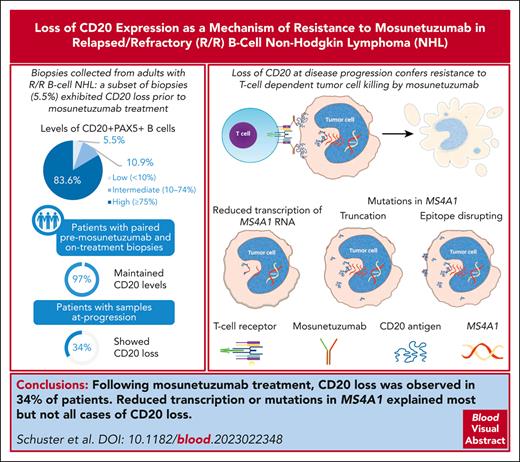
We identified CD20 loss at progression in the phase 1/2 GO29781 trial of mosunetuzumab monotherapy in B-cell NHL.
Reduced transcription or gain of truncating mutations explained most but not all cases of CD20 loss with mosunetuzumab.
Visual Abstract
CD20 is an established therapeutic target in B-cell malignancies. The CD20 × CD3 bispecific antibody mosunetuzumab has significant efficacy in B-cell non-Hodgkin lymphomas (NHLs). Because target antigen loss is a recognized mechanism of resistance, we evaluated CD20 expression relative to clinical response in patients with relapsed and/or refractory NHL in the phase 1/2 GO29781 trial investigating mosunetuzumab monotherapy. CD20 was studied using immunohistochemistry (IHC), RNA sequencing, and whole-exome sequencing performed centrally in biopsy specimens collected before treatment at predose, during treatment, or upon progression. Before treatment, most patients exhibited a high proportion of tumor cells expressing CD20; however, in 16 of 293 patients (5.5%) the proportion was <10%. Analyses of paired biopsy specimens from patients on treatment revealed that CD20 levels were maintained in 29 of 30 patients (97%) vs at progression, where CD20 loss was observed in 11 of 32 patients (34%). Reduced transcription or acquisition of truncating mutations explained most but not all cases of CD20 loss. In vitro modeling confirmed the effects of CD20 variants identified in clinical samples on reduction of CD20 expression and missense mutations in the extracellular domain that could block mosunetuzumab binding. This study expands the knowledge about the occurrence of target antigen loss after anti-CD20 therapeutics to include CD20-targeting bispecific antibodies and elucidates mechanisms of reduced CD20 expression at disease progression that may be generalizable to other anti-CD20 targeting agents. These results also confirm the utility of readily available IHC staining for CD20 as a tool to inform clinical decisions. This trial was registered at www.ClinicalTrials.gov as #NCT02500407.
Introduction
B-cell lineage markers are valuable therapeutic targets. CD20 is expressed by >95% of normal B-lymphocytes from pre-B cells until differentiation into plasma cells as well as by their malignant lymphoma counterparts.1 The restricted expression of CD20 makes it a rational therapeutic target for B-cell malignancies. CD20 has been successfully used as a therapeutic target in combination with chemotherapy; the monoclonal antibody (mAb) rituximab is the standard of care first-line therapy for diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL),2,3 and the mAb obinutuzumab is the standard of care for FL.4 These anti-CD20 agents are also included as part of the backbone for a number of treatment regimens for relapsed or refractory (R/R) B-cell non-Hodgkin lymphomas (NHLs).5-8 Despite significant progress using CD20-targeting agents for the treatment of NHL, strategies to further enhance clinical activity are being explored, including agents that promote immune engagement.
Mosunetuzumab is a first-in-class CD20 × CD3 T-cell–engaging bispecific antibody that redirects CD3-expressing T cells to eliminate malignant CD20-expressing B cells.9 Mosunetuzumab is administered as a fixed-duration regimen (eight 21-day cycles for patients who achieve a complete response [CR] and up to 17 cycles for patients who achieve partial response or stable disease). Mosunetuzumab has shown significant clinical activity with an overall response rate of 80%, CR rate of 60%, and a median duration of response of 22.8 months in adult patients with R/R FL after ≥2 lines of systemic therapy.10 Mosunetuzumab received accelerated approval for this indication, and updated results continue to demonstrate durable responses.11,12 Mosunetuzumab is currently being investigated for additional indications, including aggressive NHL,12 as well as in rational combinations.13,14 Although B-cell–directed therapies have demonstrated substantial clinical benefit, target loss has been observed as a mechanism of resistance. Loss of CD19 to chimeric antigen receptor T-cell therapies has been reported in a subset of patients across several lymphoid malignancies, including acute lymphoblastic leukemia (ALL) and large B-cell lymphomas.15-17 Similarly, loss of CD20 upon relapse after rituximab has been reported.18-22
CD20, encoded by MS4A1, is a member of the membrane-spanning 4-domain family, subfamily A (MS4A),23,24 which comprises 4 transmembrane helical domains, 2 conserved extracellular loops (a small ECL1 loop and a larger ECL2 loop), and intracellular N- and C- terminal sequences.25,26 Conserved sequences within the ECL2 loop, 170-ANPS-173, are part of a shared epitope for several anti-CD20 agents, including rituximab and obinutuzumab,26,27 whereas other anti-CD20 agents such as ofatumumab bind to sequences located in both ECL1 and ECL2.28 Understanding the mechanisms that result in innate or acquired resistance to mosunetuzumab is critical for maximizing its efficacy and directing treatment strategies. Thus, we evaluated CD20 loss across multiple NHL histologies in the phase 1/2 GO29781 mosunetuzumab monotherapy trial.
