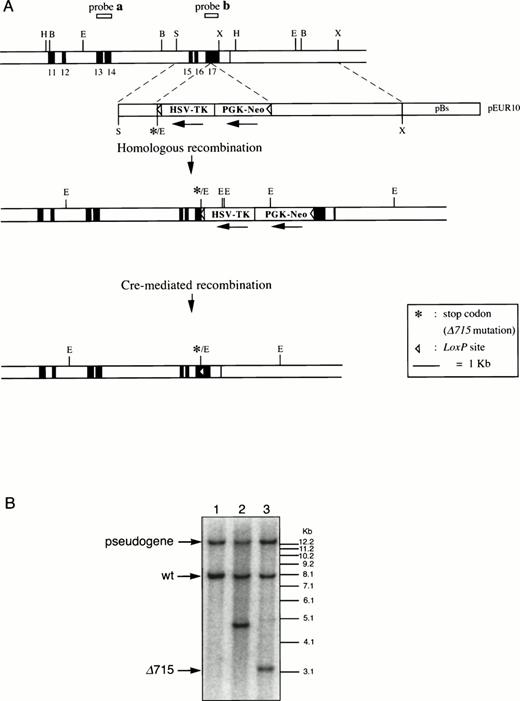
Introduction of the Δ715 mutation in thegcsfr gene by homologous and Cre-mediated recombination. (A) Genomic constructs and targeting strategy. Shown from top to bottom are DNA structures of the germ line gene to be mutated, the targeting construct, the predicted homologous recombinant, and the deletion product generated by the Cre enzyme. For the first targeting event, an external probe covering exon 13 and 14 (probe a) was used to screen colonies. Southern analysis of EcoRV digests of genomic DNA detects an 8-kb band from the wild-type allele and a 5-kb band from the targeted allele. A 13-kb band derived from an intronlessgcsfr-pseudogene present in the murine genome21 was also observed. For the second targeting event, a probe covering exon 17 (probe b) was used to screen colonies which gives bands of 13 kb, 8 kb, 4.8 kb, and 3.2 kb for the pseudogene, wild-type allele, targeted allele and Cre-recombined allele, respectively. B, BamHI; E,EcoRV; H, HindIII; S, Sma I; X, Xba I. (B) Southern blot analysis of EcoRV digests of genomic DNA from ES cells using probe b on a wild-type clone (lane 1), a homologous recombined clone (lane 2), and a subsequent Cre-recombined clone (lane 3), showing the predicted sizes.
Introduction of the Δ715 mutation in thegcsfr gene by homologous and Cre-mediated recombination. (A) Genomic constructs and targeting strategy. Shown from top to bottom are DNA structures of the germ line gene to be mutated, the targeting construct, the predicted homologous recombinant, and the deletion product generated by the Cre enzyme. For the first targeting event, an external probe covering exon 13 and 14 (probe a) was used to screen colonies. Southern analysis of EcoRV digests of genomic DNA detects an 8-kb band from the wild-type allele and a 5-kb band from the targeted allele. A 13-kb band derived from an intronlessgcsfr-pseudogene present in the murine genome21 was also observed. For the second targeting event, a probe covering exon 17 (probe b) was used to screen colonies which gives bands of 13 kb, 8 kb, 4.8 kb, and 3.2 kb for the pseudogene, wild-type allele, targeted allele and Cre-recombined allele, respectively. B, BamHI; E,EcoRV; H, HindIII; S, Sma I; X, Xba I. (B) Southern blot analysis of EcoRV digests of genomic DNA from ES cells using probe b on a wild-type clone (lane 1), a homologous recombined clone (lane 2), and a subsequent Cre-recombined clone (lane 3), showing the predicted sizes.
Introduction of gcsfr-Δ715 by homologous recombination.
E14 ES cells (107) were transfected with 25 μg linearized pEUR10 by electroporation using a Progenetor II, PG200 Hoefer Gene pulser (Hoefer Scientific Instruments, San Francisco, CA) set at 350 V/cm, 1,200 μF, 10 ms. The next day, cells were transferred to culture medium containing 200 μg/mL G418 (GIBCO-BRL), with G418-resistant colonies picked on day 6 or 7 after electroporation. Genomic DNA of these colonies was digested with EcoRV, transferred to nylon membranes, and hybridized to probes a and b andneo. Correctly targeted clones were subjected to cytogenetic analysis and clones with a normal karyotype were used for the second targeting step to remove the Neo-Tk cassette. To this end, 107 cells were transfected with 25 μg of Cre-encoding plasmid DNA by electroporation as described above. Two days later, the cells were resuspended at densities of 1 to 5 × 106cells per plate. From day 6 to 10, cells were cultured in the presence of 2 μmol/L ganciclovir (Syntex, Puteaux, France), and resistant colonies were picked on day 13. Genomic DNA, digested withEcoRV and BamHI, was analyzed by Southern blot using probe a, b, and neo to select clones in which the Neo-Tk cassette had been excised correctly.
