In a previous study of acute leukemia, we have shown thatWT1 gene mutations occur in both myeloid and biphenotypic subtypes, where they are associated with refractoriness to standard induction chemotherapy. We have now extended this study to a total of 67 cases (34 acute myeloid leukemia [AML], 23 acute lymphoblastic leukemia [ALL], 10 acute undifferentiated leukemia [AUL]/biphenotypic) and find that WT1 mutations occur in 14% of AML and 20% of biphenotypic leukemia, but are rare in ALL (one case). In contrast to the findings in Wilms' tumor, where mutations in the WT1 gene usually behave according to Knudson's two hit model for tumor suppressor genes, seven of eight leukemia-associated WT1 mutations are heterozygous, implying a dominant or dominant-negative mode of action in hematopoietic cells. In AML, the presence of a WT1 mutation is associated with failure to achieve complete remission and a lower survival rate. These data (1) confirm that WT1 mutations underlie a similar proportion of cases of AML to that seen in Wilms' tumors and (2) show for the first time that WT1 mutations can contribute to leukemogenesis of lymphoid as well as myeloid origin, suggesting that its normal role in hematopoiesis lies at a very early progenitor stage. The relationship of WT1 mutation to chemoresistance merits further investigation.
THE WILMS' TUMOR GENE, WT1, encodes a zinc finger protein, which can function as a transcription factor (reviewed in Reddy and Licht1). WT1 was originally identified as a gene involved in genetic predisposition to the childhood kidney cancer, Wilms' tumor, and is a paradigm for the relation of normal developmental processes to tumorigenesis.2 Distinct germline WT1 mutations underlie two congenital malformation syndromes, which predispose to Wilms' tumor: complete deletion of one allele in the association of Wilms' tumor with aniridia (Wilms' tumor, aniridia, genitourinary abnormalities, mental retardation [WAGR] syndrome) and missense mutations in the zinc finger region in Denys-Drash syndrome (DDS), a triad of genital malformation, nephrotic syndrome, and Wilms' tumor. Expression of WT1 is highest during embryogenesis, where it is found in multipotent progenitor cells of a restricted range of tissues, mainly in the genitourinary system.3 In the adult, expression of this tissue-specific gene continues in specific cell types of the kidney and gonad and, at much lower levels, in the bone marrow, where it is confined to CD34+ progenitor cells.4,5 Although murine knock-out experiments show that WT1 is essential for the development of the genitourinary system,6 there is no obvious effect on the hematopoietic system, suggesting functional redundancy. Nevertheless, many leukemias, like Wilms' tumors, exhibit overexpression of WT14 (and reviewed in Pritchard-Jones and King-Underwood7). We have shown recently that WT1 mutations occur in acute leukemias at a frequency similar to that found in sporadic Wilms' tumors.5 However, the type of mutation suggested a different mechanism of action of mutant WT1 in differentiating hematopoietic cells compared with metanephric blastema.
WT1 is known to function as a transcriptional regulator, with the capability of either activating or repressing transcription, depending on the cellular context, the target promoter, and the type of WT1 isoform.1 WT1 is subject to alternative splicing at two sites, involving the 17 amino acids which comprise exon 5, and a three amino acid insertion (lysine-threonine-serine, KTS) between the third and fourth zinc fingers. The number of potential isoforms is increased to 16 due to the possibility of RNA editing in exon 6, changing a Leucine to a Proline, and the use of an alternative initiation codon.1,8 The splice variant whose effect has been most studied is the KTS insertion in the zinc finger region, which alters the DNA binding affinity and specificity of WT1. Intriguingly, +KTS isoforms have been shown to colocalize with splicing factors in the nucleus and WT1 does possess RNA binding activity.9,10Therefore, WT1 is not only a dichotomous regulator in the sense of switching between transcriptional activation and repression, but may also influence gene expression through a role in RNA processing. The role of this multifunctional protein in hematopoietic differentiation is not yet understood, although possible target genes such as colony-stimulating factor have been identified in vitro.11However, the findings of hematopoietic-specific mechanisms of controlling WT1 function, such as tissue-specific enhancers within the WT1 gene12,13 and variations in exon 5 splicing,14 suggest that WT1 has an important role in the hematopoietic system.
Mutations in the WT1 gene underlie 5% to 10% of sporadic Wilms' tumors (reviewed in Little and Wells15). Although the majority follow Knudson's two hit hypothesis for tumor suppressor genes, it is now clear that a substantial minority (≈ 30%) of Wilms' tumors retain one normal WT1 allele, suggesting that in some cases, heterozygous mutation is sufficient for tumorigenesis. Five different types of mutation are commonly found in Wilms' tumors: large deletions of part or all of the gene (often germline), nonsense or frameshift mutations producing a truncated protein, missense mutations affecting amino acids in the zinc fingers critical for DNA binding, missense mutations affecting the putative activation or repression domains, and mutations preventing correct splicing. Approximately 75% of the WT1 mutations found in sporadic Wilms' tumor produce a truncated protein, whereas missense mutations in the zinc finger region predominate in DDS.1 15
We undertook this study to investigate whether WT1 mutations are confined to leukemias of specific lineage origin and whether the types of mutations might shed light on the function of WT1 in hematopoiesis. We found that although WT1 mutations occur mainly in acute myeloid leukemias, they are also found in undifferentiated, biphenotypic, and lymphoblastic leukemias, suggesting a role for WT1 in very early hematopoiesis, before determination of the lymphoid/myeloid split. This is supported by the finding that the level of expression is highest in leukemias with immature phenotypes.16 Expression of WT1 is downregulated during differentiation of leukemic cell lines, and transfection studies show that WT1 can cause cell cycle arrest and alter apoptotic responses (reviewed in Reddy and Licht1 and Pritchard-Jones and King-Underwood7). This may reflect a role in the control of normal hematopoiesis, which can be abrogated by mutations in the gene and form part of the pathway towards leukemogenesis.
