The serine protease, thrombin, is both a potent agonist for platelet aggregation and a mitogen inducing the proliferation of other cell types. Many cellular responses to thrombin are mediated by a G-protein–coupled thrombin receptor (protease-activated receptor-1, PAR-1). This represents the prototype of a new family of proteolytically cleaved receptors that includes PAR-2 and the recently identified PAR-3. Like PAR-1, PAR-3 is a potential thrombin receptor. Their similar gene structure, mechanism of activation, and colocalization to 5q13 raises the question of a common evolutionary origin and of their belonging to a clustered gene family. Construction of a physical map of the 5q13 region by pulsed-field gel electrophoresis (PFGE) has allowed us to identify six potential CpG islands and to establish a linkage of the PAR genes. Southern blot analysis showed that they were in a cluster on a 560-kb Asc I fragment, in the order PAR-2, PAR-1, and PAR-3. PAR-1 and PAR-2 genes were contained within the identical 240-kb Not I fragment, thus confirming a tight linkage between them. The localization of other CpG islands suggested that more PAR-family genes may be present.
THROMBIN is a serine protease that is a potent agonist for platelet aggregation as well as being a mitogenic agent for the proliferation of various cell types.1-3 Many cell responses are mediated by a thrombin receptor (known as protease-activated receptor-1, PAR-1) that is a member of a G-protein–coupled receptor family. It contains seven transmembrane helical domains and is activated by a two-step mechanism.4Thrombin cleaves PAR-1 at a site within the amino-terminal extension. Then, the newly exposed N-terminus acts as a tethered ligand that interacts secondarily with other domains of the same receptor to initiate signal transduction.5,6 PAR-1 is the prototype of a new putative family of proteolytically cleaved receptors that includes PAR-2 and a recently identified receptor, PAR-3.7,8 PAR-2 is not expressed by platelets, but by vascular endothelial cells, keratinocytes, and T cells. Trypsin and other “physiological” proteases activate PAR-2 by proteolytic cleavage. Like PAR-1, PAR-3 is expressed on human platelets and megakaryocytes and is a new potential thrombin receptor.9
The human genes for PAR-1, PAR-2, and PAR-3 have been cloned and localized to 5q13, by in situ hybridization (FISH) or using radiation hybrid panels.7,10-12 This region is contiguous to the proximal breakpoint in the 5q− syndrome.13 Comparison of the three human genes for PAR-1, PAR-2, and PAR-3 suggests a similar genomic organization. These genes are of limited complexity. They contain two exons, the majority of the coding sequence being encoded by the larger second exon.7,11 14 In addition, PAR-2 is structurally closely related to PAR-1 with 30% amino acid homology.
The similar gene structure, mechanism of activation of PAR-1 and PAR-2, and the recent discovery of PAR-3, raises the question of whether the three genes have a common evolutionary origin, and might be part of a gene family whose members are clustered on the long arm of human chromosome 5. The linkage of closely related genes has been shown on several occasions as, for example, for members of the hematopoietic growth factor,15 purinoceptor (P2Y),16 and mucin families.17 18 Construction of a physical map of the 5q13 region by pulsed-field gel electrophoresis (PFGE) has allowed us to establish a linkage of the PAR genes and to establish their order within this PAR locus.
MATERIALS AND METHODS
Sources of human genomic DNA.
Lymphoblastoid (3360, 3362) and erythroblastic K562 cell lines exhibit a normal karyotype for chromosome 5. White blood cells from a healthy female volunteer were also used as a source of normal DNA (BJ). Multiple sources of DNA were chosen because of the variable degrees of methylation of DNA in cultured cells.19
Probes for PAR-1, PAR-2, and PAR-3. Genomic DNA was prepared from whole human blood according to a described QIAGEN procedure (QIAamp Blood Kit, Courtaboeuf, France). Polymerase chain reaction (PCR) amplification with Taq DNA Polymerase (Promega, Charbonnières, France) was performed for 40 cycles at 95°C for 30 seconds, at the hybridization temperature (50°C for PAR-1 and PAR-3, 45°C for PAR-2) for 30 seconds, and at 72°C for 1 minute using: (1) for PAR-1, sense primer 5′-GAA TCA AAA GCA ACA AAT GCC-3′ (Oli 42) and antisense primer 5′-CTA AGT TAA CAG CTT TTT GTA-3′ (Oli 43); (2) for PAR-2, sense primer 5′-GAA CCA ATA GAT CCT CTA AA-3′ (Oli 23) and antisense primer 5′-AAT AGG AGG TCT TAA CAG T-3′ (Oli 24); and (3) for PAR-3, sense primer 5′-GTG ACC CTG TGG ATG CTT TT-3′ (Oli 38) and antisense primer 5′-CAG CTA CTT GGG AGG CTG A-3′ (Oli 39). Each pair of oligonucleotides permitted the amplification of the coding sequence within the second exon of the corresponding gene.4,7 8These primers amplify a 1190-, 1109-, and 1041-nucleotide long fragment of PAR-1, PAR-2, and PAR-3, respectively, according to the published cDNA sequences of the human receptors.