Generation of gcsfr mutant mice.
ES cell clones with the gcsfr-Δ715 mutation showing a normal karyotype were injected into blastocysts of C57BL/6 mice. The resulting male chimeras were mated to FVB females to generate heterozygous mutant and wild-type F1 mice. Heterozygouswt/Δ715 mice were intercrossed to obtaingcsfr-Δ715/Δ715 mice. DNA was isolated from tail segments and analyzed on Southern blots as described above.
Blood and bone marrow (BM) sampling.
Blood samples were collected either from the tail vein or from the retro-orbital venous plexus at fixed time points to avoid variation due to circadian rhythms.22 Blood cell counts were performed on a Sysmex-K1000X automated counter (Toa Medical Electronics Co Ltd, Kobe, Japan). To obtain enriched white blood cell suspensions, erythrocytes were lyzed in ice-cold isotonic NH4Cl solution (0.15 mol/L NH4Cl, 10 mmol/L KHCO3, 0.1 mmol/L EDTA pH 7.4) for 10 minutes, spun down, and resuspended in Hanks' balanced salt solution (HBSS)/10% FCS. To obtain BM cell suspensions, femurs and tibias were crushed in a mortar in HBSS/10% FCS. Cells were passed through a 100-μm sieve, spun down, and resuspended, resulting in monocellular suspensions containing 98% to 99% viable cells, as determined by trypan blue exclusion. For differential countings, blood and BM smears or cytospins were fixed in methanol, May-Grünwald-Giemsa (MGG) stained. Three hundred blood cells or 500 BM cells were analyzed in a Zeiss Axioscope microscope (Carl Zeiss B.V., Weesp, The Netherlands).
Progenitor cell assays.
BM cells were prepared as described above. Cells were plated at a density of 2 × 104 cells per mL per dish in triplicate in methyl cellulose medium (Methocult M3230; StemCell Technologies Inc,Vancouver, BC, Canada) containing 30% fetal bovine serum (FBS), 1% bovine serum albumin (BSA), 0.1 mmol/L 2-mercaptoethanol, 2 mmol/L L-glutamine with increasing concentrations of G-CSF (Amgen, Thousand Oaks, CA). Colony formation was monitored on days 7-8 of culture. Colonies containing 30 cells or more were scored. Cytological examination of colony cells was performed on MGG-stained cytospins.
Flow cytometric and Western blot analysis of G-CSF-R.
Expression levels of G-CSF-R on neutrophilic cells were measured by flow cytometry. To this end, G-CSF was biotinylated using D-biotinoyl-ε-aminocaproic acid-N-hydroxysuccinimide ester (Biotin-7-NHS; Boehringer, Mannheim, Germany). Free biotin was removed by gel-filtration on Sephadex G-25. BM cells (106) were incubated in 96-well plates for 60 minutes at room temperature in 25 μL PBA (phosphate-buffered saline with 1% BSA and 0.1% NaN3) and 0.2 μg/mL biotinylated G-CSF, either in the absence or the presence of a 100-fold molar excess of nonbiotinylated G-CSF. Subsequently, cells were incubated for 30 minutes at 4°C with phycoerythrin-conjugated streptavidin (SA-PE; Caltag Laboratories, Burlingame, CA). Cells were subjected to flow cytometric analysis on a FACScan (Becton Dickinson, Sunnyvale, CA). A window was set on the basis of forward and sideward light scatter to restrict the analysis to neutrophilic cells. Western blot analysis was performed according to established procedures, using rabbit antisera against the C-terminal region of murine G-CSF-R (sc-694; Santa Cruz Biotechnology Inc, Santa Cruz, CA) and STAT3 (sc-482; Santa Cruz).
BrdU incorporation in blood neutrophils.
Mice were injected subcutaneously with 250 μg/kg G-CSF for 6 days. On day 4, 150 μL of 5-bromo-2'-deoxyuridine (BrdU) (Sigma; 10 μg/mL in phosphate-buffered saline [PBS]) was injected intraperitoneally. Blood smears were made at various times and stained for BrdU according to standard procedures using a mouse monoclonal antibody (MoAb) against BrdU (kindly provided by W. Dinjens, Department of Pathology, Erasmus University, Rotterdam, The Netherlands) and fluorescein isothiocyanate (FITC)-conjugated goat–anti-mouse-Ig(H+L). The percentage of BrdU+ neutrophils was determined on a Zeiss Axioscope microscope using combined phase contrast illumination to recognize neutrophils on the basis of their segmented nuclei and fluorescence microscopy using a filter-set for FITC-detection.
