Abstract
To overcome the low efficiency of gene transfer into hematopoietic cells, we developed a novel system for selective expansion of transduced cells. To this end, we constructed a chimeric cDNA (GCRER) encoding the fusion protein between the granulocyte colony-stimulating factor receptor (G-CSFR) and the hormone-binding domain (HBD) of the estrogen receptor (ER) as a selective amplifier gene. Use of the intracellular signaling pathway of G-CSFR was considered to be appropriate, because G-CSF has the ability not only to stimulate the neutrophil production, but also to expand the hematopoietic stem/progenitor cell pool in vivo. To activate the exogenous G-CSFR signal domain selectively, the estrogen/ER-HBD system was used as a molecular switch in this study. When the GCRER gene was expressed in the interleukin-3 (IL-3)–dependent murine cell line, Ba/F3, the cells showed IL-3–independent growth in response to G-CSF or estrogen. Moreover, the Ba/F3 cells transfected with the Δ(5-195)GCRER, whose product lacks the extracellular G-CSF–binding domain, did not respond to G-CSF, but retained the ability for estrogen-dependent growth. Further, murine bone marrow cells transduced with the GCRER or Δ(5-195)GCRER gene with retroviral vectors formed a significant number of colonies in response to estrogen, as well as G-CSF, whereas estrogen did not stimulate colony formation by untransduced murine bone marrow cells. It is noteworthy that erythroid colonies were apparently formed by the bone marrow cells transduced with the GCRER gene in the presence of estrogen without the addition of erythropoietin, suggesting that the signals from the G-CSFR portion of the chimeric molecules do not preferentially induce neutrophilic differentiation, but just promote the differentiation depending on the nature of the target cells. We speculate that when the selective amplifier genes are expressed in the primitive hematopoietic stem cells, the growth signal predominates and that the population of transduced stem cells expands upon estrogen treatment, even if some of the cells enter the differentiation pathway. The present study suggests that this strategy is applicable to the in vivo selective expansion of transduced hematopoietic stem cells.
HEMATOPOIETIC STEM CELLS are ideal targets for gene therapy of certain heritable and acquired hematologic/immune diseases, because they have the ability to self-renew and the transferred gene is expected to remain in the genome for the lifespan of the patient.1 Although retroviral vectors are widely used to deliver genes into hematopoietic stem cells, the transduction efficiency has been too low to obtain the expression level required for improvement of clinical manifestations.2 There are several approaches to this problem. Developing a vector with increased transduction efficiency or the improvement of transduction methods is one way of dealing with it.3 Second, selection and enrichment of transduced hematopoietic stem cells ex vivo may be a feasible approach. For example, by delivering a selectable marker gene such as a cDNA for a cell-surface antigen4-7 or a drug-resistant gene together with a therapeutic gene into target cells, the transduced cells can be purified with a fluorescence-activated cell sorter (FACS) or by culturing in the presence of a selectable toxic drug. However, such methods need a large number of target cells to obtain a sufficient number of transduced cells. The third strategy is to give the transduced cells a growth or survival advantage in vivo for selective expansion. In the case of adenosine deaminase (ADA) deficiency8,9 or Fanconi anemia (FA),10 correction of the patient hematopoietic cells with the normal gene imparts a survival advantage to transduced cells. However, since in most other disorders therapeutic genes do not have such characteristics, cotransduction with some other selectable marker gene would be required. Multidrug-resistance gene-1 (mdr-1) is considered a possible candidate. Hematopoietic stem cells are transduced with the mdr-1 gene for chemoprotective gene therapy in cancer cases in which bone marrow toxicity of chemotherapeutic drugs is a dose-limiting side effect,11,12 and phase I clinical trials have already been initiated after obtaining the approval of the Recombinant DNA Advisory Committee (RAC) in the United States. The mdr-1 gene may also be applicable to the in vivo selection of transduced hematopoietic stem cells. It was reported that the administration of taxol to mice transplanted with mdr-1–transduced bone marrow cells resulted in the enrichment of the transduced cells.13 However, this method is not suitable for nonmalignant diseases, because it requires the administration of anticancer drugs to the patients.
With the aim of developing a safer and more generally applicable method for selective expansion of transduced hematopoietic stem cells in vivo, we explored the possibility of using a growth signal from exogenous cytokine receptor to stimulate growth of transduced hematopoietic cells. Several cytokine receptors are considered to be involved in the growth stimulation of primitive hematopoietic stem/progenitor cells, including granulocyte colony-stimulating factor receptor (G-CSFR),14 c-kit receptor,15,16 and FLT3/FLK2.17,18 Among these receptors, G-CSFR seems to be the most appropriate as a tool for expansion of transduced stem cells in clinical trials, because recombinant human G-CSF (rhG-CSF) has already been widely used in clinical settings without any untoward side effects and a large amount of information on the G-CSF/G-CSFR system is currently available.19,20 The intracellular signal from G-CSFR is believed to be triggered through dimerization of the receptor molecules which is caused by G-CSF binding.21 22 In the present study, to activate the exogenously expressed G-CSFR selectively without using a natural ligand (G-CSF), we fused the G-CSFR molecule to the hormone-binding domain (HBD) of estrogen receptor (ER); that is, a chimeric gene (GCRER) between G-CSFR cDNA and the ER-HBD cDNA was constructed, whose product was expected to be dimerized upon estrogen treatment and to transmit the signal from the intracellular domain of fused G-CSFR to the nucleus. We introduced this chimeric gene into an interleukin-3 (IL-3)–dependent cell line or murine bone marrow cells to examine whether it can be used as a selective amplifier gene for the purpose of expanding the transduced hematopoietic cells.