After PCR amplification of genomic DNA, the PCR products were electrophoretically separated on a 1% low-melting agarose gel. Bands of appropriate size were cut out, purified (Promega), and cloned into p-GEM T vector according to the manufacturer's recommendations (Promega). DNA from positive colonies was sequenced using the same primers. The DNA corresponding to the probe was extracted from the vector using restriction enzymes.
Probes were labeled with [α32P] dCTP by random priming using standard procedures, and purified by gel filtration on a Sephadex G50 column (Pharmacia Biotech, Saclay, France). PAR-3 DNA contains Alu repeats.8 As a result, the purified labeled probe was resuspended with 200 μg of sonicated human DNA in sodium phosphate buffer, pH 7.2, to a final concentration of 0.1 mol/L. The mixture was incubated at 65°C for 90 minutes. The probe was used directly without further treatment.
Classical Southern blotting analysis.
Human genomic DNA was prepared from whole human blood using the QIAamp Blood Kit as recommended by the manufacturer. DNA was cut withEcoRI, BamHI, HindIII, Pst I, andXba I. Fragments were separated by conventional electrophoresis in phosphate buffer (90 mmol/L Tris-phosphate, 2 mmol/L EDTA, pH 8.0) in an 0.8% agarose gel and transferred to Hybond-N+ nylon membrane (Amersham, Les Ulis, France) by capillary blotting.
Genomic DNA preparation and restriction enzyme digestion.
Heparinized blood (10 mL) was subjected to two incubations with 30 mL red cell lysis buffer (155 mmol/L NH4Cl, 10 mmol/L KHCO3, 1 mmol/L EDTA, pH 7.4). Each time, cells were left for 15 minutes on ice, followed by centrifugation for 15 minutes at 1,800g. Cultured cells were washed in phosphate-buffered saline (PBS).
High-molecular-weight human DNA was prepared in 1% low-melting-point agarose plugs from a cell suspension (3.5 × 107cells/mL) in PBS to give a final concentration of 106 cells or 10 μg DNA per block.20 The plugs were treated with a solution containing 0.5 mol/L EDTA, pH 8.0, 200 μg/mL proteinase K, and 1% sarkosyl, at 50°C for 48 hours and then washed once with TE buffer (10 mmol/L Tris-HCl, pH 7.0, 1 mmol/L EDTA) and twice with TE buffer containing 40 μg/mL phenylmethylsulfonyl fluoride (PMSF) at 50°C. Agarose blocks were equilibrated in a large excess of the appropriate restriction enzyme buffer for 1 hour. Restriction enzyme digestion was carried out on blocks (10 μg DNA) in a 400 μL volume with rare cutting enzymes using the buffer conditions recommended by the suppliers (Ozyme-Biolabs New England, Montigny le Bretonneux, France; or Promega). For complete digestion, two applications of 100 U and 50 U of restriction enzyme were used. Partial digests were obtained by using variable amounts of restriction enzyme (0.5 or 2 U/μg of DNA). Enzyme digestion was stopped by several washes with cold TE. Each DNA preparation was tested for the absence of nuclease activity by incubating a block without added restriction enzyme.
Pulsed-field gel electrophoresis (PFGE).
After digestion, blocks were equilibrated in running buffer and loaded onto 1% agarose gels (20 × 20 cm) in TBE buffer (90 mmol/L Tris-HCl, pH 8.3, 90 mmol/L boric acid, and 2 mmol/L EDTA) and run in a contour-clamped homogeneous electric field apparatus (CHEF)21 at a constant temperature of 12°C and a constant voltage of 170 V.
Three kinds of electrophoretic conditions were used: (1) molecular size resolution optimal between 50 and 400 kb: pulse time of 30 seconds for 24 hours, ramp from 30 to 40 seconds for 24 hours, and then 40 seconds for 24 hours; (2) molecular size resolution optimal between 300 and 1,000 kb: ramp from 110 to 140 seconds for 64 hours; (3) molecular size resolution optimal between 800 and 2,000 kb: pulse time of 120 seconds for 24 hours and 240 seconds for 36 hours. Size markers were λ phage concatemers, Midrange I PFG marker (Biolabs) and chromosomes ofSaccharomyces cerevisiae (strain AB 1380). The electrophoresis gels were stained with ethidium bromide and photographed.
Southern blotting and hybridization.