RESULTS
Generation of gcsfr-Δ715 mice.
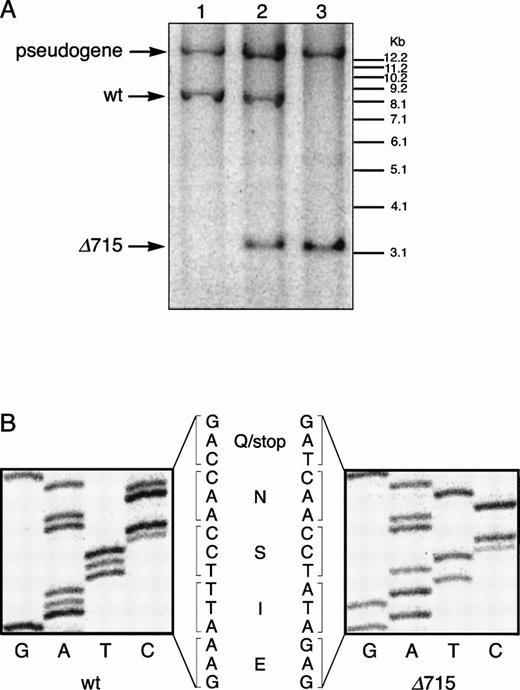
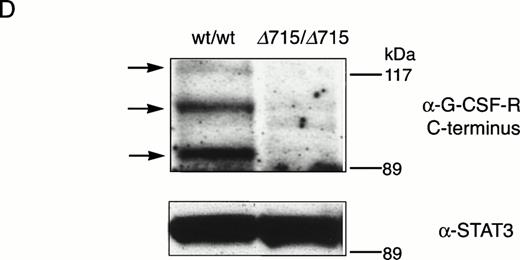
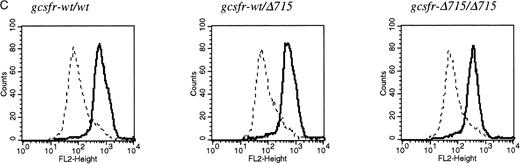
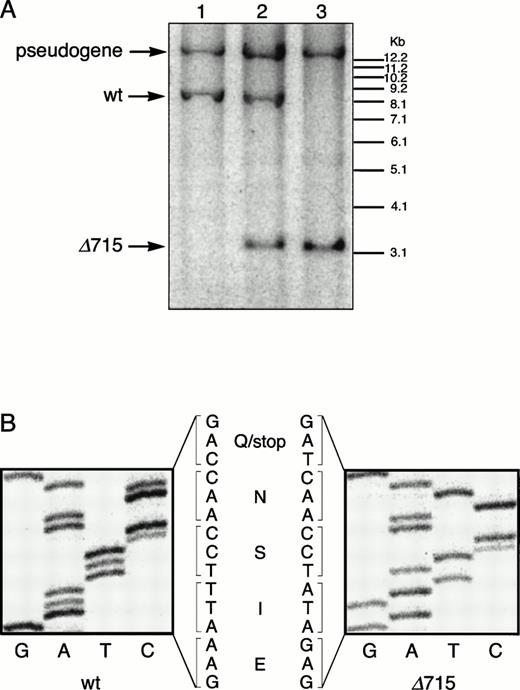
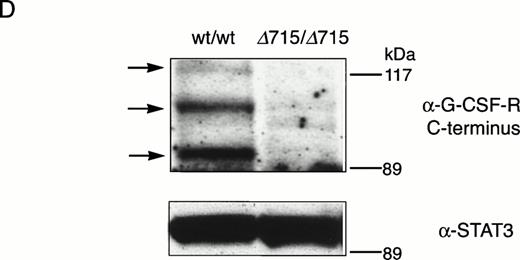
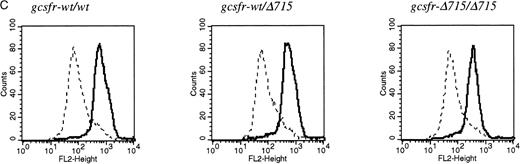
The region of GCSFR subject to mutations in SCN is located in exon 17 and contains 5 Gln residues (716, 718, 720, 726, and 731), the codons of which (CAA or CAG) are changed into stop codons by C-to-T substitutions (TAA or TAG).17 The most frequent mutation in SCN involves Gln 716, which is equivalent to Gln715 in mice. Agcsfr-Δ715 mutation to change Gln715 into a stop codon was generated using PCR, with an additional EcoRV site created by two silent mutations to facilitate analysis. To introduce the gcsfr-Δ715 mutation into ES cells, and to subsequently remove the Neo-Tk selection cassette, we used a two-step gene-targeting strategy as outlined in Fig 1A. In the first step, about one in three clones showed homologous recombination (Fig 1B, lane 2), and after Cre-excision, 90% of the clones were recombined (Fig 1B, lane 3). Two independently isolated ES cell clones were injected into blastocysts and the resulting chimeric mice were crossed with FVB-mice. Germ line transmission of the mutant allele was detected for one of the two clones. When heterozygous wt/Δ715 mice were intercrossed, about 25% of the F2 animals carried the mutation on both alleles, indicating that viability was not compromised by the mutation. Both wt/Δ715 and Δ715/Δ715animals developed normally, and no differences in weight, size, or fertility were observed. Southern blot analysis of tail-DNA from mice (Fig 2A) and nucleotide sequence analysis (Fig 2B) showed the presence and correct positioning of the mutation. Flow cytometric analysis of BM neutrophils using biotinylated G-CSF (Fig 2C) indicated that full-length and truncated G-CSF-R proteins are expressed at equal densities. Western blot analysis with antibodies reactive with the G-CSF-R C-terminus (Fig 2D) confirmed the absence of the C-terminus in Δ715/Δ715 mice.
