TO THE EDITOR:
The cyclic adenosine monophosphate response element-binding protein (CREBBP) gene is located on chromosome 16p13 and encodes a histone acetyltransferase having the same name that is involved in transcriptional regulation and cell cycle control.1,2 The translocation t(8;16)(p11;p13)[KAT6A::CREBBP] results in the disruption of CREBBP as well as its fusion to KAT6A, another gene important in transcription control. This fusion is sufficient for leukemogenesis and leads to a rare but well described type of acute myeloid leukemia (AML) with consistent biologic characteristics and a distinct gene expression profile.3-8 Although generally associated with inferior outcomes among adults, including a recent adjustment made by the European LeukemiaNet toward the adverse-risk group, there are variable reports regarding the prognostic significance of this fusion among pediatric patients.4,7,9 This prognostic variability is partially related to the high association of KAT6A::CREBBP with congenital leukemia as well as the phenomenon of spontaneous regression in a majority of these patients.4,10-17 Because of the poor outcomes associated with this alteration on North American protocols, patients aged >90 days with KAT6A::CREBBP have been reclassified as high risk in the active Phase 3 Children’s Oncology Group (COG) trial for de novo AML, AAML1831 (NCT04293562). Little is known about the prognostic significance of CREBBP sequence variants, including pathogenic single nucleotide variants (SNVs) and insertion/deletions (indels). In this letter, we report the prevalence and prognostic significance of all alterations involving CREBBP in pediatric patients with AML and their impact on co-occurring lesions, specifically t(8;21)[RUNX1::RUNX1T1].
CREBBP variant status was determined in a total of 2216 patients enrolled in 4 successive COG trials for de novo pediatric AML (NCT00003790, NCT00070174, NCT01407757, and NCT01371981) and associated with comprehensive clinical and cytogenetic information. Fusions involving CREBBP were prospectively obtained via conventional cytogenetics and retrospectively confirmed via RNA sequencing. Indels and SNVs were retrospectively interrogated via next generation sequencing18 and their pathogenicities were established in accordance with American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines. For patients with transcriptome data available, unsupervised clustering was done and their results were compared with those of patients without CREBBP alterations. The research was approved by the appropriate review boards and conducted in accordance with the Declaration of Helsinki. The Kaplan-Meier method was used to determine overall survival (OS) and event-free survival (EFS). The significance of predictor variables was tested using the log-rank statistic. The significance of observed difference in proportions was analyzed using Fisher exact test, and observed differences in medians were analyzed using the Kruskal-Wallis test. The Cox proportional hazards model was used to estimate a hazard ratio for EFS in a multivariable analysis.
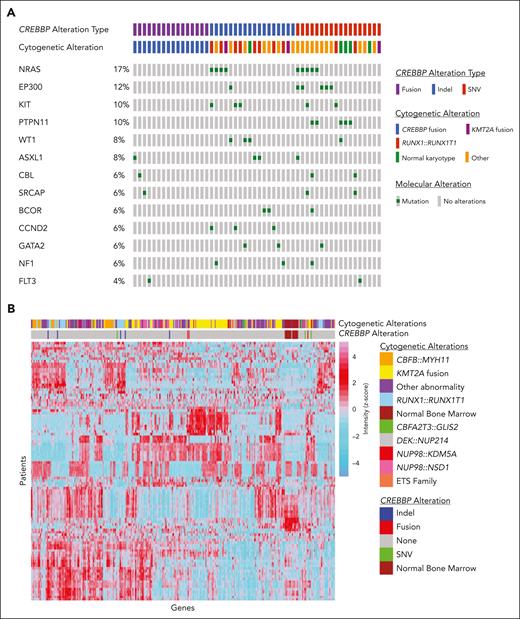
We identified 52 (2.3%) patients with a pathogenic alteration involving CREBBP, which is slightly higher than the incidence in publicly available adult AML databases (1.1%).19,20 This higher incidence is largely driven by fusions involving CREBBP (CREBBP/fus), which made up 31% (n = 16) of our cohort (compared with 12.5% in the adult database). The remaining 36 patients (69%) in our cohort had a CREBBP mutation (CREBBP/mut; supplemental Table 1, available on the Blood website). Eighteen of these were due to an indel (CREBBP/indel), including 15 leading to frameshift mutations and 3 leading to deleterious inframe insertions or deletions (Figure 1A). The other 18 were due to a SNV (CREBBP/SNV), including 17 deleterious missense or nonsense mutations and 1 splice-site mutation (Figure 1A). A single patient had 2 pathogenic missense mutations. Patients with CREBBP/fus were younger than patients with CREBBP/indel or CREBBP/SNV (median ages of 2.6 vs 8.7 vs 7.4 years, respectively; P = .056), including 4 patients with a CREBBP/fus diagnosed in the first 90 days of life. There was a higher prevalence of RUNX1::RUNX1T1 in patients with CREBBP/indel (n = 8) than in patients with CREBBP/SNV (n = 1) (44.4% vs 5.6%, respectively; P = .018; Figure 1A). In contrast, there was no association between CREBBP/SNV and a specific cytogenetic abnormality. There was a similar prevalence of cooccurring contemporarily defined high-risk lesions among patients with CREBBP/indel (n = 4; CBFA2T3::GLIS2, KMT2A::AFF1, MLLT10::PICALM, and NUP98::HOXA9) and those with CREBBP/SNV (n = 5; CBFA2T3::GLIS2, FUS::ERG, NUP98::KDM5A, ETV6::MNX1, and high allelic ratio FLT3-ITD). There was a paucity of cooccurring genomic mutations in patients with CREBBP/fus (Figure 1A). In contrast, genomic mutations most frequently cooccurring in patients with CREBBP/mut included mutations in RAS (25%), KIT (13.9%), and WT1 (11.1%). The frequency of these mutations were the same between patients with CREBBP/SNV and patients with CREBBP/indel and had a similar distribution to that of patients without CREBBP mutations. Transcriptome data were analyzed, and, consistent with previous reports, unsupervised clustering showed tight clustering of CREBBP/fus as well as a similar but distinct gene expression profile to that of KMT2A rearrangements.3,4 CREBBP/indel clustered together, which was likely driven by the cooccurrence of RUNX1::RUNX1T1, given its strong gene expression profile.21 In contrast, CREBBP/SNV did not show a specific clustering pattern (Figure 1B).
Molecular and transcriptome profiles based on CREBBP status. (A) Cytomolecular status and cooccurring mutational profile of patients based on CREBBP alterations, (B) Unsupervised clustering was performed across the entire transcriptome patient data set (n = 1079). Variance stabilizing transformation was performed (R function, vst), and scaled values were used to calculate the variance for each gene across all samples (R function, var). The genes were sorted based on variance, and the top 100 genes with highest variance were clustered (R function, ward.D2) and presented visually in a heatmap generated using pheatmap function in R.
Molecular and transcriptome profiles based on CREBBP status. (A) Cytomolecular status and cooccurring mutational profile of patients based on CREBBP alterations, (B) Unsupervised clustering was performed across the entire transcriptome patient data set (n = 1079). Variance stabilizing transformation was performed (R function, vst), and scaled values were used to calculate the variance for each gene across all samples (R function, var). The genes were sorted based on variance, and the top 100 genes with highest variance were clustered (R function, ward.D2) and presented visually in a heatmap generated using pheatmap function in R.
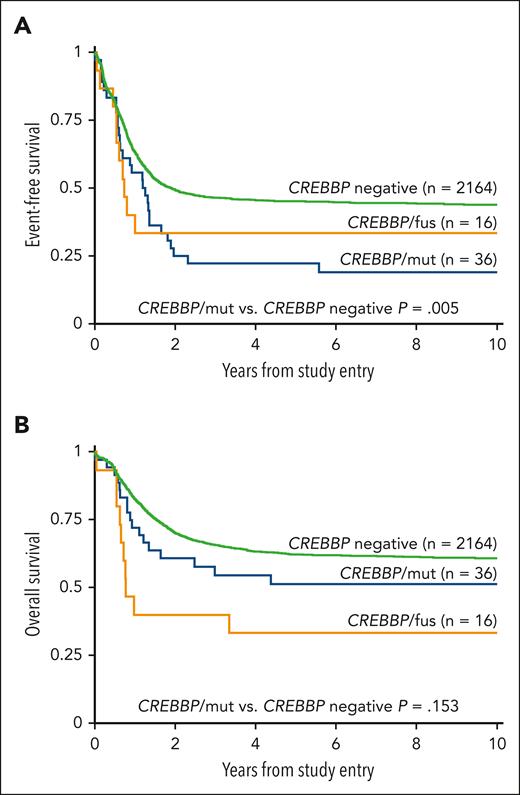
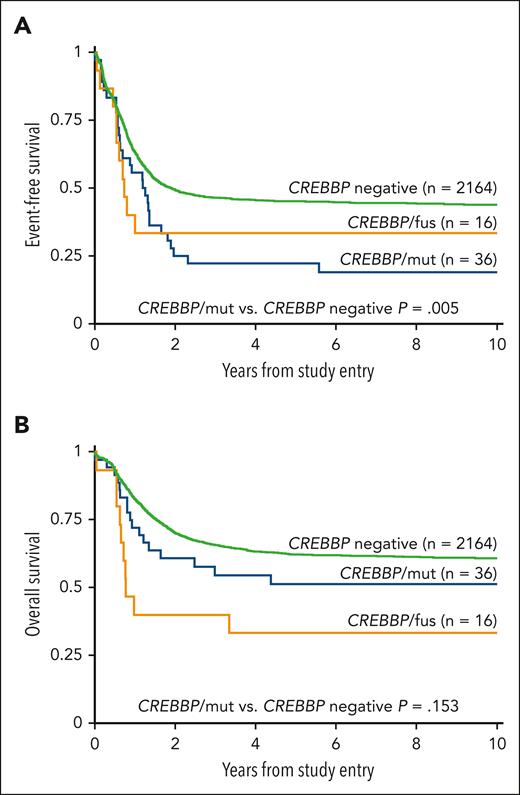
Evaluation of outcomes based on the type of alteration demonstrated a similar EFS between patients with CREBBP/fus and patients with CREBBP/mut (5-year EFS was 33.3% vs 22.2%, respectively; P = .801; Figure 2A). Patients with CREBBP/mut had a significantly worse EFS compared with patients without (5-year EFS was 22.2% vs 45.2%, respectively; P = .005; Figure 2A), and this inferior EFS is similar to that of patients with contemporarily defined high-risk but without CREBBP/mut (5-year EFS is 25.2%; P = .929). Furthermore, this poor EFS was maintained in patients with CREBBP/indel with cooccurring RUNX1::RUNX1T1, which was significantly