Methods
Samples
Patient biopsy specimens were collected in the phase 1/2 GO29781 (NCT02500407) trial of mosunetuzumab monotherapy for adults with R/R B-cell NHL who had received ≥2 prior therapies. CD20 expression was not part of the inclusion criteria, but a biopsy and associated pathology report were required. Full details of the study design have been previously reported.29
Before study drug administration (pre-mosunetuzumab), biopsy specimens were collected from patients receiving fixed-dosing (group A, 0.05 mg to 2.8 mg) or cycle 1 step-up dosing (group B, 0.4/1/2.8 mg to 1/2/60 mg) and from consenting patients while on treatment and/or at progression (optional). For nonarchival specimens, biopsy specimens were collected from safely accessible sites per investigator determination. Pre-mosunetuzumab biopsy specimens collected before the last dose of a prior anticancer therapy and first dose of mosunetuzumab were considered archival, or they were considered fresh if collected after the prior therapy but before initiating mosunetuzumab. On-treatment biopsy specimens were collected between cycle 1, day 1 and cycle 3, day 15; biopsy specimens at progression were collected at/after a progression event. When possible, tumor sites were of sufficient size to allow for 2 biopsy specimens, and paired biopsy specimens were taken from the same lesions when feasible.
IHC
Duplex immunohistochemistry (IHC) was performed centrally at Cell Carta to detect CD20 (clone L26 mouse mAb, Ventana) and PAX5 (clone DAK-PAX5 mouse mAb, Dako). Tumor area was defined by a pathologist, and image analysis (VisoPharm) was used to determine the proportion of CD20+PAX5+ cells in the PAX5+-defined tumor area as a continuous variable. Tumor specimens with <5% PAX5+ nuclear staining and bone marrow biopsies were excluded from analyses.
RNAseq
Expression of MS4A1 was measured by RNA sequencing (RNAseq) performed by Q2 Solutions using MiSeq. RNA was extracted using RNeasy FFPE Kit (Qiagen, Hilden, Germany). Purified RNA was used to create complementary DNA libraries that were assayed using TruSeq (Illumina) RNAseq.
Raw reads were quality controlled and aligned to the human reference genome (NCBI Build 38) using GSNAP.30,31 Transcript annotation was based on the Ensembl genes database (release 77). To quantify gene expression levels, the number of reads mapped to the exons of each RefSeq gene was calculated. Raw counts were normalized to reads per million using a robust library size estimation (DESeq2) and then log2-transformed after the addition of a pseudocount.
WES
MS4A1 mutation profiling was performed by whole-exome sequencing (WES) on 206 tumor samples and matched normal peripheral blood mononuclear cell samples at Q2 Solutions, using Exome Seq All exon (v6). Genomic DNA was isolated from cell pellets using the QIAamp DNA Mini Kit (Qiagen). Libraries, prepared using the Swift Accel-NGS 2S Hyb DNA library kit and the Swift 2S SureSelectXT Compatibility Module per LAB_13_3273 (Swift Biosciences), were sequenced using an Illumina sequencing-by-synthesis platform for an average coverage before deduplication of 50× and 200× per sample, respectively. FASTQ reads were aligned to the human reference genome (NCBI Build 38) using GSNAP.30,31 Duplicate reads were marked using PicardTools, and indels were realigned using the GATK IndelRealigner tool (Broad Institute). Variations were called using LoFreq32 on all exon intervals padded by 10 base pairs on both ends. One sample was removed because of poor quality, leaving 205 for analysis.
In vitro modeling
CD20 CRISPR knockout DLBCL (SU-DHL-16) and mantle cell lymphoma (MCL; MAVER-1) cell lines and a CD20– B-ALL cell line (REH) were engineered to express wild type (WT) or mutant MS4A1 and characterized as described in the supplemental Methods (available on the Blood website). The influence of these mutations on T-cell–mediated activation and tumor cell killing was evaluated using a proof-of-concept CD20 × CD3 bispecific antibody (2H7/UCHT1), as described by Sun et al,9 hereafter referred to as CD20 × CD3.
Killing assay
Isolated CD8+ T cells from healthy donors (supplemental Methods) were mixed with tumor target cells at a ratio of 3:1 and incubated for 48 hours at 37°C, 5% CO2. After incubation with the CD20-CD3 bispecific or nontargeting control antibody (CD3-NIST), target cells were stained and analyzed by spectral flow cytometry for reduction in CD19+ B cells.
Results
Local and central CD20 assessment in diagnostic samples
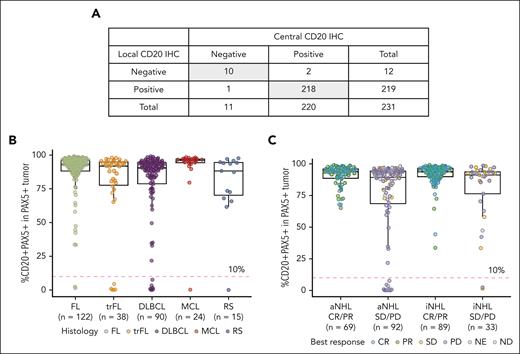
Although NHL are expected to express CD20, confirmation by local IHC is often performed at diagnosis and in the R/R setting for patients previously treated with an anti-CD20–containing regimen. A dual IHC assay was used to localize the expression of CD20 specifically on B cells, which were identified by PAX5 nuclear staining within the tumor area. Comparison of local CD20 IHC, based on pathology reports provided with biopsy specimens collected before treatment from 231 patients, and centrally assessed dual CD20+PAX5+ IHC results revealed 98% concordance (Figure 1A). This was anticipated because the CD20 detecting antibody, clone L26, which recognizes an intracellular epitope at the C terminus of CD20 that is not obstructed by therapeutic mAb binding, is also used for the dual IHC assay and routinely in clinical practice. As additional evidence to support the comparability of assays, samples from 126 patients in which both dual and single-plex IHC assays were run in parallel showed high concordance (r = 0.93; supplemental Figure 1A).