Transmission and expression of mutant G-CSF-R. (A) Southern blot of EcoRV digests of tail DNA from awt/wt mouse (lane 1), a wt/▵715 mouse (lane 2), and a ▵715/▵715 mouse (lane 3) hybridized with probe b (Fig 1). (B) Nucleotide sequence analysis of PCR-amplified genomic DNA from a wt/wt and a▵715/▵715 mouse cloned in pBs. The sequence of the▵715-derived clone shows the C-to-T substitution changing CAG (Gln715) into TAG (stop715) and the silent base pair substitutions that generated the EcoRV site (GATATC). (C) Flow cytometry analysis of biotinylated G-CSF binding to BM cells from wt/wt,wt/▵715, and ▵715/▵715 mice. Cells were incubated in the absence (solid line) or presence (broken line) of a 100-fold molar excess of nonlabeled G-CSF followed by incubation with PE-conjugated streptavidin. (D) Western blot analysis of 1 × 106 BM cells from wt/wt and▵715/▵715 mice using a rabbit antiserum raised against the 20 C-terminal amino acids of the murine G-CSF-R. The three bands indicated with arrows represent the different glycosylation forms of murine G-CSF-R5; their absence in the▵715/▵715 lanes indicates that the C-terminus is truncated in the mutant mice. Reprobing with anti-STAT3 confirms equivalent loading in both lanes.
Transmission and expression of mutant G-CSF-R. (A) Southern blot of EcoRV digests of tail DNA from awt/wt mouse (lane 1), a wt/▵715 mouse (lane 2), and a ▵715/▵715 mouse (lane 3) hybridized with probe b (Fig 1). (B) Nucleotide sequence analysis of PCR-amplified genomic DNA from a wt/wt and a▵715/▵715 mouse cloned in pBs. The sequence of the▵715-derived clone shows the C-to-T substitution changing CAG (Gln715) into TAG (stop715) and the silent base pair substitutions that generated the EcoRV site (GATATC). (C) Flow cytometry analysis of biotinylated G-CSF binding to BM cells from wt/wt,wt/▵715, and ▵715/▵715 mice. Cells were incubated in the absence (solid line) or presence (broken line) of a 100-fold molar excess of nonlabeled G-CSF followed by incubation with PE-conjugated streptavidin. (D) Western blot analysis of 1 × 106 BM cells from wt/wt and▵715/▵715 mice using a rabbit antiserum raised against the 20 C-terminal amino acids of the murine G-CSF-R. The three bands indicated with arrows represent the different glycosylation forms of murine G-CSF-R5; their absence in the▵715/▵715 lanes indicates that the C-terminus is truncated in the mutant mice. Reprobing with anti-STAT3 confirms equivalent loading in both lanes.
Numbers of circulating neutrophils are reduced in gcsfr-Δ715 mice.