MATERIALS AND METHODS
Clinical details.
Sample 101 was from a 12-year old female with acute, morphologically undifferentiated leukemia (AUL). Cytochemistry was negative, immunophenotyping was positive for CD7, CD33, CD34, and terminal deoxynucleotidyl transferase (TdT). Cytogenetic analysis was not performed. After 6 weeks of standard induction chemotherapy for acute lymphoblastic leukemia (ALL), bone marrow aspirate showed persistent excess blasts. At this point, she was referred to our center where treatment was changed to induction therapy for acute myeloid leukemia (AML). Cytogenetic analysis of her leukemic blasts showed poor chromosome morphology, but four of nine cells had deletions of 6q and abnormalities of 11p—deletion of part of the short arm of one chromosome 11 and additional material on the short arm of the other. The sample showing WT1 mutation was taken at this point. She achieved a technical remission and proceeded to sibling bone marrow transplant (BMT). Unfortunately, she experienced a bone marrow relapse 5 months posttransplant and died.
Sample 126 was from a 6-year old boy presenting with typical T-ALL, confirmed by immunophenotyping, cytochemistry, and molecular studies of T-cell receptor gene rearrangement. He was treated with standard ALL therapy to which he responded promptly. He suffered a lymph node and bone marrow relapse 21 months from diagnosis. Cytogenetic analysis at first diagnosis was unsatisfactory and the relapse specimen showed a normal karyotype.
Sample 146 was taken at diagnosis from a 17-year old male with acute promyelocytic leukemia. Cytogenetic analysis showed the characteristic t(15;17) with additional trisomy 8 and monosomy 21. He had a slow early response to standard induction therapy, achieving remission after 4 months. Consolidation therapy included autologous BMT. He relapsed 16 month from diagnosis, achieved a brief second remission, but died of disease 25 months from diagnosis.
Sample 87 was taken at first relapse from a 29-year old female with a revised diagnosis of AML (M0). She had initially been treated with ALL therapy to which she had a complete response. Four years from initial diagnosis, she relapsed in the bone marrow and skin. Blasts were morphologically undifferentiated, but were positive for myeloid markers (CD13, 15, and 33). Cytogenetic analysis showed 47XX + 19. She died of septicemia on day 10 of reinduction chemotherapy.
Leukemic samples were collected as bone marrow or peripheral blood with circulating blasts into preservative-free heparin. The mononuclear cell fraction was isolated using Lymphoprep (Nycomed, Oslo, Norway), and cell pellets were stored at −70°C. DNA and RNA were isolated from cryopreserved cells collected by leukopheresis for some samples. DNA and RNA were extracted as described.5 The T-ALL cell line, CCRF-CEM, which expresses high levels of WT1, was used as a positive control for reverse transcriptase-polymerase chain reaction (RT-PCR).
PCR and automated fluorescent sequencing.
Exons 2 to 10 and the 3′ end of exon 1 were amplified as previously described.5,17 In addition, the portion of exon 1 containing the initiation codon was amplified using primers 256 and 532,18 with the addition of 0.2 U of cloned Pfu DNA polymerase (Stratagene, Cambridge, UK) per 50 μL reaction. Products of 50 μL PCR reactions were cut out of low-melting point agarose and purified using Wizard PCR Preps (Promega, Southampton, UK) and then precipitated with ethanol, redissolving in 12 μL. A total of 2 μL was run on a gel to check recovery, and 2 to 5 μL was used as the template for sequencing using the ABI Prism Dye Terminator Cycle Sequencing kit with AmpliTaq FS (Perkin Elmer ABI, Warrington, Cheshire, UK) using a modified method.19 Sequencing products were run on a 373 Sequencer or a 310 Genetic Analyser (Perkin Elmer ABI). The results were analyzed using Factura software and by eye to identify heterozygotes. Samples containing mutations were resequenced using a second independent PCR product as a template.
RNA analysis.
Northern blots were hybridized with a 32P-labeled 1.8-kbEcoRI fragment of WT33 using standard methods.
RT-PCR was performed as previously described5 using primers in exon 6 and 10 to amplify a 602/611-bp product from RNA from the samples with mutations and from a positive control (CEM cells). Two independent RT-PCR products from each sample were purified and sequenced in both directions as above.
Statistical analyses.
For patients with AML at first diagnosis, the probabilities of achieving first remission, disease-free survival, and overall survival were analyzed by log-rank comparisons between Kaplan-Meier curves.20
RESULTS
Sequencing.