MATERIALS AND METHODS
Cell lines. The IL-3–dependent mouse pro-B cell line, Ba/F3 (kindly provided by Dr J. Ihle, St Jude Children's Research Hospital, Memphis, TN), was maintained in RPMI 1640 medium (GIBCO-BRL, Grand Island, NY) supplemented with 10% fetal calf serum (FCS; Bioserum, Victoria, Australia), 1% penicillin/streptomycin (Irvive Scientific, Santa Ana, CA), and 10 U/mL recombinant mouse IL-3 (rmIL-3; the culture supernatant of C3H10T1/2 cells transfected with the mouse IL-3 expression plasmid). The ecotropic packaging cell lines, GP + E-86 (kindly provided by Dr A.D. Miller, Fred Hutchinson Cancer Research Center, Seattle, WA) and BOSC23 (ATCC CRL 11554), were maintained in Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL) containing 10% FCS and 1% penicillin/streptomycin.
Plasmid construction. All enzymes used, except for PmeI, were purchased from Takara Shuzo (Kyoto, Japan). A mammalian expression vector pCMX-MfasER23 (kindly provided by Dr A. Kakizuka, Kyoto University, Kyoto, Japan), which contains the sequence encoding the HBD of rat ER (315 amino acids of the carboxyl terminus) between BamHI and EcoRI sites, was digested with BamHI, treated with Klenow fragment of Escherichia coli DNA polymerase I, and then ligated with a Pme I linker (GTTTAAAC; New England Biolabs Inc, Beverly, MA). After digestion with Pme I (New England Biolabs) and HindIII, the Pme I(blunt)-HindIII fragment containing the CMV promoter, ER-HBD, and the vector portion was separated by agarose gel electrophoresis and electroelution. On the other hand, the plasmid pBluescript-GCR (kindly provided by Dr S. Nagata, Osaka University, Osaka, Japan) carrying the cDNA for mouse G-CSFR between the HindIII and Xba I sites was digested with Xba I (at nt 2689), treated with mung bean nuclease, and then digested with HindIII (at nt 162) to yield a 2,527-bp HindIII-Xba I(blunt) fragment containing the G-CSFR cDNA. This fragment was ligated with the Pme I(blunt)-HindIII fragment to obtain pCMX-GCRER. The construct was confirmed by sequence analysis.
pCMX-Δ(5-195)GCRER was constructed from pCMX-GCRER by deleting the sequence corresponding to the G-CSF–binding domain.24 In brief, pCMX-GCRER was digested with HindIII (at nt 162) and Kpn I (at nt 1829), and the two fragments (6,338 bp and 1,667 bp) were separated by agarose gel electrophoresis and electroelution. The 1,667-bp HindIII-Kpn I fragment was further digested with Taq I (at nt 265 and nt 838), and then the HindIII-Taq I fragment (103 bp) and the Taq I-Kpn I fragment (991 bp) were separated and ligated with the 6,338-bp Kpn I-HindIII fragment to obtain pCMX-Δ(5-195)GCRER. As a negative control, pCMX-Δ(5-195,324-597,702-812)GCRER, whose product grossly lacks both of the extracellular domain and the intracellular domain of G-CSFR, was also constructed. After digestion of pCMX-Δ(5-195)GCRER with HindIII and Pme I, the Pme I(blunt)-HindIII fragment (6,175 bp) containing the CMV promotor, ER-HBD, and the vector portion was separated by agarose gel electrophoresis. To obtain the 5′ insert fragment, pCMX-Δ(5-195)GCRER was digested with HindIII (at 162) and Kpn I (at 1256), and the HindIII-Kpn I fragment (1,094 bp) was further digested with Xho II (at 650). The HindIII-Xho II fragment (488 bp) containing a part of extracellular domain, transmembrane domain, and 75 amino acids of juxtamembrane part of the intracellular domain of G-CSFR cDNA was then separated. On the other hand, pCMX-Δ(5-195)GCRER was digested with HindIII (at 162) and EcoRI (at 740, 1075, and 3168). The EcoRI-EcoRI fragment (2,093 bp) was separated by agarose gel electrophoresis and further digested with Stu I (at 1451 and 1783). The Stu I(blunt)-Stu I(blunt) fragment (332 bp) was ligated with a Sma I linker (CCCGGG; Takara Shuzo). After digestion with Sma I and Xho II, the Xho II-Sma I(blunt) fragment (314 bp) was separated. Finally, the three fragments described [Pme I(blunt)-HindIII, HindIII-Xho II, Xho II-Sma I(blunt)] were ligated to obtain pCMX-Δ(5-195,324-597,702-812)GCRER. The products of pCMX-GCRER, -p C M X - ;gD ( 5 - 1 9 5 ) G C R E R , a n d p C M X - ;gD ( 5 - 1 9 5 , 3 2 4 - 5 9 7 , 7 0 2 -812)GCRER are illustrated in Fig 1. Furthermore, the plasmid pCMX-GCR carrying the cDNA of wild-type G-CSFR was constructed as follows. The plasmid pCMX-MfasER was digested with EcoRI, treated with Klenow fragment of E coli DNA polymerase I, and then digested with HindIII. The plasmid vector portion was obtained by electroelution after agarose gel electrophoresis and then ligated with the HindIII-Xba I(blunt) fragment of pBluescript-GCR which contains the cDNA of wild-type G-CSFR.
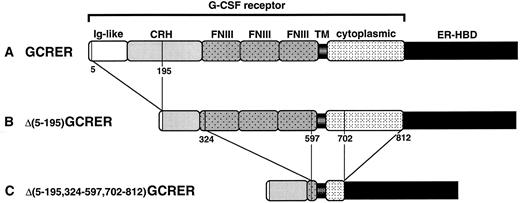
Schematic diagram of the chimeric molecules: (A) GCRER, (B) Δ(5-195)GCRER, and (C) Δ(5-195,324-597,702-812)GCRER. The amino acid numbers at the boundaries of deleted portions in G-CSFR are indicated. CRH, cytokine receptor-homologous domain; FNIII, fibronectin type III domain; TM, transmembrane domain.
Schematic diagram of the chimeric molecules: (A) GCRER, (B) Δ(5-195)GCRER, and (C) Δ(5-195,324-597,702-812)GCRER. The amino acid numbers at the boundaries of deleted portions in G-CSFR are indicated. CRH, cytokine receptor-homologous domain; FNIII, fibronectin type III domain; TM, transmembrane domain.