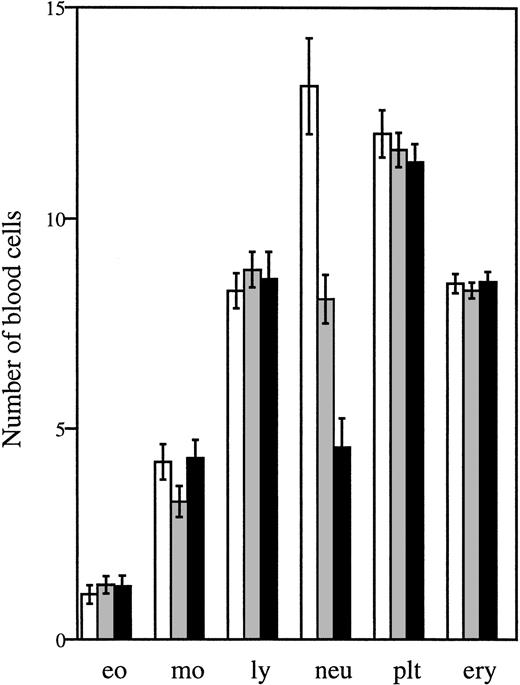
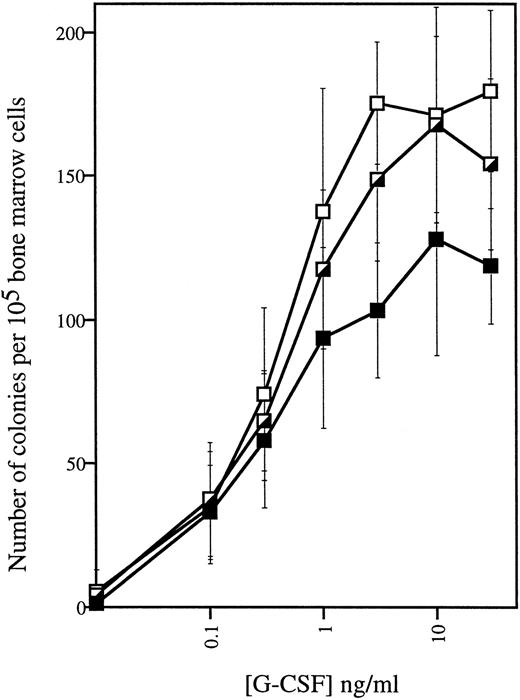
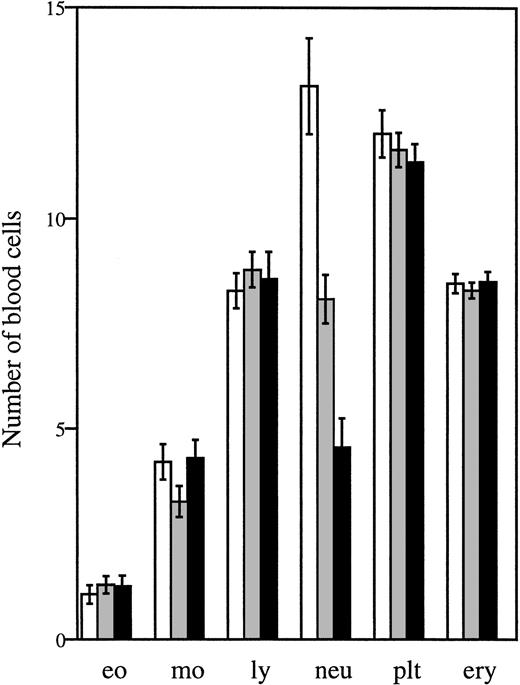
Analysis of peripheral blood samples showed that the average number of circulating neutrophils was significantly reduced in both homozygous and heterozygous mutant mice, ie, approximately 60% lower inΔ715/Δ715 mice and 30% to 40% lower inwt/Δ715 as compared to wt/wt animals (Fig 3). In contrast, other blood cell lineages were not affected by the mutation. Cellularity per bone and the frequencies myelomonocytic cells at various stages of maturation (Table 1) were similar in all genotypes, suggesting that the C-terminal truncation of G-CSF-R does not result in an early block of myeloid maturation in BM. Granulocyte colony-forming cells (CFU-G) were not reduced but slightly elevated inΔ715/Δ715 mice as compared withwt/wt animals, and G-CSF concentrations required to reach maximal colony formation were similar (Fig 4). The CFU-G colonies of all genotypes contained morphologically mature neutrophils (data not shown).
Numbers of circulating blood cells. Blood was collected from tail veins of 5- to 6-week-old mice and analyzed. Data are the means of 27 wt/wt (□), 40 wt/▵715(▧), and 29 ▵715/▵715 (▪) mice and error bars indicate standard error of mean. eo, eosinophils × 107/L; mo, monocytes × 107/L; ly, lymphocytes × 109/L; neu, neutrophils × 108/L; plt, platelets × 1011/L; ery, erythrocytes × 1012/L.