Characterization of CD20 status in pre-mosunetuzumab tumor biopsy specimens by IHC. (A) Comparison of CD20 status determined by local CD20 staining per pathology reports vs central assessments using the dual CD20+PAX5+ IHC assay; shaded cells represent concordance between IHC assessments. (B) Percent CD20+PAX5+/PAX5+ cells in the tumor area before mosunetuzumab across NHL histologies from the GO29781 study. (C) CD20+PAX5+ staining relative to best overall response determined by investigator-assessed PET/CT imaging using the International Harmonization Project response criteria.33 CT, computed tomography; ND, not determined; NE, not estimable; PD, progressive disease; PET, positron emission tomography; PR, partial response; SD, stable disease.
Characterization of CD20 status in pre-mosunetuzumab tumor biopsy specimens by IHC. (A) Comparison of CD20 status determined by local CD20 staining per pathology reports vs central assessments using the dual CD20+PAX5+ IHC assay; shaded cells represent concordance between IHC assessments. (B) Percent CD20+PAX5+/PAX5+ cells in the tumor area before mosunetuzumab across NHL histologies from the GO29781 study. (C) CD20+PAX5+ staining relative to best overall response determined by investigator-assessed PET/CT imaging using the International Harmonization Project response criteria.33 CT, computed tomography; ND, not determined; NE, not estimable; PD, progressive disease; PET, positron emission tomography; PR, partial response; SD, stable disease.
CD20 expression
A non-Gaussian distribution of CD20+PAX5+ B cells was observed in biopsy specimens collected before mosunetuzumab (Figure 1B). Most biopsy specimens (245 of 293 [83.6%]) exhibited high levels (≥75%) of CD20+PAX5+ B cells in the tumor area, a smaller subset (n = 32 [10.9%]) displayed intermediate levels (10%-74%), and a minor fraction (n = 16 [5.5%]) had low levels (<10%). A similar distribution was observed across histologies; however, CD20+PAX5+ levels <10% were seen more frequently in patients with aggressive NHL (aNHL; DLBCL, 7/90 [8%]; MCL, 1/24 [4%]; transformed FL [tFL], 5/38 [13%]; and Richter syndrome [RS], 1/15 [7%]) than in those with indolent NHL (iNHL; FL, 2/122 [2%]; Figure 1C).
Of the 293 pre-mosunetuzumab biopsy specimens, 220 (75%) were fresh, and 73 (25%) were archival. A similar distribution of CD20+PAX5+ cells and relationship to response was observed in both biopsy types, however, most samples with <10% CD20+PAX5+ cells were from fresh (14 of 16) vs archival biopsy specimen (2 of 16; supplemental Figure 1B).
Although there was no significant correlation between the proportion of CD20+PAX5+ cells before mosunetuzumab and best response, clinical activity was not observed in 5.5% of patients with levels <10% CD20+ (aNHL, n = 14; and iNHL, n = 2; Figure 1C). The proportion of CD20+PAX5+ cells in tumor specimens and its relationship with response was confirmed when refining the analysis to include only patients treated at an interim expansion dose (cycle 1, day 1 = 1 mg; cycle 1, day 8 = 2 mg; and cycle 1, day 15 = 13.5 mg) or at the recommended phase 2 and registration dose (cycle 1, day 1 = 1 mg; cycle 1, day 8 = 2 mg; and cycle 1, day 15 = 60 mg; cycle 2, day 1 = 60 mg; and cycle 3, day 1 = 30 mg; supplemental Figure 1C).
Changes in CD20 levels during mosunetuzumab therapy
Modulation of CD20 by mosunetuzumab was evaluated by monitoring the proportion of CD20+PAX5+ cells in pre-mosunetuzumab biopsy specimens compared with on treatment (cycle 1, day 15 to cycle 3, day 1) or at/after progression using sequential biopsy specimens from 64 patients. In 7 patients (FL, n = 2; DLBCL, n = 2; tFL, n = 2; and RS, n = 1), the proportion of CD20+PAX5+ cells before mosunetuzumab was <10% and remained so in sequentially collected paired biopsy specimens; 6 of these 7 patients progressed early, before completing a second treatment cycle (supplemental Figure 2A). In patients with CD20 values >10% who had paired pre-mosunetuzumab and on-treatment samples, 29 of 30 (97%) maintained CD20 levels, whereas only 1 patient with the lowest pre-mosunetuzumab CD20 level in this group (32% CD20+PAX5+) had a reduction in CD20+PAX5+ cells to <10% (Figure 2A). By contrast, in 32 patients with sequential pre-mosunetuzumab and at-progression biopsy specimens, 11 (34%) showed loss of CD20 to levels <10% (Figure 2B). These included 5 patients who provided 3 biopsy specimens (pre-mosunetuzumab, on-treatment, and at-progression), of whom all maintained CD20 on treatment and 1 had CD20 loss at progression (supplemental Figure 2B). CD20 loss was observed across histologies but was more frequent in aNHL (7 of 11 [64%]; 2 with DLBCL, 3 tFL, 1 MCL, and 1 RS) than iNHL (4 of 11 [36%]; 4 FL) (Figure 2B).