Transfection of Ba/F3 cells. IL-3–dependent Ba/F3 cells were transfected with plasmid DNA by electroporation using a Gene Pulser apparatus (BioRad Laboratories, Richmond, CA). After washing with cold phosphate-buffered saline (PBS) twice and with OPTI-minimum essential medium I (OPTI-MEMI; GIBCO-BRL) once, 1 × 107 cells were suspended in 0.2 mL of OPTI-MEMI. Ten micrograms of plasmid DNA [pCMX-GCRER, pCMX-Δ(5-195)GCRER, pCMX-Δ(5-195,324-597,702-812)GCRER, or pCMX-GCR] and 1 μg of a plasmid carrying the brasticidin S-resistant gene (bsr) as a selectable marker gene were added to the cell suspension and then incubated on ice for 10 minutes. The cell mixture was exposed to a 200-V pulse with a capacitance of 960 μF, and returned to ice. After incubation on ice for 5 minutes, cells were diluted with 10 mL of RPMI 1640/10% FCS containing 10 U/mL rmIL-3 and cultivated in a 10-cm dish (Falcon 3003; Becton Dickinson, Lincoln Park, NJ). After 24 hours of incubation, the transfected cells were divided into 24-well plates (Falcon 3047; 5 × 105 cells/well). For selection, blasticidin S hydrochloride (Funakoshi, Tokyo, Japan) was added to the culture medium at a final concentration of 10 μg/mL. After an appropriate period, the number of wells in which blasticidin S-resistant cells appeared was scored. The blasticidin S-resistant cells in each well were divided into three wells, in two of which the cells were cultured with or without 10−7 mol/L β-estradiol (Sigma, St Louis, MO) in the absence of rmIL-3. The number of wells in which β-estradiol–dependent cell growth appeared was scored. Cloning of the β-estradiol–responsive Ba/F3 cells was performed by limiting dilution.
Western blot analysis. Rabbit polyclonal immunoglobulin G (IgG; Santa Cruz Biotechnology, Santa Cruz, CA), which recognizes the epitope corresponding to amino acids 580 to 599 at the carboxyl terminus of the ER of mouse and rat origin, and rabbit polyclonal IgG (Santa Cruz Biotechnology), which recognizes the epitope corresponding to amino acids 793 to 812 at the carboxyl terminus of G-CSFR of mouse and rat origin, were used for immunoblotting. Approximately 1 × 107 untreated or transfected Ba/F3 cells were dissolved in 200 μL of lysing buffer containing 50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L NaF, 1% NP-40, 1% aprotinin, and 1 mmol/L phenylmethylsulfonyl fluoride. After rotating at 4°C for 20 minutes, the cell lysates were centrifuged at 12,000g for 10 minutes and the aliquots of supernatants containing 30 μg of protein were mixed with 2× Laemmli sample buffer (120 mmol/L Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.004% bromophenol blue [BPB], 10% 2-mercaptoethanol). After boiling at 100°C for 3 minutes, the samples were electrophoresed on a 7.5% SDS polyacrylamide gel, and then electroblotted to a polyvinylidene fluoride (PVDF) membrane (Nihon Millipore, Yamagata, Japan). The blotted membrane was rinsed with TBST (10 mmol/L Tris-HCl [pH 8.0], 150 mmol/L NaCl, 0.05% Tween-20) and blocked in TBST containing 4% bovine serum albumin (BSA; Boehringer Mannheim, Mannheim, Germany) at room temperature for 60 minutes, washed with TBST twice, and incubated with 10 mL of TBST, which contains 1 μg of antibody (either for G-CSFR or ER), for 60 minutes at room temperature. The membrane was then washed with TBST four times and incubated with 10 mL of TBST containing 1.3 μL of 100 μg/mL of alkaline phosphatase–conjugated second antibody [anti-rabbit IgG(Fc), affinity-purified from goat antisera; Promega, Madison, WI] for 60 minutes at room temperature. After washing with TBS (10 mmol/L Tris-HCl [pH 8.0], 150 mmol/L NaCl) once, the membrane was incubated in 10 mL of alkaline phosphatase buffer (50 mmol/L Tris-HCl [pH 9.5], 100 mmol/L NaCl, 5 mmol/L MgCl2 ) containing 66 μL of nitroblue tetrazolium (NBT; Promega) and 33 μL of BCIP (Promega) as the color development substrate for appropriate time. The membrane was washed with water and subsequently air-dried.
Short-term cell proliferation assay. A quantity of 4 × 103 untreated or transfected Ba/F3 cells in 100 μL of RPMI 1640, 10% FCS, and 1% penicillin/streptomycin was cultured in the presence or absence of 10 U/mL of rmIL-3, 1 nmol/L (20 ng/mL) of rhG-CSF (kindly provided by Chugai Pharmaceutical, Tokyo, Japan), or various concentrations of β-estradiol (Sigma) in 96-well microtiter plates (Falcon 3072). Cell proliferation assay was periodically performed using Cell Proliferation Kit II (XTT; Boehringer Mannheim) essentially according to the manufacturer's instructions. In brief, 50 μL per well of XTT-labeling mixture was added. After the incubation at 37°C for 4 hours, the spectrophotometrical absorbance was measured at the wave length of 490 nm and 620 nm.
Long-term cell proliferation assay. A representative clone of Ba/F3 cells transfected with pCMX-Δ(5-195)GCRER was cultured in the presence of 10 U/mL of rmIL-3 or 10−7 mol/L of β-estradiol. The rmIL-3– and β-estradiol–containing cultures were diluted to the cell concentration of 5,000 cells/mL every 3 and 4 days, respectively. Twenty-one days later, the cells in the β-estradiol–containing cultures were washed three times with PBS and further cultured in the presence or absence of β-estradiol. XTT assays were performed as described earlier.