In this study, 36 samples from 33 patients were analyzed for the presence of WT1 mutations by direct sequencing of PCR products covering exons 2 to 10 and most of the coding sequence of exon 1. These samples included 13 ALL, 17 AML, five biphenotypic, and one undifferentiated leukemia from 10 adults and 23 children. Samples from patients at diagnosis and at subsequent relapse were analyzed in three cases of AML. One sample was from the relapse of a patient included in our earlier study.5
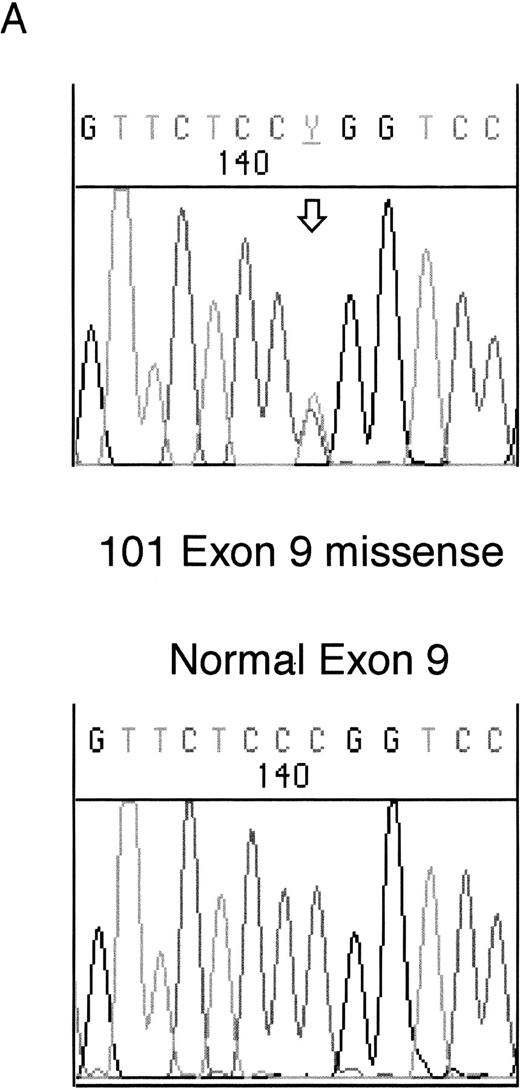
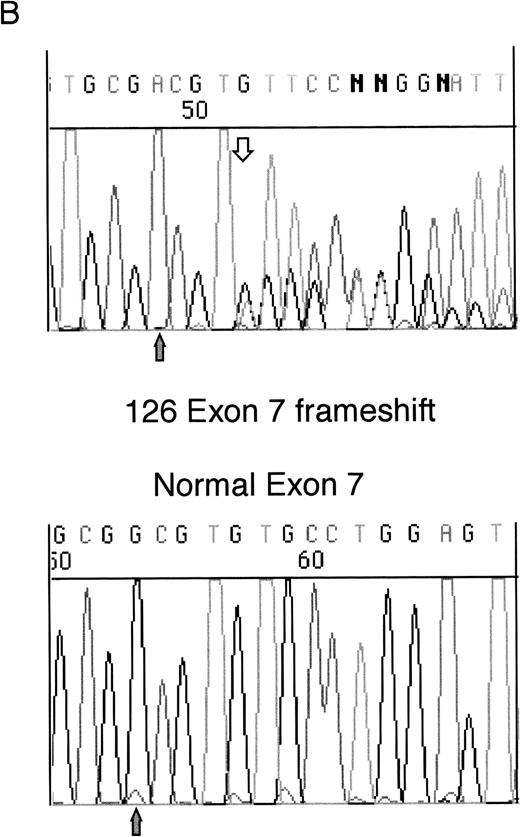
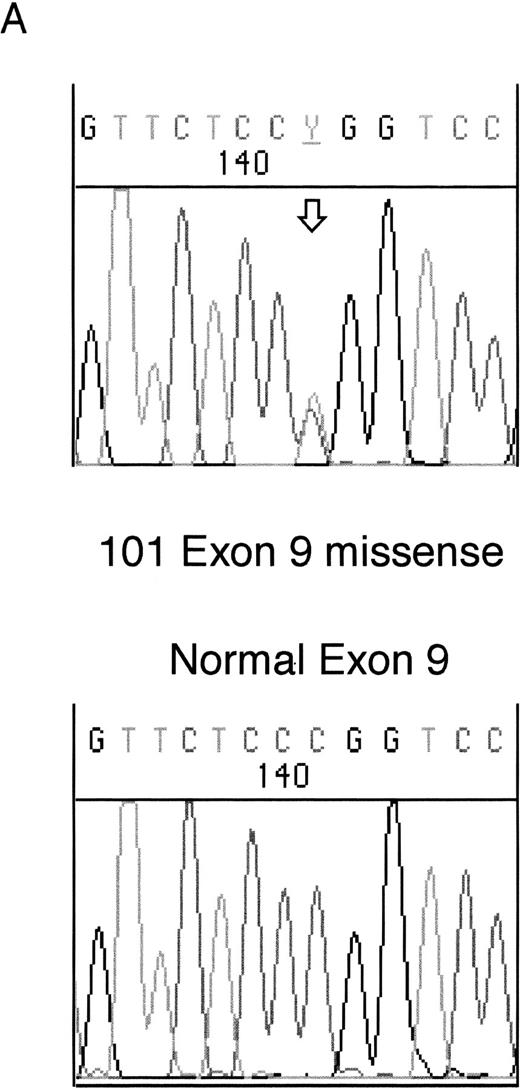
Heterozygous mutations were found in four patient samples (11%); two were frameshifts and two were missense mutations. Examples of each of these types of mutation are shown in Fig 1. None of the patients with samples taken at both diagnosis and relapse had WT1 mutations. All of the samples with mutations consisted of bone marrow or leukopheresed peripheral blood mononuclear cells with 93% to 100% blasts so that contamination by normal DNA was assumed to be negligible.