Changes in CD20 status after treatment with mosunetuzumab detected by CD20+PAX5+ dual IHC. (A) Changes in the proportion of CD20+PAX5+ cells in serial biopsy specimens collected on treatment. (B) CD20 status assessed in serial biopsy specimens collected at or after progression based on progression event. (C) Time to response in paired biopsy samples by CD20 status before mosunetuzumab or at progression; numbers in each row represent percent of CD20. Symbols indicating visits are colored black or pink to represent CD20 status at that visit: black (negative), pink (positive).
Changes in CD20 status after treatment with mosunetuzumab detected by CD20+PAX5+ dual IHC. (A) Changes in the proportion of CD20+PAX5+ cells in serial biopsy specimens collected on treatment. (B) CD20 status assessed in serial biopsy specimens collected at or after progression based on progression event. (C) Time to response in paired biopsy samples by CD20 status before mosunetuzumab or at progression; numbers in each row represent percent of CD20. Symbols indicating visits are colored black or pink to represent CD20 status at that visit: black (negative), pink (positive).
Paired biopsy specimens were collected during dose escalation in nonfractionated dosing cohorts (A1-A8) and during dose escalation and expansion in step-up–dosing cohorts (B1-B11; supplemental Table 1). On-treatment reduction to <10% was observed in 1 patient in cohort B5 (0.8/2/6 mg). A reduction in CD20+PAX5+ cells (but not to <10%) was also seen in 2 patients, 1 in cohort B7 (1/2/13.5 mg) and 1 in cohort B11 (1/2/60/30 mg) (supplemental Figure 2C). At progression, a decrease in CD20+PAX5+ to <10% was observed in cohorts B2 (0.8/2/4.2 mg), B7 (1/2/13.5 mg), B9 (1/2/27 mg), and B11 (1/2/60/30 mg), with most cases seen at the highest dose tested (supplemental Figure 2D). The majority of paired biopsy specimens evaluated were from patients receiving higher doses (cohorts B7-B11), which included 15 on-treatment and 25 at-progression biopsy pairs.
Time to progression varied across different patterns of CD20 loss. For patients with pre-mosunetuzumab CD20– biopsy specimens (n = 7), progression-free survival (PFS) was short (median, 42 days; range, 23-102 days). For patients with CD20-retention in the absence of an objective response (n = 12), PFS was similar to that seen in patients who were CD20– before mosunetuzumab (median, 40 days; range, 21-78 days). This contrasted with patients who achieved an objective response and retained CD20 at progression (n = 8); PFS ranged from 134 to 913 days (median, 485 days). For patients with CD20 loss at progression, PFS was longer if an objective response had been achieved (n = 5; median, 164 days; range, 106-259 days) compared with those not achieving a response (n = 6; median, 75 days; range, 36-167 days). Shorter PFS can in part be explained by CD20 loss before mosunetuzumab or at progression, however, a subset of patients was refractory and exhibited limited PFS despite maintaining CD20 levels (Figure 2C; supplemental Figure 2E).
Effect of mutations on CD20 loss at progression
WES was performed on 205 biopsy specimens from 156 patients; 107 from patients with a single biopsy specimen and 98 from patients with ≥2 biopsy specimens collected at different times during treatment. CD20 mutations were identified in 13 patients (18 different variants; Table 1).
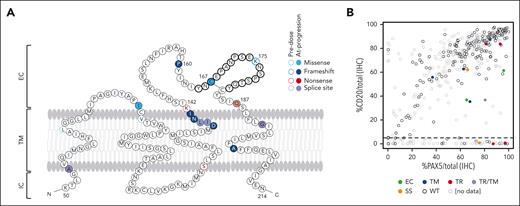
Fourteen CD20 variants were found in pre-mosunetuzumab biopsy specimens from 10 of 154 patients (8 with only pre-mosunetuzumab biopsy specimens + 2 with mutations before mosunetuzumab and at progression); CD20 was detectable by IHC in all 10 patients. Overall, 9 of 14 variants were mutations in the transmembrane domains (5 of 9 were missense) and 4 of 14 were in the extracellular loop including 1, K175E, located in ECL2 that is part of the shared epitope for anti-CD20 therapeutics (Figure 3A). Among patients with paired pre-mosunetuzumab and posttreatment biopsy specimens, CD20 mutations were observed in 5 of 55 patients, with loss of CD20 (<10%) seen at progression in 4 of the 5 patients. One patient acquired a missense mutation at progression at C167G located in the therapeutic binding site of ECL2, with only a minor reduction in CD20 (from 63% to 45%) despite potential to disrupt the predicted internal disulfide bridge in this extracellular domain (supplemental Figure 3). The type and location of CD20 mutations appeared to affect CD20 expression. Alterations identified in the transmembrane domain tended not to alter expression (Figure 3B). Two splice variants with initiation sites in the transmembrane domain were identified: 1 was associated with CD20 loss, whereas the other was observed before mosunetuzumab and at progression, which was characterized by CD20 loss. The effects of truncating mutations were variable with CD20 levels reduced to <10% in 2 patients (Q187∗ and P160fs), whereas protein expression appeared unaffected in another 3 patients (Figure 3B). Variants identified in the extracellular domain reduced CD20 levels but not to <10%.