Retrovirus vector production. Retrovirus vector plasmids, pMX-GCRER and pMX-Δ(5-195)GCRER, were constructed by inserting the cDNA for GCRER or Δ(5-195)GCRER into the multicloning site of pMX, a Molony murine leukemia virus (MoMLV)-based retrovirus vector generated from pBabe-X by replacing 5′LTR and the packaging signal with those of the MFG vector.25 Virus producer-cell lines were made by cotransfecting the ecotropic packaging cell line, GP + E-86, with pMX-GCRER or pMX-Δ(5-195)GCRER and the bsr expression plasmid by the standard calcium phosphate coprecipitation method. After 48 hours, the transfected cells were selected in DMEM containing 10% FCS, 1% penicillin/streptomycin, and 10 μg/mL blasticidin S hydrochloride. Approximately 20 to 30 blasticidin S-resistant colonies were picked up and the supernatant of each colony was examined by RNA dot blot assay. In brief, the viral particles in the supernatant were precipitated with polyethylene glycol, and then viral RNA was extracted after treatment with lysis buffer (0.5% SDS, 0.6 mol/L NaCl, 10 mmol/L EDTA, 10 mmol/L Tris-HCl [pH 7.4]). The RNA samples were blotted to the nylon membrane (Hybond-N+; Amersham Life Science, Buckinghamshire, England) using 96-well dot blot apparatus (Micro-Sample Filtration Manifold; Schleicher & Schuell, Keene, NH). The membranes were hybridized with the 32P-labeled cDNA probe, which encodes ER, and the viral titer of each sample was roughly estimated by comparing the dot intensity with those of the standard samples. The clone with the highest viral titer (107-8 particles/mL) was selected as a vector producer cell line. At 80% confluence, the medium of vector producer cells was replaced with Iscove's modified Dulbecco's medium (IMDM; GIBCO-BRL), 20% FCS (Bio Whittaker, Walkersville, MD), and 1% penicillin/streptomycin. After 24 hours, viral supernatant was collected and filtered through 0.45-μm filters and either used fresh or frozen at −80°C. The LacZ expression retrovirus vector was produced as a control vector by transient transfection of the BOSC23 ecotropic packaging cell line with pGK LacZ (obtained from RIKEN DNA BANK, Ibaraki, Japan). The viral supernatant was similarly treated as described earlier.
Transduction of murine bone marrow cells. Six-week-old C57BL/6Njcl mice (Saitama Experimental Animal Center, Saitama, Japan) were injected with 150 mg/kg 5-fluorouracil (5-FU; Wako Pure Chemical Industries, Osaka, Japan) intraperitoneally, and 2 days later, the bone marrow cells were flushed from femoral shafts with IMDM containing 5% FCS. The bone marrow mononuclear (BMMN) cells were collected by density centrifugation using Lympholyte-M (Cedarlane, Ontario, Canada). Approximately 5 × 106/mL BMMN cells were prestimulated with 100 ng/mL of recombinant rat SCF (rrSCF; kindly provided by Amgen, Inc, Thousand Oaks, CA) and 100 U/mL of recombinant human IL-6 (rhIL-6; kindly provided by Ajinomoto, Yokohama, Japan) in IMDM/20% FCS at 37°C for 48 hours in a humidified atmosphere of 5% CO2 in air. Subsequently, the cells were resuspended in 2 mL of viral supernatant containing rrSCF and rhIL-6 at a concentration of 5 × 105 cells/mL, transferred to the six-well plates (Falcon 1146) precoated with 20 μg/cm2 of human fibronectin fragment (CH-296; kindly provided by Takara Shuzo, Otsu, Japan),3 and then incubated at 37°C in a humidified atmosphere of 5% CO2 in air for 72 hours. During this transduction period, viral supernatant was changed six times. As an untransduced control, BMMN cells were similarly treated without using viral supernatant.
In vitro clonogenic progenitor assay. The transduced and untransduced BMMN cells were harvested, and 1 × 105 cells each were plated in 35-mm dish (Falcon 1008) with 1 mL of culture medium containing IMDM, 1.2% methylcellulose (1500 cp; Wako), 20% FCS, 1% deionized BSA (Sigma), and 1 × 10−4 mol/L 2-mercaptoethanol in the presence or absence of 10 ng/mL of rhG-CSF or 10−7 mol/L β-estradiol. In some experiments, 100 ng/mL of rrSCF, 100 U/mL of rhIL-6, 10 ng/mL of rhG-CSF, and 2 U/mL of recombinant human erythropoietin (rhEpo; Chugai Pharmaceutical) were added to the cultures. After 10 days of incubation at 37°C in a humidified atmosphere of 5% CO2 in air, colonies were scored using an inverted microscope. Colony type was determined by the morphology of colony-constituting cells, and erythroid colonies were identified by their reddish color due to hemoglobinization.
Polymerase chain reaction analysis of colonies for transgenes. Colonies in methylcellulose cultures were harvested using a P-10 micropipet (Gilson Medical Electronics, Villiers-le-bel, France) under an inverted microscope, and were suspended in a 100 μL of cell lysing solution consisting of 1 × TNE (100 mmol/L Tris-HCl [pH 7.4], 1 mol/L NaCl, 10 mmol/L EDTA), 0.5% SDS, and 0.1 mg/mL of proteinase K (Boehringer Mannheim). Samples were incubated for 2 hours at 60°C followed by a 10-minute incubation at 95°C to inactivate proteinase K. DNA was then phenol-extracted, ethanol-precipitated, and resuspended in 50 μL of water. Polymerase chain reaction (PCR) was performed to amplify a 476-bp fragment containing the fusion site of GCRER using a proximal primer (TCCAGCGTGCC ATCAATCAC) and a distal primer (GCAGCTCTCATGTCTCCTGA) at the portion of G-CSFR and ER-HBD, respectively. To check the PCR performance, mouse endogenous β-actin genomic DNA fragment was amplified using a proximal primer (GTGGGCCGCTCTAGGCACCAA) and a distal primer (TTCTACAATGAGCTGCGTGT). Ten microliters each of DNA samples was mixed with the same volume of 2× PCR buffer. The final concentration of the PCR buffer was as follows: 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2 , 200 μmol/L, dNTPs, 500 nmol/L proximal primer, 500 nmol/L distal primer, and 1.0 U/40 μL reaction mixture of recombinant Taq DNA polymerase (TaKaRa Ex Taq; Takara Shuzo). PCR amplification was conducted using the TaKaRa PCR thermal cycler MP. The 40-cycle PCR (60 seconds denaturation at 94°C, 60 seconds annealing at 55°C, and 120 seconds extension at 72°C) was finished with a final 5-minute extension at 72°C and a 4°C soak. PCR fragments were separated on a 2% agarose gel electrophoresis and detected by ethidium bromide staining.