Numbers of circulating blood cells. Blood was collected from tail veins of 5- to 6-week-old mice and analyzed. Data are the means of 27 wt/wt (□), 40 wt/▵715(▧), and 29 ▵715/▵715 (▪) mice and error bars indicate standard error of mean. eo, eosinophils × 107/L; mo, monocytes × 107/L; ly, lymphocytes × 109/L; neu, neutrophils × 108/L; plt, platelets × 1011/L; ery, erythrocytes × 1012/L.
G-CSF-induced colony growth in vitro. BM cells were plated in methylcellulose-containing media supplemented with various quantities of G-CSF. Hematopoietic colonies containing 30 cells or more were scored after 7 to 8 days. Data are the means of four animals for each genotype and error bars represent standard deviation.gcsfr-wt/wt, (▪); gcsfr-wt/▵715, (┌);gcsfr-▵715/▵715, (□).
G-CSF-induced colony growth in vitro. BM cells were plated in methylcellulose-containing media supplemented with various quantities of G-CSF. Hematopoietic colonies containing 30 cells or more were scored after 7 to 8 days. Data are the means of four animals for each genotype and error bars represent standard deviation.gcsfr-wt/wt, (▪); gcsfr-wt/▵715, (┌);gcsfr-▵715/▵715, (□).
G-CSF treatment induces a hyperproliferative response in gcsfr-Δ715 mice.
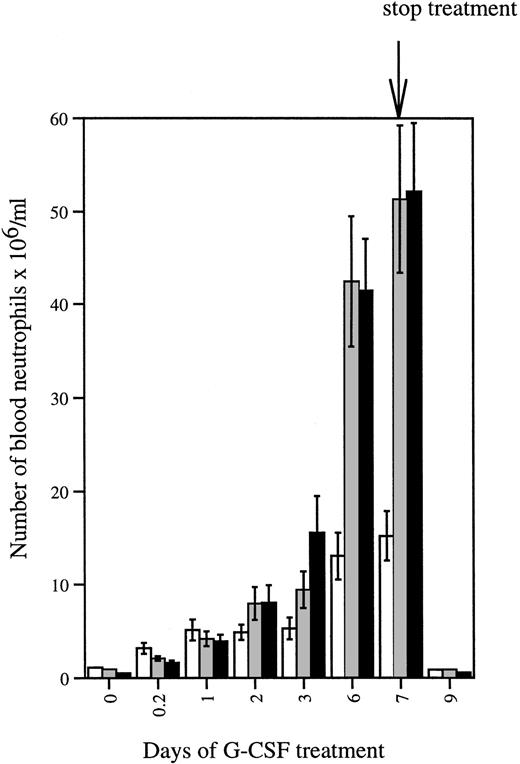
We then evaluated the in vivo responses of the mutant mice to daily injections of G-CSF (250 μg/kg) for 7 days (Fig 5). On day 2, blood neutrophil numbers of the mutant mice reached levels comparable to wt/wtmice. Thereafter, the numbers of blood neutrophils began to significantly exceed those of wild-type controls. On day 7, average peripheral neutrophil counts of wt/Δ715 andΔ715/Δ715 mice were threefold to fourfold higher than those of wt/wt animals (Fig 5). In a separate experiment, in which Δ715/Δ715 (n = 4) andwt/wt mice (n = 4) received G-CSF daily for 21 days, the average peripheral blood neutrophil counts in mutant mice increased to 200 × 106/mL versus 22 × 106/mL in wild-type mice.
Neutrophil counts in mice treated with G-CSF. Mice were injected subcutaneously with G-CSF (250 μg/kg/d) for 7 days. Blood was collected daily and analyzed as described in Materials and Methods. Data are from 7 to 11 animals for each genotype. Error bars represent standard error of mean. gcsfr-wt/wt, (□);gcsfr-wt/▵715, (▧); gcsfr-▵715/▵715, (▪).
Neutrophil counts in mice treated with G-CSF. Mice were injected subcutaneously with G-CSF (250 μg/kg/d) for 7 days. Blood was collected daily and analyzed as described in Materials and Methods. Data are from 7 to 11 animals for each genotype. Error bars represent standard error of mean. gcsfr-wt/wt, (□);gcsfr-wt/▵715, (▧); gcsfr-▵715/▵715, (▪).