CD20 variants identified by WES before mosunetuzumab and at progression affect CD20 status. (A) Mutations identified in tumor biopsies from patients treated with mosunetuzumab; missense (light blue), frameshift (dark blue), nonsense (red), and splice variants (purple). Predicted epitope for rituximab and obinutuzumab binding is bolded. (B) CD20 levels assessed as the percent CD20+ cells/total nucleated cells relative to PAX5+ cells/total nucleated cells for all biopsy specimens sequenced. Dots represent single samples colored by position and type of variant. EC, extracellular; IC, intracellular; SS, splice site; TM, transmembrane; TR, truncating. Panel A is adapted from Rushton et al.21
CD20 variants identified by WES before mosunetuzumab and at progression affect CD20 status. (A) Mutations identified in tumor biopsies from patients treated with mosunetuzumab; missense (light blue), frameshift (dark blue), nonsense (red), and splice variants (purple). Predicted epitope for rituximab and obinutuzumab binding is bolded. (B) CD20 levels assessed as the percent CD20+ cells/total nucleated cells relative to PAX5+ cells/total nucleated cells for all biopsy specimens sequenced. Dots represent single samples colored by position and type of variant. EC, extracellular; IC, intracellular; SS, splice site; TM, transmembrane; TR, truncating. Panel A is adapted from Rushton et al.21
MS4A1 expression at progression
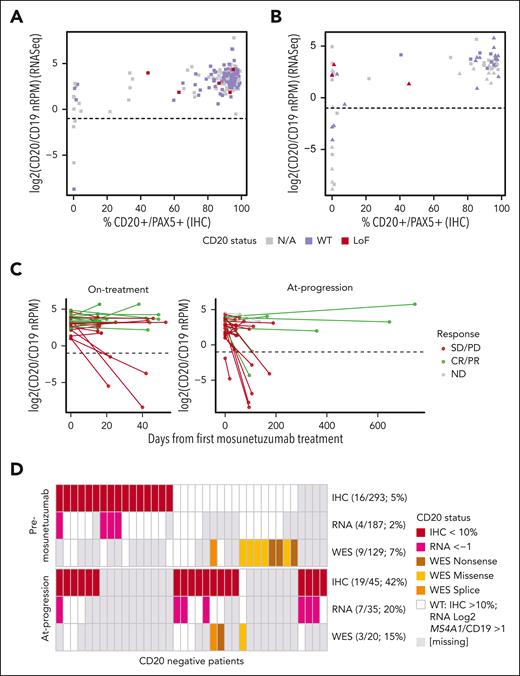
The correlation between RNAseq-based gene expression of MS4A1 (normalized by CD19) and protein based on CD20 IHC was examined in patients with both measurements available (186 pre-mosunetuzumab biopsy specimens and 65 on-treatment or at-progression biopsy specimens). Generally, RNA levels correlated with protein levels (r = 0.57); however, in 10 of 186 (5.5%) pre-mosunetuzumab samples, MS4A1 was expressed, but CD20 protein was not detected (Figure 4A). RNA and protein concordance was high in samples collected on treatment and at progression (r = 0.64). However, discordance between RNA and protein levels (CD20 IHC negative but RNA positive) was observed in 11 of 65 (16%) on-treatment/at-progression biopsy specimens (Figure 4B). For a subset of patient samples with RNAseq, WES, and IHC data available (n = 119), alterations in MS4A1 were assessed relative to MS4A1 RNA and CD20 protein levels. Discordance between RNA and protein could be attributed to variants within MS4A1 in 2 patients, but for the remaining 6 patients with available WES data showing WT MS4A1 (3 patients before and 3 patients after mosunetuzumab), mutations could not account for the discordance (Figure 4A-B).
MS4A1 (the gene that encodes CD20) transcript profiling. (A) MS4A1 RNAseq-based expression relative to CD20+PAX5+ protein IHC staining in pre-mosunetuzumab tumor biopsies, and (B) in posttreatment tumor biopsies obtained on treatment (square) vs at progression (triangle). Loss of function (LoF) variants in CD20 identified by WES (red) and WES confirmed CD20 WT (purple). (C) Changes in the levels of MS4A1 in paired tumor biopsy specimens collected on treatment (left) or at progression (right) by best overall response (CR/PR, green; SD/PD, red). (D) Summary samples with multiple CD20 biomarker measurements (IHC, RNASeq, and WES) in samples collected before mosunetuzumab (top) or at progression (ie, at progression and on treatment) (bottom). LoF, loss of function; N/A, not available; nRPM, normalized reads per million.
MS4A1 (the gene that encodes CD20) transcript profiling. (A) MS4A1 RNAseq-based expression relative to CD20+PAX5+ protein IHC staining in pre-mosunetuzumab tumor biopsies, and (B) in posttreatment tumor biopsies obtained on treatment (square) vs at progression (triangle). Loss of function (LoF) variants in CD20 identified by WES (red) and WES confirmed CD20 WT (purple). (C) Changes in the levels of MS4A1 in paired tumor biopsy specimens collected on treatment (left) or at progression (right) by best overall response (CR/PR, green; SD/PD, red). (D) Summary samples with multiple CD20 biomarker measurements (IHC, RNASeq, and WES) in samples collected before mosunetuzumab (top) or at progression (ie, at progression and on treatment) (bottom). LoF, loss of function; N/A, not available; nRPM, normalized reads per million.