Gels were depurinated for 15 minutes in 0.25 N HCl, denatured by two 30-minute treatments with 1.5 mol/L NaCl, 0.5 mol/L NaOH, and then neutralized by two 30-minute treatments with 0.5 mol/L Tris-HCl, pH 7.5, 3 mol/L NaCl, before being blotted onto charged Hybond-N+ nylon membrane overnight with 20× SSC buffer (1 × SSC = 0.15 mol/L NaCl, 0.015 mol/L trisodium citrate). After transfer, DNA was fixed on the membrane by drying for 1 hour at 37°C and then cross-linked using UV light.
Prehybridizations were performed at 65°C in prehybridization buffer (6× SSC, 5× Denhardt's, 0.5% sodium dodecyl sulfate [SDS]) for 3 hours. Hybridizations were performed overnight at 65°C in hybridization buffer (6× SSC, 5× Denhardt's, 0.5% SDS, 10% dextran sulfate, 750 μg/mL ssDNA) for all probes. Filters were washed twice in 2× SSC, 0.1% SDS, for 30 minutes at 65°C. Higher stringency washes of 0.1× SSC, 0.1% SDS for the PAR-3 probe were performed at 70°C. Autoradiography of filters was at −80°C between two intensifying screens.
RESULTS
Classical Southern blot analysis.
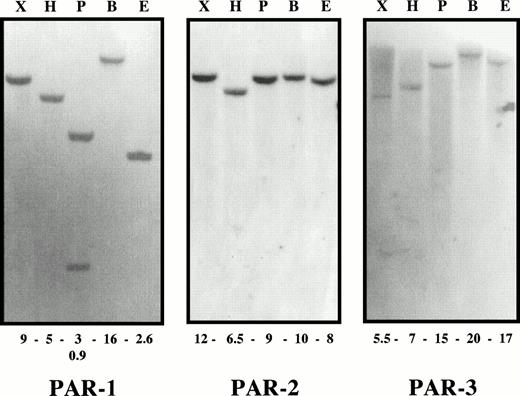
To verify the specificity of each probe, we first performed Southern blot analysis of human genomic DNA digested with EcoRI,BamHI, HindIII, Pst I, and Xba I. Results showing hybridization with the PAR-1, PAR-2, and PAR-3 probes are illustrated in Fig 1. Whatever the enzyme used, a single cross-hybridizing fragment was seen for each probe thereby establishing that PAR-1, PAR-2, and PAR-3 are present as single-copy locus genes and that they do not cross-hybridize with each other. For each of the EcoRI and Xba I digestions, the three probes gave patterns similar to those reported by Schmidt et al11,12 using total human genomic DNA. However, Schmidt et al12 also found an additional cross-hybridizing ∼5-kbEcoRI fragment in YAC DNA hybridized to the PAR-1 probe, suggested by the investigators to represent either incompletely digested DNA, rearrangements within the YAC, or a homologous gene or pseudogene. This 5-kb fragment was not found by us.
Classical Southern blot analysis. Human genomic DNA was digested by Xba I (X), HindIII (H), Pst I (P),BamHI (B) and EcoRI (E). Successive hybridizations were performed with PAR-1, PAR-2, and PAR-3 probes. The fragment size is given in kilobases below each lane. Whatever the enzyme used, a single cross-hybridizing fragment was seen for the three probes. For thePst I digest, PAR-1 hybridized with two fragments because of the presence of a Pst I site in the PAR-1 coding sequence.
Classical Southern blot analysis. Human genomic DNA was digested by Xba I (X), HindIII (H), Pst I (P),BamHI (B) and EcoRI (E). Successive hybridizations were performed with PAR-1, PAR-2, and PAR-3 probes. The fragment size is given in kilobases below each lane. Whatever the enzyme used, a single cross-hybridizing fragment was seen for the three probes. For thePst I digest, PAR-1 hybridized with two fragments because of the presence of a Pst I site in the PAR-1 coding sequence.
General considerations.
PFGE analysis was used to establish a physical map of the region containing the PAR family genes. High-molecular-weight DNAs from two lymphoblastoid cell lines (3360, 3362), an erythroblastic cell line (K562), and human lymphocytes (BJ) were analyzed. The selected restriction enzymes fell into two categories: (1) those recognizing (G+C)-rich sites included in CpG islands (Not I, Asc I,Sgf I, BssHII, SacII) and (2) those mainly recognizing sites outside CpG islands (Mlu I, Nru I,Xho I, Sfi I). A series of 30 blots with single or double digests allowed us to construct a restriction map of the region containing the PAR cluster in relation to the putative CpG islands. The sizes of the various restriction fragments detected by each of the PAR-1, PAR-2, and PAR-3 probes are given in Tables 1, 2 and3.