worse than that of patients with RUNX1::RUNX1T1 but without CREBBP/indel (5-year EFS was 12.5% vs 67.7%, respectively; P = .001). When patients with cooccurring high-risk lesions were excluded from the analysis, the remaining patients with CREBBP/mut (n = 27) who would otherwise be considered low-risk maintained their poor EFS (5-year EFS was 29.6%). We performed a Cox regression analysis and found CREBBP/mut was an independent predictor of an inferior EFS in the presence of contemporarily defined cooccurring low- or high-risk cytomolecular alterations (hazard ratio, 1.71; 95% confidence interval, 1.18-2.47; P = .005; supplemental Table 2). Despite their poor EFS, patients with CREBBP/mut had a comparable OS with those without CREBBP disruptions (5-year OS was 51.4% vs 62.4%, respectively; P = .153, Figure 2B), demonstrating that these patients could successfully be salvaged following relapse. In contrast, patients with CREBBP/fus had an inferior OS (5-year OS was 33.3%; P = .002). Strikingly, all patients with CREBBP/fus that experienced a relapse ultimately died from their disease. Of the 5 long-term survivors with CREBBP/fus, 2 were diagnosed within the first 90 days of life and might have ultimately benefited from spontaneous regression.
Correlation of clinical outcomes with CREBBP status. (A) EFS based on CREBBP status, (B) OS based on CREBBP status.
Correlation of clinical outcomes with CREBBP status. (A) EFS based on CREBBP status, (B) OS based on CREBBP status.
We report the prevalence of CREBBP/fus in a large cohort of patients with pediatric AML with an incidence similar to that reported in a prior analysis.4 This is also the first report, to our knowledge, to comprehensively report the prevalence of CREBBP/mut in pediatric AML. Furthermore, we show that these patients have a poor EFS, regardless of the alteration type. This was especially notable in those patients with CREBBP/indel with a cooccurring RUNX1::RUNX1T1. The favorable EFS typically conferred by RUNX1::RUNX1T1 was abrogated by the cooccurrence of CREBBP/indel. Finally, the presence of a CREBBP/mut maintained independent prognostic significance for an inferior EFS, within the context of a Cox regression analysis.
Translocations between CREBBP and KAT6A in patients aged >90 days are considered as high risk in the active COG phase 3 trial. Survival of patients with relapsed CREBBP/fus AML is dismal and warrants novel interventions. Histone deacetylase inhibitors have shown preclinical promise in CREBBP mutated tumors, and further studies in AML are warranted.22,23 Given the comparably poor EFS and high salvage rates associated with CREBBP/mut AML, intensification of upfront treatment, including hematopoietic stem cell transplant, should be considered in this population.
Acknowledgments
This work was supported by grants from the St Baldrick’s Foundation; the National Cancer Institute's National Clinical Trials Network Operations Center (U10CA180886) and National Clinical Trials Network Statistics & Data Center (U10CA180899). The authors also thank Bayer, Jazz Pharmaceuticals, and Astellas Pharma Global Development for their support of CCG2961, AAML03P1, AAML0531, and AAML1031.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: A.J.L., R.E.R., J.L.S., and S.M. designed the research; A.J.L., K.H., R.E.R., J.L.S., S.M., T.A.A., R.B.G., P.K., E.A.K., and X.M. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: E.A.K. reports funding from Roche/Genentech for travel andaccommodation expenses. The remaining authors declare no competing financial interests.
Correspondence: Adam J. Lamble, Division of Hematology/Oncology, University of Washington, Seattle Children's Hospital, Seattle, WA; e-mail: adam.lamble@seattlechildrens.org.
References
Author notes
The online version of this article contains a data supplement.