RESULTS
Transfection of Ba/F3 cells with the chimeric genes. When the Ba/F3 cells were cotransfected with the pCMX-GCRER and the bsr expression plasmid, the blasticidin S-resistant cell growth was observed in 11 out of 23 wells in the presence of IL-3. In seven of these 11 wells, the transfected Ba/F3 cells proliferated in response to β-estradiol in the absence of rmIL-3. As for the pCMX-Δ(5-195)GCRER plasmid, the blasticidin S-resistant Ba/F3 cells appeared in three of 11 wells, and all of them responded to β-estradiol treatment. Three estradiol-responsive clones were selected by limiting dilution from each transfectants, and used for further analyses. On the other hand, when Ba/F3 cells were cotransfected with pCMX-Δ(5-195,324-597,702-812) GCRER and the bsr expression plasmid, the blasticidin S-resistant cell growth was observed in 12 out of 24 wells, but none of them responded to β-estradiol treatment.
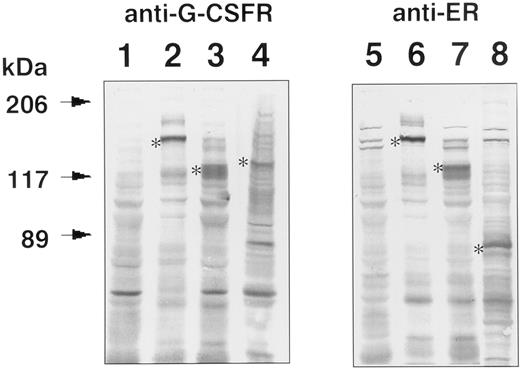
Expression of GCRER, Δ(5-195)GCRER, Δ(5-195,324-597,702-812)GCRER, or wild-type GCR in transfected Ba/F3 cells. The transgene expression was analyzed by Western blotting using anti–G-CSFR or anti-ER antibody (Fig 2). Parental Ba/F3 cells were negative for G-CSFR and ER (lanes 1 and 5), while the chimeric protein, derived from GCRER or Δ(5-195)GCRER gene, was detected in transfected Ba/F3 cells by both antibodies. The molecular size of the chimeric protein, GCRER, estimated from the SDS polyacrylamide gel, was approximately 140 kD (lanes 2 and 6). The size decreased to approximately 120 kD (lanes 3 and 7) when 191 amino acids were deleted from the extracellular domain [Δ(5-195)GCRER]. The band of wild-type mouse G-CSFR expressed in transfected Ba/F3 cells (lane 4) was compatible with the reported molecular size (125 to 135 kD) with N-glycosylation of the extracellular domain.24 As for the control plasmid pCMX-Δ(5-195,324-597,702-812) GCRER, the transgene product was examined using anti-ER antibody, and the molecular size of the chimeric protein was approximately 80 kD (lane 8).
Western blot analysis for GCRER, Δ(5-195)GCRER, Δ(5-195,324-597,702-812)GCRER, and GCR expression in the transfected Ba/F3 cells. Cell lysates of parent Ba/F3 cells (lanes 1 and 5) and the Ba/F3 cells transfected with pCMX-GCRER (lanes 2 and 6), pCMX-Δ(5-195)GCRER (lanes 3 and 7), pCMX-Δ(5-195,324-597,702-812)GCRER (lane 8), and pCMX-GCR (lane 4) were size-fractionated in SDS polyacrylamide gels, and electroblotted to PVDF membranes. Chimeric molecules were detected using anti–G-CSFR antibody (lanes 1, 2, 3, and 4) or anti-ER antibody (lanes 5, 6, 7, and 8). The bands were visualized using alkaline phosphatase–conjugated second antibody and 5-bromo-4-chloro-3-indoyl-phosphate (BCIP)/NBT color development substrate. *Transgene products.
Western blot analysis for GCRER, Δ(5-195)GCRER, Δ(5-195,324-597,702-812)GCRER, and GCR expression in the transfected Ba/F3 cells. Cell lysates of parent Ba/F3 cells (lanes 1 and 5) and the Ba/F3 cells transfected with pCMX-GCRER (lanes 2 and 6), pCMX-Δ(5-195)GCRER (lanes 3 and 7), pCMX-Δ(5-195,324-597,702-812)GCRER (lane 8), and pCMX-GCR (lane 4) were size-fractionated in SDS polyacrylamide gels, and electroblotted to PVDF membranes. Chimeric molecules were detected using anti–G-CSFR antibody (lanes 1, 2, 3, and 4) or anti-ER antibody (lanes 5, 6, 7, and 8). The bands were visualized using alkaline phosphatase–conjugated second antibody and 5-bromo-4-chloro-3-indoyl-phosphate (BCIP)/NBT color development substrate. *Transgene products.