In the BM, an overall increase was seen in the percentage of neutrophilic cells after G-CSF treatment (Table 1), as has been described before.23 In contrast to wt/wt mice, which were similar to wt/Δ715 mice, Δ715/Δ715mice showed a clear elevation in the early stages of neutrophilic development.
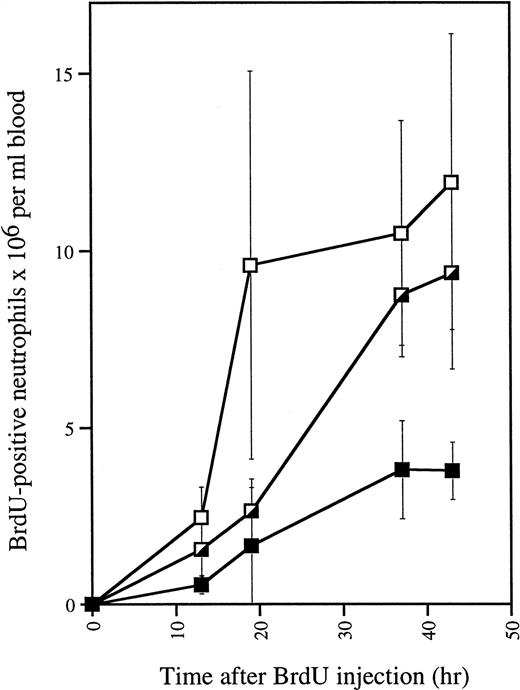
To investigate whether increased production of neutrophils could be the cause of the neutrophilia in G-CSF–treated mutant mice, we measured proliferative responses in vivo by means of BrdU. The numbers of BrdU-labeled neutrophils generated in G-CSF–treatedwt/Δ715 and Δ715/Δ715 mice significantly exceeded those of the wild-type controls (Fig 6). These data indicate that myeloid precursors of both heterozygous and homozygous mutant animals show a hyperproliferative response to G-CSF.
In vivo BrdU incorporation in blood neutrophils of G-CSF–treated mice. Mice (n = 3 for each genotype) were injected daily with G-CSF. After 4 days, 150 μL of BrdU (10 μg/mL in PBS) was injected intraperitoneally and blood was collected and analyzed at various times. Smears were made for May-Grünwald Giemsa and BrdU staining. Error bars represent standard deviation. gcsfr-wt/wt, (▪); gcsfr-wt/▵715, (┌); gcsfr-▵715/▵715, (□).
In vivo BrdU incorporation in blood neutrophils of G-CSF–treated mice. Mice (n = 3 for each genotype) were injected daily with G-CSF. After 4 days, 150 μL of BrdU (10 μg/mL in PBS) was injected intraperitoneally and blood was collected and analyzed at various times. Smears were made for May-Grünwald Giemsa and BrdU staining. Error bars represent standard deviation. gcsfr-wt/wt, (▪); gcsfr-wt/▵715, (┌); gcsfr-▵715/▵715, (□).
DISCUSSION
Mutations in the GCSFR gene truncating the C-terminal region of the receptor protein are found in a minority (about 15% to 20%) of SCN patients. However, because cases with disease progression to AML almost invariably harbor GCSFR mutations, this category of patients is of particular clinical importance. In addition, while previous studies have established that the C-terminal region of the G-CSF-R is required for G-CSF–induced neutrophilic differentiation in cell lines, the functional consequences of the mutations for neutrophil development in vivo and their potential role in leukemogenesis have remained unclear. To directly address these issues, we developed a mouse model in which we introduced an equivalent gcsfrmutation (Δ715) by homologous and Cre-mediated recombination.
The gcsfr-Δ715 mutation resulted in reduced basal neutrophil levels, but there was no significant reduction in numbers of band and segmented neutrophils, neutrophil precursors, and in vitro colony-forming progenitor cells in the BM. This suggests that truncation of the G-CSF-R C-terminus affects neutrophil development at a late stage of maturation, resulting in an impaired transit from BM segmented cell to circulating neutrophil. In contrast to the fullgcsfr-knock out, in which heterozygous mice have normal neutrophil levels in the blood, mice heterozygous for theΔ715 mutation had numbers of circulating neutrophils intermediate to homozygous and wild-type animals. These findings show that truncated G-CSF-R proteins interfere with the differentiation-inducing function of wild-type G-CSF-R in a dominant-negative manner. In myeloid cell lines (32D or L-GM), mitogenic signaling from G-CSF-R is greatly enhanced as a result of truncation of the C-terminal region.7 Our in vivo data of mice treated with G-CSF essentially show a similar phenomenon. The observation that continuous G-CSF administration induced a hyperproliferation in both heterozygous and homozygous mice indicates that truncated receptors also here dominate over wild-type receptors. In view of the increased neutrophil production in G-CSF–treated mutant mice, it remains somewhat puzzling as to why nontreated mutant mice have reduced levels of mature neutrophils in their blood. A possible explanation is that in vivo the absence of the C-terminal domain impairs differentiation only at low concentrations of G-CSF. The fact that no difference between Δ715/Δ715 and wt/wt mice is found in the neutrophilic compartment in BM suggests that a late maturation defect or an impaired transition of BM neutrophils to blood is responsible for the neutropenia. However, when more receptors are crosslinked, a threshold might be reached by which a differentiation signal overcomes other signaling events so that the BM neutrophils enter the bloodstream. Alternatively, high levels of G-CSF might be responsible for the transition of relatively immature BM neutrophils to blood.