Summary of the Restriction Fragments Hybridized by the PAR-1, PAR-2, and PAR-3 Probes Using Lymphoblastoid (L), Erythroblastic K562 (E), and BJ DNA
| . | PAR-1 . | PAR-2 . | PAR-3 . |
|---|---|---|---|
| Asc I | |||
| L | 560-150 | 560-150 | 560-150 |
| E | 650-700 | 650-700 | 650-700 |
| BJ | 560-151-700-151 | 560-151-700-151 | 560-151-700-151 |
| Asc I/Mlu I | |||
| L | 310 | 310 | ND |
| BJ | 390-151-700-151 | 390-151-700-151 | 140-151-310-151-700-151 |
| Mlu I | |||
| L | 560 | 560 | 400-550 |
| BJ | 560 | 560 | ND |
| Asc I/Not I | |||
| L and BJ | 240 | 240 | 320 |
| Not I | |||
| L and BJ | 240-150 | 240-150 | ND |
| Asc I/Nru I | |||
| L and BJ | 240 | 240 | 320 |
| Nru I | |||
| L and BJ | 380-150 | 380-150 | 370 |
| Mlu I/Nru I | |||
| L and BJ | 380 | 380 | ND |
| MluI/Not I | |||
| L and BJ | 240 | 240 | ND |
| Not I/Nru I | |||
| L and BJ | 240 | 240 | ND |
| . | PAR-1 . | PAR-2 . | PAR-3 . |
|---|---|---|---|
| Asc I | |||
| L | 560-150 | 560-150 | 560-150 |
| E | 650-700 | 650-700 | 650-700 |
| BJ | 560-151-700-151 | 560-151-700-151 | 560-151-700-151 |
| Asc I/Mlu I | |||
| L | 310 | 310 | ND |
| BJ | 390-151-700-151 | 390-151-700-151 | 140-151-310-151-700-151 |
| Mlu I | |||
| L | 560 | 560 | 400-550 |
| BJ | 560 | 560 | ND |
| Asc I/Not I | |||
| L and BJ | 240 | 240 | 320 |
| Not I | |||
| L and BJ | 240-150 | 240-150 | ND |
| Asc I/Nru I | |||
| L and BJ | 240 | 240 | 320 |
| Nru I | |||
| L and BJ | 380-150 | 380-150 | 370 |
| Mlu I/Nru I | |||
| L and BJ | 380 | 380 | ND |
| MluI/Not I | |||
| L and BJ | 240 | 240 | ND |
| Not I/Nru I | |||
| L and BJ | 240 | 240 | ND |
List of the BssHII Fragments From the Lymphoblastoid (L) and Erythroblastic K562 (E) Cell Lines and From BJ DNA Hybridized by the Three PAR Gene Probes
| DNA . | PAR-1 . | PAR-2 . | PAR-3 . |
|---|---|---|---|
| L | 110* | 130* | |
| E | 110 | 130 | 280 |
| 190 | 220 | 400 | |
| 310 | 270 | 480 (db) | |
| 480 | 370 | 590 | |
| 590 | 400 | 650 | |
| 650 | |||
| BJ | 110 | 130 | 280 |
| 220 | 400 |
| DNA . | PAR-1 . | PAR-2 . | PAR-3 . |
|---|---|---|---|
| L | 110* | 130* | |
| E | 110 | 130 | 280 |
| 190 | 220 | 400 | |
| 310 | 270 | 480 (db) | |
| 480 | 370 | 590 | |
| 590 | 400 | 650 | |
| 650 | |||
| BJ | 110 | 130 | 280 |
| 220 | 400 |
(db) indicates a double band.
Seen in Fig 2.
| Double Digests on a Lymphoblastoid Cell Line . | ||
|---|---|---|
| SacII/Not I | 110* | 130* |
| SacII/Asc I | 110 | 130 |
| SacII/Nru I | 110 | 130 |
| SacII/BssHII | 110* | 130* |
| BssHII/Not I | 110* | 130* |
| BssHII/Nru I | 110 | 130 |
| BssHII/Mlu I | 110 | 130 |
| BssHII/Asc I | 110 | 130 |
| Double Digests on a Lymphoblastoid Cell Line . | ||
|---|---|---|
| SacII/Not I | 110* | 130* |
| SacII/Asc I | 110 | 130 |
| SacII/Nru I | 110 | 130 |
| SacII/BssHII | 110* | 130* |
| BssHII/Not I | 110* | 130* |
| BssHII/Nru I | 110 | 130 |
| BssHII/Mlu I | 110 | 130 |
| BssHII/Asc I | 110 | 130 |
Abbreviation: ND, not determined.
Seen in Fig 2.