Changes in the transcriptional level of MS4A1 were evaluated in paired biopsy specimens. As with protein (IHC), a reduction in RNA levels was observed in some patients on treatment (4 of 29 [13.8%]) and some at progression (6 of 28 [21.4%]), particularly those without an objective response. A decrease in MS4A1 transcripts was seen in 2 patients with objective responses at progression, but only 1 decreased to below the transcriptional threshold (log2 MS4A1/CD19, –1.0; Figure 4C).
Multiple mechanisms regulating CD20 were identified. Before mosunetuzumab treatment, CD20 loss was associated, in some cases, with transcriptional downregulation but not with identified exonic mutations, whereas in cases in which CD20 mutations were detected, CD20 protein expression was retained (Figure 4D). In contrast, CD20 loss at progression was observed in patients with evidence of transcriptional downmodulation or with mutations identified by WES. There was also evidence of reduced CD20 levels in the absence of downregulated transcription or exonic mutations in CD20 (Figure 4D). UpSet plots showing the various CD20 assays used and their overlap are provided (supplemental Figure 4).
In vitro modeling of CD20 mutations
To validate the effects of identified variants, isogenic cell lines were engineered to express WT or mutant MS4A1 for in vitro assessment of protein expression, localization, and sensitivity to T-cell–mediated killing (supplemental Figure 5A). Mutations identified at or near the extracellular loops with potential to influence mosunetuzumab binding (P160fs, Q187∗, K175E, and C167G) were expressed in CD20 CRISPR knockout NHL cell lines SU-DHL-16 (DLBCL), MAVER-1 (MCL), and a CD20– B-ALL cell line (REH). CD20 protein was not detected in cell lines harboring the frameshift or truncating mutations P160fs and Q187∗ but was observed for both C167G and K175E variants (Figure 5A). Extracellular staining followed by flow cytometry (2H7 clone; overlapping binding site with mosunetuzumab) did not detect the surface expression of any of the CD20 mutants; however, intracellular staining using an antibody directed to the cytoplasmic domain (H1 clone) confirmed the expression profiles observed by western blot (Figure 5B). To assess the possibility that mutations interfered with extracellular antibody binding, the expression pattern of CD20 mutant protein was assessed by immunofluorescence microscopy using an intracellular antibody. Membrane localization was detected for both C167G and K175E (Figure 5C), confirming that these mutations do not alter protein localization but do interfere with extracellular anti-CD20 binding. Similar patterns of expression and membrane localization were observed in engineered MAVER-1 and REH cell lines (supplemental Figure 5B-C).
In vitro assessment of the effects of MS4A1 variants on CD20 expression and CD20 × CD3 activity. (A) CD20 protein expression by western blot in the SU-DHL-16–engineered cell lines. (B) Detection of CD20 by flow cytometry in SU-DHL-16–engineered cell lines. Intracellular expression was detected after permeabilization using anti-CD20 antibody targeting the C-terminus (H-1, BD-561174), and extracellular expression was detected using anti-CD20 antibody targeting ECL2 (2H7, BD-555623). (C) Immunofluorescence detection of CD20 using an intracellular antibody (ABCAM-78237): red fluorescence = CD20; blue fluorescence = 4′,6-diamidino-2-phenylindole (DAPI). (D) (left) CD20 × CD3–directed CD8-dependent cell killing of SU-DHL-16–engineered cell lines after 48 hours of treatment assessed by spectral flow cytometry. (Right) CD20 × CD3–directed CD8+ T-cell activation after 48 hours of treatment. T-cell activation monitored by flow cytometry by detecting activation markers CD69 and CD25. Flow cytometry data are represented as mean ± SD of 3 replicates. EV, empty vector; CD20-CD3, proof-of-concept CD20 × CD3 bispecific molecule; MFI, median fluorescence intensity; N, non-targeting control/CD3 antibody; parent, parental cell line; SD, standard deviation.
In vitro assessment of the effects of MS4A1 variants on CD20 expression and CD20 × CD3 activity. (A) CD20 protein expression by western blot in the SU-DHL-16–engineered cell lines. (B) Detection of CD20 by flow cytometry in SU-DHL-16–engineered cell lines. Intracellular expression was detected after permeabilization using anti-CD20 antibody targeting the C-terminus (H-1, BD-561174), and extracellular expression was detected using anti-CD20 antibody targeting ECL2 (2H7, BD-555623). (C) Immunofluorescence detection of CD20 using an intracellular antibody (ABCAM-78237): red fluorescence = CD20; blue fluorescence = 4′,6-diamidino-2-phenylindole (DAPI). (D) (left) CD20 × CD3–directed CD8-dependent cell killing of SU-DHL-16–engineered cell lines after 48 hours of treatment assessed by spectral flow cytometry. (Right) CD20 × CD3–directed CD8+ T-cell activation after 48 hours of treatment. T-cell activation monitored by flow cytometry by detecting activation markers CD69 and CD25. Flow cytometry data are represented as mean ± SD of 3 replicates. EV, empty vector; CD20-CD3, proof-of-concept CD20 × CD3 bispecific molecule; MFI, median fluorescence intensity; N, non-targeting control/CD3 antibody; parent, parental cell line; SD, standard deviation.