Much effort is currently being directed at the elucidation of the signal transduction mechanisms activated by hematopoietin receptors and their contribution to different cellular responses, such as proliferation, differentiation, and cell survival. Multiple signaling events are activated via distinct intracellular subdomains of these receptors. An important question that arises is whether these signaling mechanisms exert instructive functions in the regulation of differentiation or merely control proliferation and survival of cells that are already programmed to differentiate.24 Evidence in support of a differentiation regulatory function of hematopoietin receptors has come from a variety of in vitro studies demonstrating that in certain receptors distinct cytoplasmic subdomains exist that are indispensable for induction of differentiation but not for transduction of mitogenic signals.7,8,25 26 32D cells with truncated G-CSF-R, in the absence or presence of wild-type receptors, do not differentiate in response to G-CSF, but continue proliferation, while 32D cells with only wild-type receptors mature into segmented neutrophil. We found that BM cells expressing truncated G-CSF-R differentiated in vitro to morphologically mature neutrophils in response to G-CSF. This difference could be linked to the fact that 32D cells are immortilized and that some mechanisms inducing differentiation are not strong enough in these cells to overcome the proliferation signal. Alternatively, primary cells lacking the C-terminus of G-CSF-R may have alternative stimuli driving their differentiation into neutrophil which are not present in 32D cells.
Our model has established that, under basal conditions, truncation of the G-CSF-R does give rise to neutropenia. Importantly, in view of the fact that only one allele is affected in SCN patients, heterozygous mice also showed reduced neutrophil levels. However, the neutropenia seen in these mice is not as severe as in SCN. A possible explanation for this is that the alternative mechanisms controlling neutrophil development in mice are not, or less efficiently, operational in humans. A similar discrepancy has been noted in X-linked immune disease involving the Bruton's tyrosine kinase gene.27Alternatively, the GCSFR mutation by itself may not be sufficient to cause a severe neutropenia because multiple genetic defects are required to attain the full SCN phenotype.
In the SCN patients evaluated to date, GCSFR mutations had been acquired and were restricted to the granulocytic lineage. Thus, neutrophilic progenitor cells harboring GCSFR mutations attained the ability to clonally expand, which likely reflects an early step in leukemogenesis. The hyperproliferation of neutrophil precursor cells in Δ715 mice may explain the vast clonal expansion of mutant cells in patients. The majority of patients with neutropenia, including those harboring GCSFR mutations, are now routinely treated with G-CSF to reduce the risk of bacterial infections. An important question that can now be addressed in our mouse model is whether G-CSF treatment accelerates leukemic progression. If so, it will become important to routinely identify SCN patients withGCSFR mutations and to consider alternative treatment strategies, for instance allogeneic BM transplantation, for this group of patients.
ACKNOWLEDGMENT
We are grateful to Pim van Schalkwijk and Ton Boijmans for technical assistance; Jan de Wit and Winand Dinjens for technical advice; Kirsten van Lom for differential BM and blood counts; Annelies van ‘t Hof, Els van Bodegom, and Helen Koek for assistance in the animal house; and Karola van Rooyen for graphical assistance. Frank Grosveld is gratefully acknowledged for support and advice, and Marieke Von Lindern for critical reading of the manuscript.
Supported by a PIONEER grant from the Netherlands Organization for Scientific Research NWO (M.H.A.H., C.A., I.P.T.), an EMBO Long Term Fellowship (A.C.W.), and the Dutch Cancer Society (I.P.T.).
Address reprint requests to Ivo P. Touw, PhD, Institute of Hematology, Erasmus University Rotterdam, PO Box 1738, 3000 DR Rotterdam, The Netherlands; e-mail: touw@hema.fgg.eur.nl.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.