Sites in the 5q13 region containing the PAR-1, PAR-2, and PAR-3 genes were sequentially mapped using the data generated by the single and double digests. Some enzymes gave little useful information because the fragments were too large (Sgf I) or, in contrast, too small (Sfi I and Xho I). Nevertheless, numerous variably methylated sites for Asc I, Not I, BssHII,SacII, Mlu I, and Nru I were found in the different sources of DNA studied. The variable degree of DNA methylation generated partial digestion fragments that were useful in both the assignment and the confirmation of sites.
Physical linkage and relative positions of the PAR-1, PAR-2, and PAR-3 genes.
The first Southern blot analysis demonstrated that large-sized DNA fragments (>240 kb) were in most cases positive with cDNA probes for PAR-1 and PAR-2 (Fig 2A). A summary of these fragments is given in Table 1. The smallest common fragment cohybridizing with PAR-1 and PAR-2 is a 240-kb fragment in theNot I digest (N1N2) from the lymphoblastoid cell line (also see Fig 3). A 110-kb fragment was detected by PAR-1 and a 130-kb fragment by the PAR-2 probe in samples digested by BssHII (B6B7, B5B6) andSacII (S3S4, S2S3) alone or in combination with NotI (Tables 1-3, Fig 2). The sum of the size of these two fragments is in good agreement with the estimated size of 240 kb for the Not I fragment that houses the PAR-1 and PAR-2 genes and indicates that these two fragments are contiguous.
PFGE analysis of DNA from a lymphoblastoid cell line to localize the PAR-1 and PAR-2 genes. Fragment size was assigned using the λ multimer ladders (λ). (A) The largest fragments obtained withAsc I (A), Nru I (Nr), and Not I (N) are common to PAR-1 and PAR-2 whereas the smallest obtained with SacII (S) are specific for each probe. PFGE conditions: ramp from 110 to 140 seconds for 64 hours. (B) By varying the electrophoretic conditions (pulse time of 30 seconds for 24 hours, ramp from 30 to 40 seconds for 24 hours and then 40 seconds for 24 hours), we achieved an improved separation of the different 110-kb and 130-kb fragments. Whichever the combination of enzymes used (Not I, BssHII [B],SacII), each probe gave an identical pattern, indicating that the cutting sites for these enzymes are clustered.
PFGE analysis of DNA from a lymphoblastoid cell line to localize the PAR-1 and PAR-2 genes. Fragment size was assigned using the λ multimer ladders (λ). (A) The largest fragments obtained withAsc I (A), Nru I (Nr), and Not I (N) are common to PAR-1 and PAR-2 whereas the smallest obtained with SacII (S) are specific for each probe. PFGE conditions: ramp from 110 to 140 seconds for 64 hours. (B) By varying the electrophoretic conditions (pulse time of 30 seconds for 24 hours, ramp from 30 to 40 seconds for 24 hours and then 40 seconds for 24 hours), we achieved an improved separation of the different 110-kb and 130-kb fragments. Whichever the combination of enzymes used (Not I, BssHII [B],SacII), each probe gave an identical pattern, indicating that the cutting sites for these enzymes are clustered.
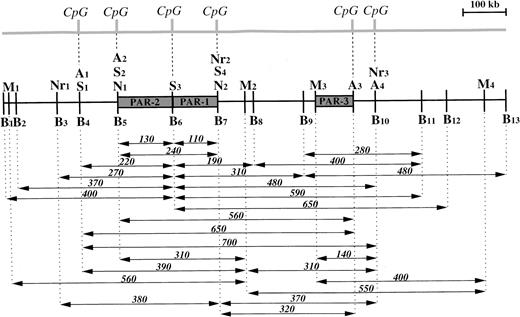
Restriction map of the region 5q13 containing the PAR-1, PAR-2, and PAR-3 genes. Sites used were Asc I (A),BssHII (B), Mlu I (M), Not I (N), Nru I (Nr), SacII (S). The majority of the fragments shown in Tables1-3 are presented in the form of arrows below the restriction map. Their sizes are given in kilobases. Some partial fragments forSacII are not illustrated. A map of the CpG islands is shown at the top of the figure.
Restriction map of the region 5q13 containing the PAR-1, PAR-2, and PAR-3 genes. Sites used were Asc I (A),BssHII (B), Mlu I (M), Not I (N), Nru I (Nr), SacII (S). The majority of the fragments shown in Tables1-3 are presented in the form of arrows below the restriction map. Their sizes are given in kilobases. Some partial fragments forSacII are not illustrated. A map of the CpG islands is shown at the top of the figure.