CD20 × CD3–dependent T-cell activation was assessed in coculture assays using purified CD8+ T cells from healthy donors and engineered SU-DHL-16 cell lines. Detection of the early activation marker CD69 by flow cytometry demonstrated minimal CD8+ T-cell activation with cell lines expressing the CD20 mutations, whereas the parental CD20-expressing cell line and the CRISPR/Cas9 CD20 WT engineered cell lines showed robust CD8+ T-cell activation (Figure 5D). Consistent with abrogated CD20 × CD3–induced T-cell activation, cell killing in coculture assays was eliminated in cell lines harboring the nonexpressing variants P160fs and Q187∗ and was reduced relative to WT CD20 for the extracellular domain variants C167G and K175E (Figure 5D). T-cell–mediated killing was eliminated in the MAVER-1 and REH cell lines engineered to express CD20 mutations (supplemental Figure 5B-C). These results demonstrate that these CD20 mutations can diminish mosunetuzumab binding and recognition by effector cells.
Discussion
CD20-targeted therapeutics in combination with chemotherapy have revolutionized NHL treatment. Most recently, targeting CD20 as a component of bispecific T-cell–engaging antibodies (which include mosunetuzumab) has been investigated as an updated strategy for treating NHL. Mosunetuzumab is administered as an off-the-shelf treatment9 using a fixed-duration regimen that minimizes extended exposure and potentially reduces cumulative safety concerns (eg, B-cell aplasia) or potential acquired resistance (eg, target loss or T-cell exhaustion). However, even with fixed-duration therapy, evidence for target loss is observed. The frequency and mechanisms of target loss with mosunetuzumab are consistent with other targeted agents including rituximab and chimeric antigen receptor T-cell therapies. Given the importance of CD20 as a clinical target and the consistent observation of target loss as an acquired mechanism of resistance, understanding the frequency, distribution, and mechanisms underlying these phenomena are of critical importance.
CD20 levels may vary in R/R NHL due in large part to prior CD20-targeting treatments. Although most patients in this study had CD20-expressing tumors, CD20 was not part of the inclusion criteria, and patients with low/no CD20 were enrolled. The distribution of CD20 was not normal, with only a minor fraction of patients with intermediate (10%-74%) or very low levels of CD20 (<10%) before mosunetuzumab administration making an empirical determination of a level of target expression required for efficacy challenging. Clinical activity was observed across the distribution of CD20+PAX5+ expression except for levels <10%, at which objective responses were not seen. Consistent observations have been reported for other CD20-targeting bispecifics.34 Although CD20 expression was generally maintained during mosunetuzumab treatment, CD20 loss was observed in 1 of 30 patients, and expression was reduced (but not to <10%) in 2 patients to intermediate levels. Reduction of CD20 was seen more frequently in patients with intermediate CD20 levels before mosunetuzumab, suggesting that CD20 loss in these patients may be due to clonal heterogeneity, and patients who harbor pre-existing CD20– clones are more sensitive to this mechanism of resistance. In contrast to changes seen during treatment, patients with paired pre-mosunetuzumab and at-progression biopsy specimens (11 of 32 [34%]) showed significant decreases in CD20 levels to <10%. These decreases were observed across multiple histologies in patients with high (7 of 11) and intermediate (4 of 11) pre-mosunetuzumab CD20+ levels. Time to progression for these patients was more rapid than for patients who achieved a response and maintained CD20. CD20 loss was also seen in patients who did not achieve clinical benefit, suggesting that mosunetuzumab alone was insufficient to drive response, and potentially, these patients could benefit from mosunetuzumab in combination with another agent.
Our data demonstrate that multiple mechanisms regulate CD20 and may influence the activity of anti-CD20 targeting agents including mosunetuzumab. These include reduced transcription and acquisition of truncating mutations, as well as acquisition of variants potentially altering the therapeutic binding epitope. Variants within MS4A1 were identified in 13 of 156 patients (∼8%) before mosunetuzumab and in samples at progression. Although rare, variants identified before mosunetuzumab likely reflect mutations acquired during prior therapies, and some, such as L66R, have been previously described.21 These missense mutations were primarily in the transmembrane or extracellular domains and not associated with CD20 loss. In contrast, at progression, 4 of 5 patients had variants that encoded stop codons or caused truncations and were associated with CD20 loss. Two missense mutations (C167G and K175E) were identified in ECL2, which contains the epitope for anti-CD20 targeting therapies; these were not associated with CD20 loss, but patients harboring them did not respond to therapy. In vitro modeling of variants identified in patient samples confirmed loss of CD20 expression due to frameshift and truncating mutations. For the ECL2 variants, expression was maintained above background and present on the cell membrane. Despite sufficient expression and appropriate cellular localization, both T-cell activation and cell killing in response to CD20 × CD3 treatment was significantly reduced.
Although we were able to confirm the effect of these variants on mosunetuzumab activity, distinguishing whether these variants were acquired resistance mutations or enrichment of low-level resident mutations was hampered by the lack of sufficient clinical material for deeper sequencing. Several variants identified before mosunetuzumab were observed in previously published analyses using different data sets,18,21 lending credibility to variants within MS4A1 contributing to resistance to anti-CD20 therapies. However, variants identified at progression were novel, and based on the conserved epitope binding site, these variants might not be unique to mosunetuzumab and may impart resistance to other anti-CD20 agents.