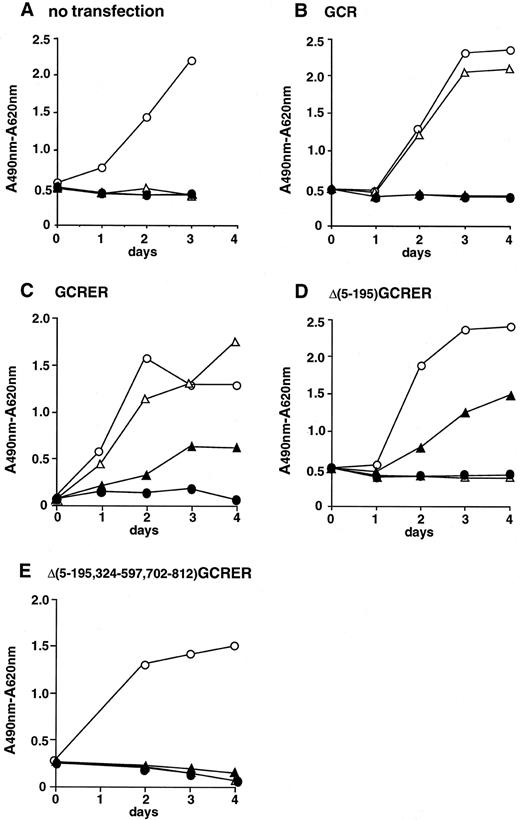
Cell proliferation assay of Ba/F3 cells transfected with the chimeric genes. rhG-CSF or β-estradiol did not stimulate the growth of parental Ba/F3 cells in the absence of rmIL-3 (Fig 3A). The Ba/F3 cells transfected with mouse G-CSFR expression plasmid, pCMX-GCR, proliferated in response to rhG-CSF, but not to β-estradiol (Fig 3B). On the other hand, the Ba/F3 cells transfected with pCMX-GCRER acquired the ability to respond not only to rhG-CSF, but also to β-estradiol in the absence of rmIL-3, although the growth-stimulating activity of β-estradiol was weaker than that of rhG-CSF (Fig 3C). In the case of pCMX-Δ(5-195)GCRER, whose product lacks the extracellular G-CSF binding domain, the transfected Ba/F3 cells did not respond to rhG-CSF, but retained the ability to respond to β-estradiol (Fig 3D). Furthermore, the Ba/F3 cells transfected with the Δ(5-195)GCRER gene proliferated continuously at the constant rate in the long-term culture in the presence of β-estradiol without adding rmIL-3, and the cells ceased to proliferate after depleting β-estradiol from the culture media (Fig 4). This finding indicates that the on/off switch of the growth signal can be efficiently controlled by β-estradiol. The Ba/F3 cells transfected with pCMX-Δ(5-195,324-597,702-812)GCRER, a negative control, did not respond to rhG-CSF or to β-estradiol (Fig 3E). Next, the growth rates of Ba/F3 clones transfected with pCMX-Δ(5-195)GCRER were assayed at various concentrations of β-estradiol (Fig 5). All three clones examined proliferated well at concentrations between 10−9 mol/L and 10−7 mol/L of β-estradiol.
Growth patterns of (A) parental Ba/F3 cells and (B) Ba/F3 cells transfected with pCMX-GCR, (C) pCMX-GCRER, (D) pCMX-Δ(5-195)GCRER, and (E) pCMX-Δ(5-195,324-597,702-812)GCRER in short-term cultures. Cell proliferation was estimated using XTT assay. (○) 10 U/mL of rmIL-3; (▵) 1 nmol/L (20 ng/mL) of rhG-CSF; (▴) 10−7 mol/L of β-estradiol; (•) none.
Growth patterns of (A) parental Ba/F3 cells and (B) Ba/F3 cells transfected with pCMX-GCR, (C) pCMX-GCRER, (D) pCMX-Δ(5-195)GCRER, and (E) pCMX-Δ(5-195,324-597,702-812)GCRER in short-term cultures. Cell proliferation was estimated using XTT assay. (○) 10 U/mL of rmIL-3; (▵) 1 nmol/L (20 ng/mL) of rhG-CSF; (▴) 10−7 mol/L of β-estradiol; (•) none.
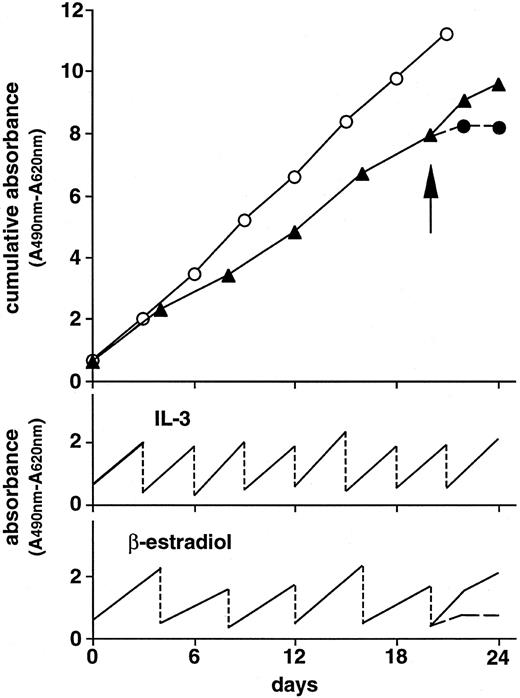
Long-term culture of the Ba/F3 cells transfected with pCMX-Δ(5-195)GCRER in the presence of 10 U/mL of rmIL-3 (○) or 10−7 mol/L of β-estradiol (▴). The rmIL-3– and β-estradiol–containing cultures were diluted to the cell concentration of 5,000 cells/mL every 3 and 4 days, respectively, as shown in lower line graphs. Upper line graph shows cumulative data during the culture period. Twenty-one days later, cells in the β-estradiol–containing cultures were washed (arrow) and further cultured in the (▴) presence or (•) absence of β-estradiol. Cell proliferation was estimated using XTT assay.
Long-term culture of the Ba/F3 cells transfected with pCMX-Δ(5-195)GCRER in the presence of 10 U/mL of rmIL-3 (○) or 10−7 mol/L of β-estradiol (▴). The rmIL-3– and β-estradiol–containing cultures were diluted to the cell concentration of 5,000 cells/mL every 3 and 4 days, respectively, as shown in lower line graphs. Upper line graph shows cumulative data during the culture period. Twenty-one days later, cells in the β-estradiol–containing cultures were washed (arrow) and further cultured in the (▴) presence or (•) absence of β-estradiol. Cell proliferation was estimated using XTT assay.
Growth rates of Ba/F3 cells transfected with pCMX-Δ(5-195)GCRER at various concentrations of β-estradiol. XTT assay was performed on days 0 and 4. The ratio of [A490nm−A620nm](day 4) to [A490nm−A620nm](day 0) at each concentration of β-estradiol was calculated. Data of three clones are shown. Arrow shows normal range of β-estradiol concentrations in human plasma. (○) Clone no. 1; (□) clone no. 2; (▵) clone no. 3.