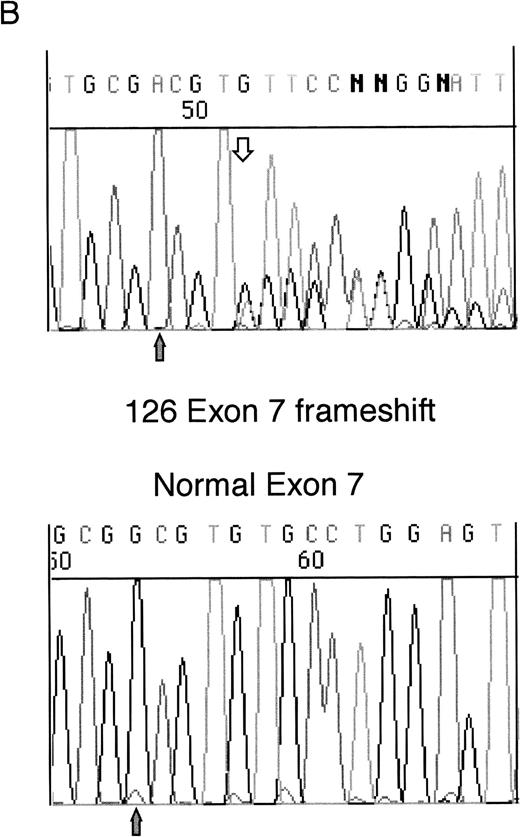
Sequencing of WT1 mutations in leukemias. (A) An example of a heterozygous point mutation (arrow). (B) An example of a heterozygous frameshift mutation in exon 7. The open arrow marks the beginning of the insertion and the shaded arrow indicates the position of the polymorphism.
Sequencing of WT1 mutations in leukemias. (A) An example of a heterozygous point mutation (arrow). (B) An example of a heterozygous frameshift mutation in exon 7. The open arrow marks the beginning of the insertion and the shaded arrow indicates the position of the polymorphism.
A heterozygous frameshift mutation was found in exon 1 in sample 87, due to an insertion of 3 bases and duplication of the preceding 7-bp. The result is a 10-bp insertion after Ala144, leading to the predicted introduction of 38 novel amino acids before a termination codon. This sample also contained a g to a in the intron 87-bp upstream of the beginning of exon 9. This heterozygous base change was not seen in any other samples. Unfortunately, the RNA from this sample was too degraded for analysis of WT1expression so that any effect on splicing could not be ascertained.
Exon 6 contained a heterozygous point mutation in sample 146. This was a missense mutation, Cys282 (TGC) to Arg (CGC). No other base changes were seen in this sample.
Sample 126 contained a heterozygous insertion of a single T in exon 7, after Arg302. This is predicted to produce a sequence encoding 13 novel amino acids after Arg302 followed by a Stop codon, which terminates the protein before the zinc finger region. This insertion is at the same site as a 4-bp insertion we previously reported in a leukemia sample (patient 216).5
The fourth mutation was found in exon 9 in sample 101. This was a heterozygous missense mutation altering Arg394 (CGG) within the third zinc finger to Trp (TGG). This mutation commonly occurs as a germline mutation in DDS and is usually reduced to homozygosity in the Wilms' tumors of DDS patients (see Reddy and Licht1 for references).
A number of samples contained apparent point mutations or frameshifts within the 5′ portion of exon 1 amplified using primers 256 and 532, which were confirmed by sequencing in both directions, but were then absent from a second or third independent PCR product. This occurred when the amplification was performed both in the presence and the absence of a proofreading polymerase, suggesting that this was not caused by typical Taq polymerase errors. A similar phenomenon was sometimes seen for the two polymorphisms present within this PCR product, with the same sample exhibiting heterozygosity or homozygosity on different occasions. This seemed to occur particularly when the sample was difficult to amplify and may be due to arbitrary amplification of only one allele when the available template concentration is low. The extreme GC-richness and repetitive nature of this region may contribute to these phenomena and make it difficult to ascertain WT1 mutations in the 5′ end of exon 1.
RNA analysis.
RNA from all four of these samples was analyzed for WT1 expression by RT-PCR and by Northern blot for sample 146. Insufficient RNA was available for Northern blot analysis for the remaining samples. Samples 101 and 146 were strongly positive for WT1 expression by RT-PCR, but WT1 mRNA was undetectable in 87 and 126, as the RNA was degraded (data not shown). Sample 146 had WT1 mRNA of the normal size on a Northern blot.