The emergence of variants conferring resistance appears low (∼8%) in our analysis, but the use of circulating tumor DNA to monitor the emergence of mutations in CD20 may provide a more robust assessment of the prevalence of this type of resistance or even serve as a tool to anticipate resistance. Alternative mechanisms of reducing CD20 levels, including transcriptional regulation, were also seen at progression; however, generally, CD20 protein and transcript levels were concordant. Low CD20 levels that could not be explained by reduced transcription or presence of exonic mutations may be due to alternative splicing resulting in translation-competent or deficient isoforms ultimately leading to reduced CD20 translation, as described by Ang et al.35
Accumulating evidence suggests that target loss is a general mechanism of resistance to CD20 targeting agents. However, this study demonstrates that it only accounts for resistance in a subset of patients. Thus, alternative mechanisms of resistance need to be elucidated. Based on our observations that patients with CD20 levels <10% fail to respond to bispecific antibody therapy, assessment of CD20 expression before treatment is warranted to accurately inform sequential treatment and to improve clinical efficacy. Despite multiple mechanisms of CD20 regulation, readily available IHC assays using the intracellular epitope-binding antibody L26 can quantify CD20 levels to inform clinical decisions.
Although our analyses are based on a limited number of patients with biomarker-evaluable specimens, this population had similar characteristics to the intent-to-treat population in the mosunetuzumab trial (supplemental Table 2). The incidence of CD20 loss may differ based on the mechanisms of action of specific anti-CD20 therapy, duration of exposure, and the susceptibility of the histology to target loss. The frequency of CD20 loss we observed reflects a dual targeting (CD20 × CD3) approach with fixed-duration exposure. To our knowledge, this is the largest reported collection of paired biopsy specimens evaluated using a central, prespecified assay to assess the contribution of CD20 expression for clinical activity of a CD20 × CD3 bispecific antibody. These data demonstrate the adequacy of local CD20 IHC for determining CD20 expression for efficacy and detail multiple potential mechanisms resulting in the observed loss of CD20 associated with resistance to a CD20 × CD3 bispecific antibody.
Acknowledgments
Third-party editorial assistance, under the direction of all authors, was provided by Fiona Fernando, contract medical writer at Ashfield MedComms, an Inizio company, and was funded by F. Hoffmann-La Roche Ltd. Study NCT02500407 is sponsored by Genentech, Inc.
Authorship
Contribution: V.M., A.G.P., E.A.L., E.C.P., M.C.W., and S.Y. contributed to the study design; V.M. generated the figures; and all authors contributed to the acquisition, analysis, and interpretation of study data; critically reviewed the manuscript; and provided final approval for publication.
Conflict-of-interest disclosure: S.J.S. reports consultancy for Acerta, Celgene, Genentech, Inc., Novartis, and Pharmacyclics; research funding from Celgene, Gilead, Janssen Research & Development, Merck, Novartis, and Pharmacyclics; and membership on scientific advisory committee for Nordic Nanovector. L.-Y.H., C.R.B., V.M., A.G.P., K.H., E.A.L., H.K. and E.P. are employees of Genentech, Inc. and hold equity/stock in F. Hoffmann-La Roche Ltd/Genentech, Inc. S.E.A. reports consultancy for BeiGene and Ipsen; research funding from Novartis Canada; and honoraria from AbbVie, AstraZeneca, Gilead, Janssen, Palladin, and F. Hoffmann-La Roche Ltd/Genentech, Inc. N.L.B. reports research funding from ADC Therapeutics, Autolus, Bristol Myers Squibb, Celgene, Forty Seven, Gilead/Kite Pharma, Janssen, Merck, Millenium, Pharmacyclics, F. Hoffmann-La Roche Ltd/Genentech, Inc., and Seattle Genetics; and membership on advisory committee for ADC Therapeutics, Foresight Diagnostics, Kite, F. Hoffmann-La Roche Ltd/Genentech, Inc., and Seattle Genetics. M.J.M. is an equity holder in Merck; reports consultancy for Bayer, Juno Therapeutics, F. Hoffmann-La Roche Ltd/Genentech, Inc., Seattle Genetics, Takeda, and Teva; research funding from Bayer, GM Biosciences, Immunovaccine Technologies, Janssen, Pharmacyclics, F. Hoffmann-La Roche Ltd/Genentech, Inc., and Seattle Genetics; honoraria from ADC Therapeutics, AstraZeneca, Bayer, Bristol Myers Squibb, Celgene, Epizyme, Immunovaccine Technologies, IMV Therapeutics, Janssen, Kite, Pharmacyclics, Regeneron, F. Hoffmann-La Roche Ltd, Seagen, Seattle Genetics, and Takeda; membership on advisory committee for Genentech, Inc. and Merck; and stipends from ADC Therapeutics, AstraZeneca, Bristol Myers Squibb, Celgene, Epizyme, IMV Therapeutics, Kite, Regeneron, and Seagen. E.C.P. is a former employee of Genentech, Inc. and current equity holder. D.W. is a former employee of Genentech, Inc. M.C.W. and S.Y. are employees of Genentech, Inc. and hold equity and patents/royalties in F. Hoffmann-La Roche Ltd. L.E.B. declares no competing financial interests.
The current affiliation for E.C.P. and D.W. is Inhibrx, La Jolla, CA.
Correspondence: Elicia Penuel, Genentech, Inc, 1 DNA Way, South San Francisco, CA 94080-4918; email: penuel.elicia@gene.com.
References
Author notes
Individual patient level data, including deidentified clinical metadata, raw RNA-seq and WES data, and processed RNA-seq and WES data are available to researchers at the European Genome-Phenome Archive (accession number EGAS50000000151). To request access to such data, researchers can contact devsci-dac-d@gene.com.
The data will be released to such requesters with necessary agreements to enforce terms such as security, patient privacy, and consent of specified data use, consistent with evolving, applicable data protection laws. Up-to-date details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents are available at https://go.roche.com/data_sharing.
Anonymized records for individual patients across more than one data source external to Roche cannot and should not be linked because of a potential increase in risk of patient reidentification.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.