With the K562 cell line, the smallest BssHII restriction fragments were monospecific for each probe (Table 2): the 110-kb (B6B7), 130-kb (B5B6) fragments recognized by PAR-1 and PAR-2, respectively, were the same as those seen with the lymphoblastoid cell line (Fig 3). A 280-kb band (B9B11) was now recognized by the PAR-3 probe. Partial fragments specific for the PAR-2 probe were also detected. Strikingly, no partial restriction fragment common to PAR-1 and PAR-2 was obtained, indicating a preferential cutting-site for BssHII between the PAR-1 and PAR-2 genes. With these partial digests, fragments above 480 kb reacted identically with PAR-1 and PAR-3 probes. This finding allows us to deduce that PAR-3 resides on a 280-kbBssHII fragment distal to the 110-kb BssHII fragment carrying PAR-1. Both probes are also linked on a common 590-kbBssHII partial digestion product (B6B11). In this partial digest, PAR-1 alone recognized a 310-kb BssHII partial hydrolysis fragment (B6B9), which therefore must be juxtaposed to the 280-kb fragment (B9B11) recognized by PAR-3. By deduction, the PAR-1 310-kb BssHII (B6B9)-specific fragment is located between the PAR-2 130-kb and PAR-3 280-kb specific fragments.
In conclusion, the above results imply that PAR-2 is contained in a 130-kb fragment adjacent to the 110-kb fragment on which PAR-1 lies. In contrast, the PAR-1 and PAR-3 genes are not contiguous, they are at least 200-kb apart (Fig 3).The order of these genes is PAR-2,PAR-1, PAR-3.
Additional restriction sites.
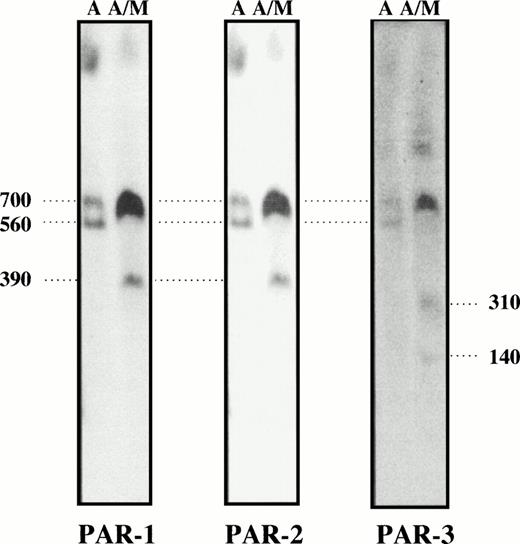
The smallest common DNA fragment cohybridizing with PAR-1, PAR-2, and PAR-3 probes is a 560-kb Asc I fragment (A2A3) for the lymphoblastoid cell line (Table1, Fig 3). Asc I/Mlu I digests generated small 310-kb (A2M2) and 390-kb (A1M2) fragments only hybridizing with PAR-1 and PAR-2 probes using lymphoblastoid cell line and BJ DNA, respectively. An additional 700-kb fragment (A1A4) is also identified by these two probes in BJ DNA (Table 1, Fig 4). PAR-3 hybridizes to this fragment, as well as to smaller 140-kb (A4M3) and 310-kb (A4M2) fragments. The sum of the 310-kb (A4M2) PAR-3 and of the 390-kb (A1M2) PAR-1/PAR-2 hybridizing fragments suggests that they are contiguous within the 700-kb (A1A4) fragment.
PFGE analysis of Asc I and AscI/Mlu I fragments from BJ DNA. The Asc I fragments (700 and 560 kb) are common for the three probes showing the existence of a PAR cluster. The double digest gave a partial 700-kb Asc I fragment, the smaller 560-kb fragment was not detected. Asc I shows marked site preference, and we obtained variable amounts of cleavage at this site in different digests. The AscI/Mlu I digest shows that PAR-3 is located at one end of the PAR cluster. PAR-1 and PAR-2 are located on a 390-kb fragment while PAR-3 is seen on a separate 310-kb fragment (and on a shorter 140-kb fragment). PFGE conditions: ramp from 110 to 140 seconds for 64 hours.
PFGE analysis of Asc I and AscI/Mlu I fragments from BJ DNA. The Asc I fragments (700 and 560 kb) are common for the three probes showing the existence of a PAR cluster. The double digest gave a partial 700-kb Asc I fragment, the smaller 560-kb fragment was not detected. Asc I shows marked site preference, and we obtained variable amounts of cleavage at this site in different digests. The AscI/Mlu I digest shows that PAR-3 is located at one end of the PAR cluster. PAR-1 and PAR-2 are located on a 390-kb fragment while PAR-3 is seen on a separate 310-kb fragment (and on a shorter 140-kb fragment). PFGE conditions: ramp from 110 to 140 seconds for 64 hours.
When using lymphoblastoid cell line or BJ DNA, PAR-1 and PAR-2 probes identified a 380-kb Nru I fragment (Nr1Nr2), whereas the PAR-3 probe detected a 370-kb band (Nr2Nr3) (Fig 3). Double digests performed with Asc I and Not I or Asc I andNru I gave a 320-kb (A3N2 or A3Nr2) fragment specific to the PAR-3 probe, whereas the PAR-1 and PAR-2 probes hybridized to a shorter 240-kb fragment (A2Nr2), corresponding to the fragmentNot I (N1N2).