Growth rates of Ba/F3 cells transfected with pCMX-Δ(5-195)GCRER at various concentrations of β-estradiol. XTT assay was performed on days 0 and 4. The ratio of [A490nm−A620nm](day 4) to [A490nm−A620nm](day 0) at each concentration of β-estradiol was calculated. Data of three clones are shown. Arrow shows normal range of β-estradiol concentrations in human plasma. (○) Clone no. 1; (□) clone no. 2; (▵) clone no. 3.
Transduction of murine bone marrow cells with the chimeric genes. As shown in Table 1, the untransduced and LacZ gene transduced BMMN cells from 5-FU–treated mice formed colonies in the presence of rhG-CSF, but the addition of β-estradiol to these control cultures did not induce apparent colony formation. On the other hand, the BMMN cells transduced with the GCRER or Δ(5-195)GCRER gene using retroviral vectors formed a significant number of colonies not only when stimulated with rhG-CSF, but also when cultured in the presence of β-estradiol. Although more colonies were formed with the Δ(5-195)GCRER gene than the GCRER gene in experiment 2, we cannot compare the data directly, since the cells were processed separately before colony formation assay. A small number of colonies were also formed by the BMMN cells transduced with the chimeric genes in the absence of exogenous stimulators, such as rhG-CSF or β-estradiol. Similar results were obtained in the two other separate experiments.
Interestingly, a significant number of erythroid colonies were formed by the BMMN cells transduced with the chimeric GCRER gene in the presence of rhG-CSF or β-estradiol without adding rhEpo (Table 2). On the other hand, the untransduced or LacZ-gene–transduced BMMN cells formed erythroid colonies only when the cells were cultured in the presence of rhEpo.
In experiment 1 of Table 1, several hematopoietic colonies were picked up to examine whether the colony-constituting cells contain the transgene by using PCR analysis. The transgene was detected in all colonies formed by the transduced bone marrow cells; that is, all of the estrogen-induced hematopoietic colonies contained the GCRER gene.
DISCUSSION
One of the main obstacles to stem cell gene therapy is the low efficiency of gene transfer into hematopoietic stem cells. To solve this problem, we have tried to develop a novel system for selective amplification of transduced hematopoietic stem/progenitor cells in vivo. In the present study, we constructed a chimeric cDNA (GCRER) encoding the fusion protein between G-CSFR and ER-HBD and its derivative [Δ(5-195)GCRER] as the selective amplifier genes, and examined their function in vitro using an IL-3–dependent cell line (Ba/F3) and murine bone marrow cells.
The chimeric genes were shown to be able to confer the ability for estrogen-dependent growth on the transduced hematopoietic progenitor cells, as well as on the transduced Ba/F3 cells. The stimulatory effect of β-estradiol on the growth of Ba/F3 cells was not transient, but rather persisted in long-term cultures. The strategy used in the present study is based on the finding that estrogen can activate fusion proteins between ER-HBD and a wide variety of heterologous proteins.26,27 There are several possible mechanisms for this activation. The HBD, a dimerization domain, and an HSP90-binding domain are located together on the C-terminal region of ER. ER-mediated dimerization may be critical for the activation of G-CSFR intracellular signaling, because the ligand-induced dimerization of G-CSFR is believed to be a key step for its natural activation.21 22 HSP90 may also contribute in some degree to this estrogen-controlled signal switching. The Ba/F3 cells transfected with the GCRER gene proliferated in response to β-estradiol, but to a lower extent than rhG-CSF. The β-estradiol–induced dimerization of the G-CSFR portion of the chimeric molecules may be less efficient than the natural ligand (G-CSF)–mediated dimerization.
G-CSF is known to act on a wide range of myeloid cells, including primitive multipotential hematopoietic stem cells and cells of neutrophilic lineage at various stages of differentiation. The growth and differentiation signals are transmitted from the intracellular regions of G-CSFR, although the strength of each signal may vary depending on the cell type and the differentiation stage. This concept is supported by previous reports on the function of exogenously expressed G-CSFR. When G-CSFR was expressed on IL-3–dependent FDC-P1 cells, the transformed cells acquired the ability for G-CSF–dependent growth and showed a slight differentiation upon G-CSF treatment, which was assessed by myeloperoxidase gene expression.28 On the other hand, an IL-2–dependent T-cell line, CTLL-2, did not grow in response to G-CSF after G-CSFR expression, although DNA synthesis was transiently stimulated by G-CSF treatment.24 In addition, when G-CSFR was expressed on a murine IL-3–dependent cell line, LGM, which was derived from the multipotential bone marrow cell clone LyD9, G-CSF treatment induced mild proliferation and then morphologic differentiation into mature neutrophils.29 30 These findings suggest that G-CSFR expressed on immature cells transmits mainly growth signal on G-CSF stimulation and that the differentiation signal predominates over the growth signal for the cells at maturing stages. In addition, it is noteworthy that erythroid colonies were apparently formed by the bone marrow progenitor cells transduced with the GCRER gene in the presence of estrogen without the addition of rhEpo, suggesting that the signals from the G-CSFR portion of the chimeric molecules do not preferentially induce neutrophilic differentiation, but just promote the differentiation depending on the nature of the target progenitor cells. The activation of such selective amplifier gene products probably induces natural proliferation and/or differentiation of the target cells. Therefore, we speculate that when the selective amplifier genes are expressed in the primitive hematopoietic stem cells, the population of transduced stem cells expands upon estrogen treatment, even if some of the cells enter the differentiation pathway. This idea is supported by the findings obtained from the clinical trials of rhG-CSF; that is, rhG-CSF administration caused the increase in the total number of hematopoietic progenitors in bone marrow and peripheral blood, and the sustained use of rhG-CSF did not bring about the exhaustion of hematopoietic activity. To confirm that the intracellular signal of G-CSFR can be used for expansion of transduced hematopoietic stem cells, we are now conducting in vivo experiments using mice whose hematopoietic system was reconstituted with the murine bone marrow cells transduced with the selective amplifier gene.