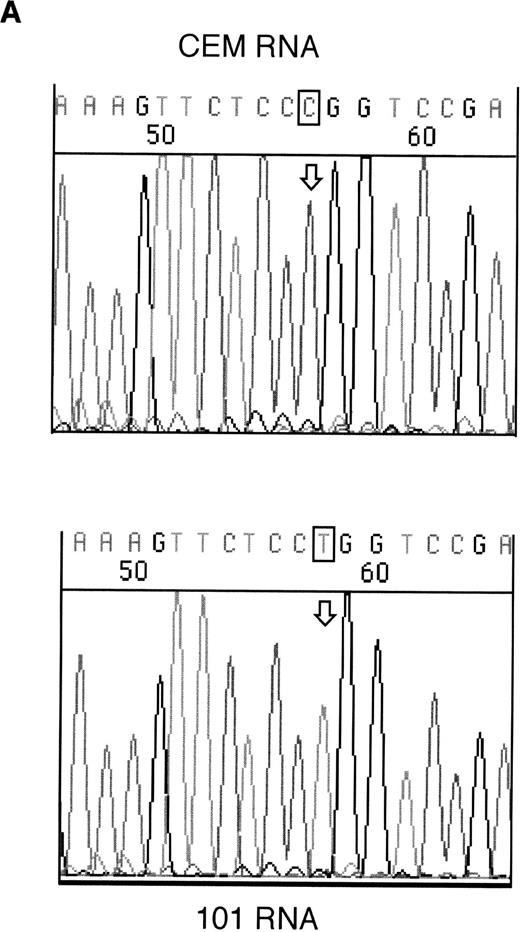
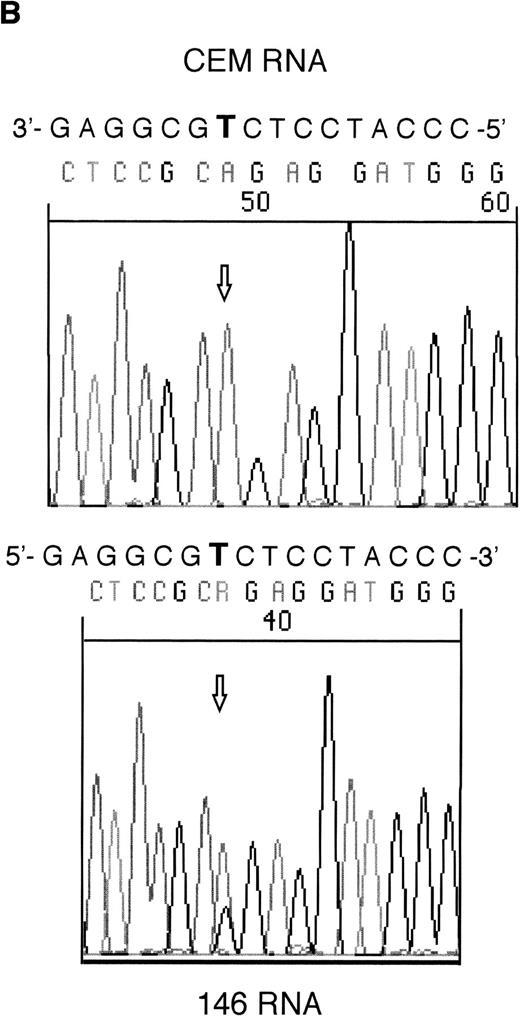
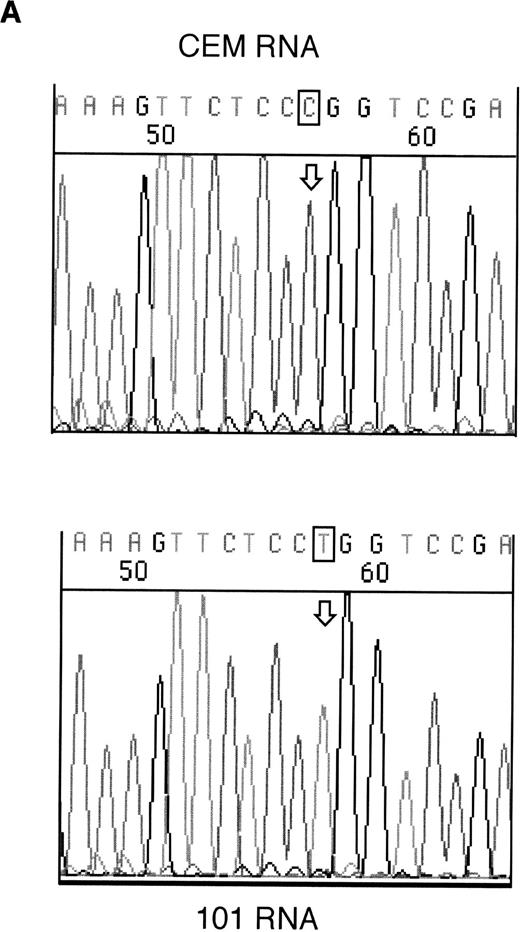
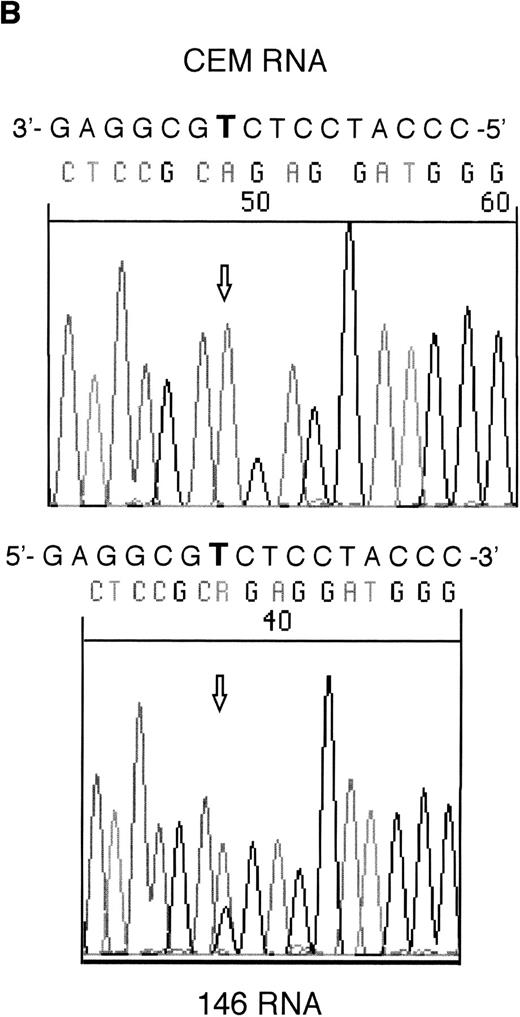
RT-PCR products from samples 146 and 101 were sequenced to assess whether the mutant alleles were expressed. Both the mutant and wild-type alleles could be seen for the exon 6 mutation in sample 146; however, expression of only the mutant allele could be detected in sample 101 (Fig 2).