These results with additional sites supplement the construction of the most probable restriction map shown in Fig 3. The map extends about 1,200 kb and satisfies all of the restriction data obtained.
Identification of CpG islands surrounding the PAR-1, PAR-2, and PAR-3 genes.
Rare cutting restriction enzymes were selected according to their ability to cut into CpG islands in a specific manner.22 The enzymes selected were Not I, Asc I, BssHII, andSacII. Lymphoblastoid cell line DNA was first cut byNot I, Sac II, and BssHII. Whatever the combination of enzymes used, PAR-1 and PAR-2 genes are always seen on the same 110-kb and 130-kb fragments (Table 3, Fig 2B). This indicates that the cutting sites for these enzymes are clustered and that they show the presence of three CpG islands, one on each side of the PAR-1 and PAR-2 genes and one located between them. With SacII andBssHII, we observed the same 220-kb fragment (S1S3 and B4B6) hybridizing to the PAR-2 probe, superimposing sites A1, B4 and S1 and so indicating the presence of a fourth CpG island. The close vicinity of the A4 and B10 sites localizes a fifth, while the A3 site suggests a sixth CpG island, since 93% of Asc I sites are within CpG islands.22 Therefore, the entire 1,200-kb map potentially contains at least six putative CpG islands (Fig 3).
Remark.
The human PAR-3 cDNA was initially cloned by screening a human small intestine cDNA library.8 In our study, the PCR and sequencing of PAR-3 were performed on human genomic DNA with Oli 38 and 39 primers covering the majority of the cDNA sequence. This implies that the major part of the coding sequence is contained in a single exon, just as for PAR-1 and PAR-2.
DISCUSSION
The protease-activated receptors are closely related with 45% sequence homology. Previous studies have shown that the PAR-1, PAR-2, and PAR-3 genes have a similar genomic organization and reside on the long arm of chromosome 5.7-12,14 Preliminary studies on radiation hybrids have shown that PAR-3 is colocalized with the other PARs in the same region of this chromosome.11 We have now used PFGE to identify potential CpG islands and to produce a map of the 5q13 region. Evidence has been obtained that the three PAR genes are arranged in a cluster within a 560-kb region and in the order PAR-2,PAR-1, PAR-3.
Central to our long-range mapping strategy was the use of several cell lines with different methylation status as a source of DNA, because “natural” partial digestions could be obtained. The detection of partial restriction fragments in common confirmed that the PAR-1, PAR-2, and PAR-3 genes were in the same genomic region (for example, see Table 1 for Asc I digests). In addition, the analysis of partial digests helped us define the order of the three genes along the chromosome. Finally, this strategy allowed us to extend the restriction map on both sides of the PAR-1, PAR-2, and PAR-3 gene cluster.
Using field inversion gel electrophoresis (FIGE) and YAC clones, Schmidt et al12 found that PAR-1 and PAR-2 genes are contained within identical Xho I (∼90-kb) and Not I (∼120-kb) restriction fragments. Using CHEF and genomic DNA, we found that PAR-1 and PAR-2 genes are contained within two specificXho I (∼70- and 50-kb, respectively; data not shown) and within the same Not I (∼40-kb) restriction fragments. These differences may be explained by the fact that YAC DNA, when compared with genomic DNA, particularly that from cultured cells,19is unmethylated but is subject to rearrangements. In addition, if there is a restriction site for Not I in the CpG island between PAR-1 and PAR-2 genes, this site may be methylated in genomic DNA and the site would not be cut. An explanation could be that the 240-kb fragment observed by us effectively corresponds to the sum of two bands of ∼120-kb when YAC unmethylated genomic DNA was the substrate and which were not resolved using FIGE. With this technique, mobility decreases with size until an inflection point above which longer molecules travel faster than shorter ones. This complex relationship makes it difficult to obtain accurate sizes.23 Thus, for optimal separation and accurate sizing we used the CHEF procedure. By varying the electrophoretic conditions we were able to separate different 110-kb and 130-kb fragments with BssHII and SacII, which for others were detected as a single ∼120-kb fragment.12
It is interesting that genes encoding other G-protein–coupled receptors, for example certain adrenergic receptor subtypes24 and P2Y receptors,16 are clustered in the genome. Conservation of structural and/or functional domains among these G-protein–coupled receptors suggests that they may have a common ancestor.25 The PAR-1 and PAR-3 genes were located on BssHII fragments that were separated by at least 200 kb and PAR-1 and PAR-2 were on contiguous BssHII fragments of combined size of 240 kb. The proximity of these three related genes on human 5q13 supports the possibility that they may have evolved by recent gene duplication and that the close linkage, particularly between PAR-1 and PAR-2, may be important in the regulation of their expression. However, further studies will be required to determine the mechanisms leading to their transcription in cells. In our study, we found SacII fragments in the same size range (315 and 400 kb) hybridized by the three probes. However, each fragment is too small to contain all three genes (Table 2). Given the results obtained with other restriction enzymes, it is probable that each probe recognizes anSacII fragment (315 kb) containing one PAR gene. This suggests that a symmetry exists in the distribution of these sites.18 This observation reinforces the previous suggestion that a single ancestral PAR gene may have given rise by successive duplications to several distinct PAR genes.