If it turns out that the differentiation signal of the current chimeric gene is too strong, we will construct a more potent selective amplifier gene, which contains the N-terminal growth-signaling domain, but lacks the C-terminal differentiation-signaling domain of the cytoplasmic region of G-CSFR cDNA.28,30 31 In addition, this modified amplifier gene will be applicable to the ex vivo expansion of transduced hematopoietic stem/progenitor cells.
To control the activity of the selective amplifier gene, the response to endogenous G-CSF or estrogen should be minimized. As shown in Fig 3D, the response to G-CSF was abolished by deleting 191 amino acids (from amino acid 5 to amino acid 195) from the extracellular domain of the chimeric molecule [Δ(5-195)GCRER]. The growth of transduced cells with this deleted form of the chimeric gene can be controlled by estrogen alone, but not by G-CSF.
As for the response to estrogen, we cannot exclude the possibility that the transduced hematopoietic stem/progenitor cells will proliferate in response to the endogenous hormone in vivo, since the mean concentration of β-estradiol in human plasma is approximately 20 to 42 pg/mL (7.3 × 10−11 to 1.54 × 10−10 mol/L) in males,32 and approximately 0.06 to 0.7 ng/mL (2.20 × 10−10 to 2.57 × 10−9 mol/L) and approximately 0.2 ng/mL (7.34 × 10−10 mol/L) in the follicular and luteal phase, respectively, in females.33 A slight response of transduced cells to this level of estrogen may be anticipated from our experiments using Ba/F3 cells. That a small number of colonies were formed by the transduced bone marrow cells without the addition of CSF or estrogen may be due to the estrogen contained in FCS or estrogenic activities in culture media.34 If this mechanism works in vivo, it is possible that the transduced hematopoietic cells will be gradually selected without the administration of estrogen. This kind of natural selection may be acceptable or even preferable in many instances. However, such a response to endogenous estrogen may cause low-level, but constitutive, activation of the chimeric gene and may be inappropriate in the long term. In that case, we will be able to use mutant ER gene, thereby reducing the responsiveness to estrogen. Moreover, there is a report of a mutant ER that is unable to bind estrogen yet retains the affinity for the synthetic ligand, 4-hydroxytamoxifen.35 36
In the present study, we proposed the novel concept of a selective amplifier gene by constructing its prototypes, GCRER and Δ(5-195)GCRER. Various modifications are possible to improve the system as described here. We are also constructing similar chimeric genes using other growth factor receptor genes such as c-kit and erythropoietin receptor genes. However, the G-CSFR gene may be the most appropriate for clinical application, since it has already been extensively studied and, more importantly, there is plenty of experience in the clinical use of rhG-CSF. The safe use of rhG-CSF in humans suggests that the signals from exogenously expressed G-CSFR–derived molecules are safer than those of other receptors. For stem-cell gene therapy, the therapeutic gene of interest will be introduced into hematopoietic stem cells in combination with the selective amplifier gene using a dicistronic vector containing an internal ribosome entry site (IRES).
ACKNOWLEDGMENT
The authors thank Dr S. Nagata for pBluescript-GCR, Dr A. Kakizuka for pCMX-MfasER, RIKEN DNA BANK for pGK LacZ, Chugai Pharmaceutical for rhG-CSF and rhEpo, Amgen for rrSCF, Ajinomoto for rhIL-6, and Takara Shuzo for CH-296.
Supported in part by grants from the Ministry of Health and Welfare of Japan, a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and the Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corp (JST).
Address reprint requests to Keiya Ozawa, MD, PhD, Department of Molecular Biology, Institute of Hematology, Jichi Medical School, 3311-1 Yakushiji, Minamikawachi-machi, Kawachi-gun, Tochigi 329-04, Japan; email: kozawa@ms.jichi.ac.jp.

 to [A490nm−A620nm](day 0) at each concentration of β-estradiol was calculated. Data of three clones are shown. Arrow shows normal range of β-estradiol concentrations in human plasma. (○) Clone no. 1; (□) clone no. 2; (▵) clone no. 3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3884/3/m_bl_0014f5.jpeg?Expires=1767762838&Signature=KgtmH6-mtKxrODZkHmTbW4NMBoK2nDA1Ijtc1fmAk115-WO5jQVE1QJC0j4gGZY00pZJscvDmStu324hJCS2ilL9jAisDfTgzn6Ka2p3H-xojbichdP3W6vgx~mbkI615GlbViX4csabNBQUOgxGTCIA9d3Nve8xBAVPWod1S3lShwcSF9~FtRWwwP1vtueM5hFATU4iiZ0JRjH6z2KzUrNvz2skLhGdWwMNcx5c6bKGGf6uYEx6a1jV7sYFM6aevs2M3qAbUTN53IYr3MZPRmk~eISfhu5vj4andgjnVcnOm~yYzDbNZ9UAKKXNliPiCmHmkVJSBsvFKSHUBQDDMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

 to [A490nm−A620nm](day 0) at each concentration of β-estradiol was calculated. Data of three clones are shown. Arrow shows normal range of β-estradiol concentrations in human plasma. (○) Clone no. 1; (□) clone no. 2; (▵) clone no. 3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3884/3/m_bl_0014f5.jpeg?Expires=1767762839&Signature=aeD4nsMtkBeOZm9Dx2QCNWgwcWktaThUPwzOjmWUzehi89WrdGIgYb0uRjPCfXo4gsbZigkBD8Tq3FPQjuNReuqDH3nYNp85q48OTJQ2SIS1JX~7xEZsx9tW5B7X-JClOpgSLWHLeqHalqZ7LN69ExoinvsD47cWt5GtMC3l0nOUWpjhSr1wP9hQEK9tkljtpZ7ucQHTznAJn8I3In7PtVXs6jSg80~Ji~8h4pEAFUKogCdMZxMSV4g43r1ll9OE02pbyPCP-mRqZYQXNJrnGAHpDx0ZgfXroEeNXHbs6jFtBYpz9g06WLcfMFtJc0oTS7jfU5lK0L2xYxVTwmSh9Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)