Sequencing of RT-PCR products from samples withWT1 mutations and normal CEM cells. (A) 101 showing expression of only the mutant allele, T (boxed), compared with the wild-type sequence, (C) in CEM. (B) Reverse sequencing of 146 showing expression of wild-type and mutant alleles. Coding sequence is shown 3′ to 5′ above the reverse sequence, with the mutated T in bold.
Sequencing of RT-PCR products from samples withWT1 mutations and normal CEM cells. (A) 101 showing expression of only the mutant allele, T (boxed), compared with the wild-type sequence, (C) in CEM. (B) Reverse sequencing of 146 showing expression of wild-type and mutant alleles. Coding sequence is shown 3′ to 5′ above the reverse sequence, with the mutated T in bold.
Relationship of WT1 mutation to chemosensitivity.
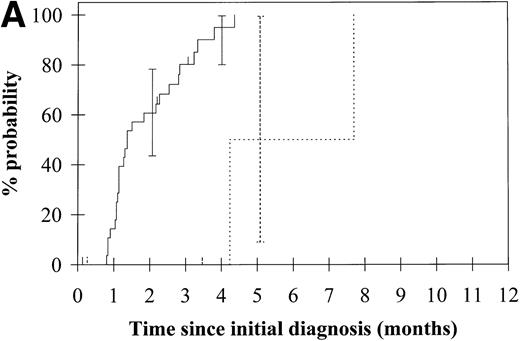
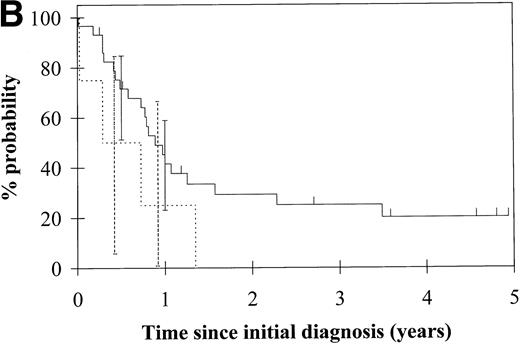
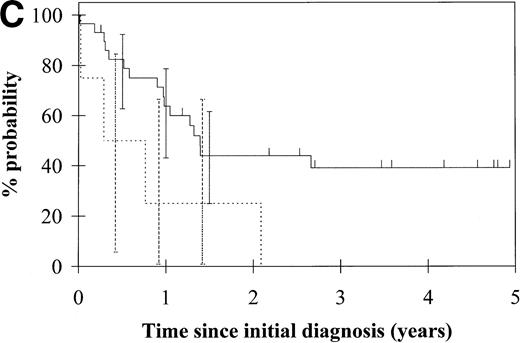
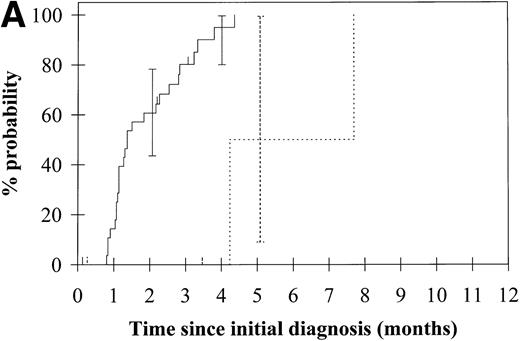
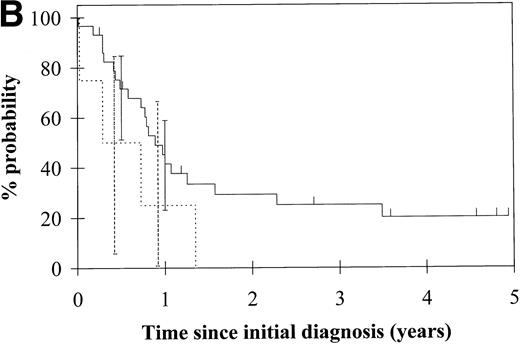
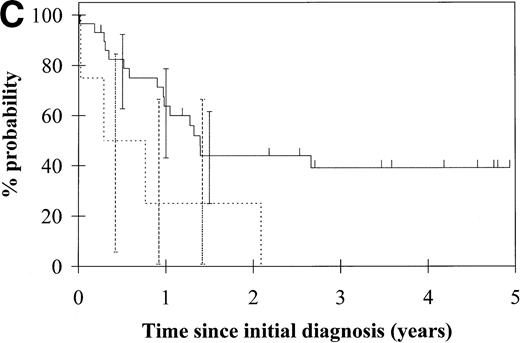
To assess the relationship of WT1 mutation to clinical outcome, we combined data from this and our previous study and confined the analysis to the largest single diagnostic group, AML at first diagnosis (n = 33). None of the four patients with WT1 mutation in this group went into remission with standard induction chemotherapy (Fig 3A). Both disease-free survival and overall survival were significantly worse in those with WT1mutation (Fig 3B and C). Although log-rank comparisons in both probability of first remission and overall survival gave significantP values (.02 and .03, respectively), the power of these calculations (.35 and .15, respectively) is low due to the small group size. We therefore cannot be certain that they are typical.
Probability of (A) first remission, (B) disease-free survival, and (C) overall survival in 33 patients with acute myeloid leukemia. (—) WT1 mutation absent, (…)WT1 mutation present. Ninety-five percent confidence intervals are marked. P values and statistical issues are discussed in the Results.
Probability of (A) first remission, (B) disease-free survival, and (C) overall survival in 33 patients with acute myeloid leukemia. (—) WT1 mutation absent, (…)WT1 mutation present. Ninety-five percent confidence intervals are marked. P values and statistical issues are discussed in the Results.
DISCUSSION
In our earlier study,5 we found WT1 mutations in samples from four patients, three with AML and one with biphenotypic leukemia, all of whom were adults. This second cohort of patients includes two cases of WT1 mutation in childhood leukemia, one of which is a T-ALL, demonstrating that WT1 mutation is not restricted to adults or to the myeloid lineage, although mutations are more frequent in these groups. In total, we have analyzed 67 patients and have found nine WT1 mutations in eight patients (12%) (summarized in Tables 1,2, and 3). This is similar to the proportion of Wilms' tumors which have WT1mutations.15 A previous study of 48 cases of childhood acute leukemia failed to show any with WT1 mutation, but this included only 15 cases of AML.21 This discrepancy may therefore be accounted for by the small sample size and the possibility that WT1 mutation is rarer in childhood than adult AML. A second study of adult leukemias (39 cases of CML, 13 of ALL, and 11 of AML) found a single case of ALL in which WT1 was aberrantly spliced to produce an in-frame fusion of zinc finger 2 onto 4.22 This deletion of zinc finger 3 has been shown previously to produce a WT1 protein with dominant oncogenic properties.23
The effects on WT1 function of the mutations we have found can be predicted by comparison with similar mutations associated with Wilms' tumor. Both of the exon 9 mutations have previously been reported in sporadic Wilms' tumors and as germline mutations in patients with DDS (Arg394 →Trp, 101) and a patient with bilateral Wilms' tumor (Arg390 → Stop, 232M).24Both of these mutations have been shown to abolish DNA binding by WT1.25,26 The mutation at Cys282 in sample 146 is only the second missense mutation to be reported in exon 6; the other was in a case of multicystic mesothelioma.27Cys282 is conserved in WT1 from rat, mouse, alligator, chick, zebrafish,28 and Xenopus,29suggesting that it has functional significance. Indeed, the similar missense mutation in the mesothelioma has been shown to convert WT1 from a transcriptional repressor to an activator. Frameshift mutations in exons 1 and 7, similar to those we have described in leukemias, have also been described in Wilms' tumors.18,30-32 Such truncated forms of WT1 are incapable of DNA binding, but are thought to act in a dominant-negative manner, interfering with the remaining wild-type allele. Indeed WT1 has been shown to self-associate and truncated forms do alter the subnuclear localization of the wild-type protein.33 As few as the first 180 amino acids are capable of interfering with WT1 transcriptional activity.34 The greater frequency of heterozygous mutations in leukemia suggests such dominant or dominant negative effects are of greater significance in hematopoietic cells than in developing kidney cells, implying the existence of hematopoiesis-specific proteins whose interaction with wild-type WT1 can be disrupted by a truncated WT1.
Like those seen in Wilms' tumors, the majority of WT1mutations seen in leukemias cause truncation of the protein; however, in contrast to Wilms' tumor, they are mainly heterozygous. Although apparent heterozygosity could in theory be caused by contaminating normal cells, we do not believe this to be the case here as all samples contained 93% to 100% leukemic blasts. Further evidence for true heterozygosity is provided by analysis of WT1 expression: wild-type mRNA should not come from normal bone marrow contaminants, as these express only very low levels of WT1 compared with leukemic blasts. Two samples (146 and 232M) were definitely heterozygous based on expression of mutant and wild-type WT1 alleles. Two of the samples with mutations appeared to express only the mutant allele (132 and 101). In these cases, mutation of the other allele outside the coding region, affecting either promoter activity or mRNA stability, or other epigenetic effects such as imprinting may contribute to the expression of only one allele. One case, 58, was a compound heterozygote with a different mutation in each WT1 allele, demonstrating that some leukemias conform to the two hit hypothesis.
Our data support the notion that heterozygous WT1 mutations are sufficient to contribute to leukemogenesis. This raises the questions of what is the normal role of WT1 in hematopoiesis and how do mutations cause cellular transformation? In both DDS and WAGR syndromes,WT1 mutations are dominant for genitourinary development, but usually recessive for tumorigenesis. In these and other individuals known to have germline WT1 mutations, hematopoiesis appears to be intact, although the number of such individuals is small and detailed analysis of the hematopoietic system and tolerance of chemotherapy have not been reported. Hence, subtle defects may have gone undetected. Similar to findings in the murine WT1 null model, WT1 appears not to be essential for hematopoiesis in man, as one patient with Wilms' tumor and no obvious hematopoietic defect is reported to have a germline homozygous WT1 missense mutation.35Whatever its role in hematopoiesis, WT1 exhibits functional redundancy in this tissue. However, Wilms' tumor patients do have an increased frequency of leukemias as second primary tumors, some of which may be due to WT1 mutation.17 36
The cell type expressing WT1 in normal bone marrow remains elusive. It appears to be confined to the CD34 positive hematopoietic progenitor cell compartment, but is present at such low levels that it is not detected consistently.7 This implies that WT1 is either expressed at very low levels by several cell types or that its expression is confined to a very infrequent cell. The latter might imply that WT1 expression is a property of the hematopoietic stem cell. The fact that expression persists at high levels in both myeloid and lymphoid acute leukemias, as well as our demonstration of WT1mutations in leukemias of both lineages, would support this view. Indeed, recently it has been shown that AML derives from a very primitive cell, close to the hematopoietic stem cell.37 It seems likely that WT1 will play a role in control of differentiation at this early stage. One conundrum is whether the high levels of WT1 expression seen in leukemias represent ectopic expression or simply reflect the arrested differentiation stage of the equivalent normal progenitor, as seems to be the case in Wilms' tumors. If we accept that WT1 is normally expressed by an infrequent hematopoietic progenitor cell, then it is more likely that leukemic expression of WT1 reflects cellular origin rather than aberrant expression.
Although the numbers are small, it appears that in primary AML, the presence of WT1 mutation is associated with a failure to respond satisfactorily to standard induction chemotherapy. Such drug resistance is compatible with WT1 having a role in cell cycle check points and apoptopic responses to cytotoxic agents. Several recent experiments have shown that in vitro modulation of WT1 expression can either inhibit or stimulate apoptosis depending on the isoforms used and the cell type, and that exon 5-containing isoforms appear to cause cell cycle arrest.1,38-40 Antisense experiments suggest that WT1 is necessary for continued proliferation and protection from apoptosis of some leukemic cell lines.41,42 The various isoforms also have differential effects on differentiation.43 We have shown that alternative splicing of the WT1 gene is controlled in a cell type-specific manner, with hematopoietic cells and acute leukemias having an excess of + exon 5 isoforms compared with fetal kidney cells.14 TheWT1 gene is also known to contain enhancer sequences, which are only active in hematopoietic cells. Because WT1 function appears to depend critically on the intracellular milieu and the relative isoform presence, it is perhaps not surprising that the type of mutation (heterozygous or homozygous) differs between leukemia and Wilms' tumor. We speculate that high levels of WT1 expression in acute leukemia promote proliferation and protect from apoptosis. Mutant WT1 may have the same effect, not through loss of function mutations as seen in Wilms' tumor, but rather through protein-protein interactions of the truncated protein, which are specific to hematopoietic cells. These could act either to alter the function of the remaining wild-type WT1 or by binding to novel protein targets. Evidence to support these hypotheses requires knowledge of the regulatory pathways which involve WT1, and how these are affected by alterations in isoform ratios; as yet, in vitro cotransfection studies have not convincingly identified any target genes regulated by physiologically relevant levels of WT1 and further studies are required.
In conclusion, we have shown that WT1 mutations occur in both myeloid and lymphoid acute leukemias and suggest that the action of mutant WT1 may be different in hematopoietic versus nephrogenic cells. The question of whether WT1 mutation is involved in leukemic initiation or progression remains unanswered. Clearly other genetic events contribute to the leukemogenic phenotype as shown by the additional cytogenetic changes in several of our cases. A greater understanding of how WT1 contributes to leukemogenesis should lead to its rational use as a panleukemic target for therapy.
ACKNOWLEDGMENT
We thank Drs J. Treleaven, R. Powles, S, Meller, S. Height, and Prof C.R. Pinkerton for access to patient samples. We also thank Dr Clive Horton, Department of Computing and Information, Royal Marsden NHS Trust, Sutton, for the Kaplan-Meier curves and analyses, and Carolanne Brown for excellent sequencing assistance supported by Breakthrough Breast Cancer.
K.P.-J. and L.K.-U. are supported by the Cancer Research Campaign and the Royal Marsden Children's Cancer Unit Fund.
Address reprint requests to K. Pritchard-Jones, MD, Institute of Cancer Research, 15 Cotswold Rd, Belmont, Sutton, Surrey SM2 5NG, UK; e-mail: kpj@icr.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.