PFGE analysis of large DNA fragments is a powerful tool to identify CpG islands that are frequently found in the promoter regions of genes and that constitute valuable markers to locate genes.22 If these PAR genes are members of a distantly related gene family, the possibility exists that other as yet unidentified members of the family exist within the cluster. It is possible that they are associated with one of the six potential CpG islands that we have identified, bearing in mind that associations with PAR-1, -2, or -3 can account for a maximum of three of them. Studies are planned in our laboratory to examine the DNA surrounding these islands and to determine if other PAR genes are also localized in this region of the chromosome.
The PAR genes are localized to 5q13, a region that is contiguous to the proximal breakpoint (q13 → q15) identified in the 5q- syndrome.13 Because of the known effect of thrombin on multiple cell types, including megakaryocytes, Boultwood et al26 and Demetrick et al13 suggested that the PAR-1 gene may be involved in the dysmegakaryocytopoiesis observed in this syndrome. Our results suggest that the recently cloned second thrombin receptor, PAR-3, could also be a potential candidate gene implicated in this disorder. Using dual-label FISH and complementary techniques involving interphase and metaphase nuclear analysis of cells from seven patients with a del (5)(q13q33), Demetrick et al13 showed that the PAR-1 gene is centromeric to the proximal breakpoint and therefore uninvolved in the region encompassed by the interstitial deletion. However, it is possible that PAR-2, PAR-3, and other potential PAR genes would be involved. In addition, it should be noted that the breakpoints in the 5q− anomaly vary somewhat among patients and that the 5q deletion is also often associated with other chromosomal abnormalities.27 So it would be interesting to analyze the rearrangements in the 5q13 region of DNA of patients with the 5q− syndrome by FISH and PFGE to clarify the involvement of PAR genes in the 5q− syndrome and related disorders.
Supported by the CNRS, Université Bordeaux II, the Conseil Régional d'Aquitaine, and the Ministère de l'Enseignement Supérieur et de la Recherche (ACC-SV No. 9). V.G.D. was a recipient of postdoctoral fellowships from the Association Sanofi Thrombose pour la Recherche and from l'ARC.
Address reprint requests to Véronique Guyonnet Dupérat, PhD, UMR 5533 CNRS, Hôpital Cardiologique, 33604 Pessac, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 2. PFGE analysis of DNA from a lymphoblastoid cell line to localize the PAR-1 and PAR-2 genes. Fragment size was assigned using the λ multimer ladders (λ). (A) The largest fragments obtained withAsc I (A), Nru I (Nr), and Not I (N) are common to PAR-1 and PAR-2 whereas the smallest obtained with SacII (S) are specific for each probe. PFGE conditions: ramp from 110 to 140 seconds for 64 hours. (B) By varying the electrophoretic conditions (pulse time of 30 seconds for 24 hours, ramp from 30 to 40 seconds for 24 hours and then 40 seconds for 24 hours), we achieved an improved separation of the different 110-kb and 130-kb fragments. Whichever the combination of enzymes used (Not I, BssHII [B],SacII), each probe gave an identical pattern, indicating that the cutting sites for these enzymes are clustered.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/1/10.1182_blood.v92.1.25.413k41_25_31/5/m_blod41341002w.jpeg?Expires=1769126878&Signature=FWDbw-Ea-xYRtOqTpwEd1S7RD5JQr3da~aP5GHT0GrwQAeVjBOwr8mCdt83FRIbJq3p~dAbi5HswoU6ihWoW5u~HvT0oTag1QnFIHC9ej7swh0X9cGd7gDhiMqjThKPrcMGH2W51k4MglaG9Qcve27lRxu66xfbGgl7ErWkgNHUKlkZtvgjluG0boew3yZt1EzOAVpqyw-xVUvjhqokjq7y51mXYpJczLPcbVqPDALr17ws7ucNIbQDweWkIBj-J6T~l~zo6QTn8ayVLlm6QXPBooZRvx1sQIiMjuQABEjHAVe5XXvywe3atClzykU18xeSDpDy5K01Nzthl3wBKGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal