Key Points
Transcriptomic analysis of PTCL tumors reveals recurrent MYCN overexpression and the presence of a MYC signature in 50% of PTCL cases.
EZH2 is a transcriptional cofactor for the MYCN-driven gene expression program, which confers sensitivity to HDAC inhibition.
Abstract
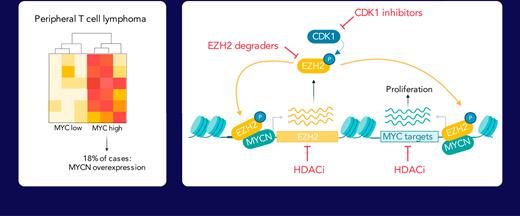
Peripheral T-cell lymphoma (PTCL) is a heterogeneous group of hematological cancers arising from the malignant transformation of mature T cells. In a cohort of 28 PTCL cases, we identified recurrent overexpression of MYCN, a member of the MYC family of oncogenic transcription factors. Approximately half of all PTCL cases was characterized by a MYC expression signature. Inducible expression of MYCN in lymphoid cells in a mouse model caused T-cell lymphoma that recapitulated human PTCL with an MYC expression signature. Integration of mouse and human expression data identified EZH2 as a key downstream target of MYCN. Remarkably, EZH2 was found to be an essential cofactor for the transcriptional activation of the MYCN-driven gene expression program, which was independent of methyltransferase activity but dependent on phosphorylation by CDK1. MYCN-driven T-cell lymphoma was sensitive to EZH2 degradation or CDK1 inhibition, which displayed synergy with US Food and Drug Administration–approved histone deacetylase (HDAC) inhibitors.
Introduction
Peripheral T-cell lymphoma (PTCL) is a heterogeneous group of rare but aggressive hematological cancers arising from the malignant transformation of mature postthymic T cells. Except for ALK-positive anaplastic large cell lymphoma (ALCL), patients with PTCL currently face an unfavorable prognosis: ∼75% of all patients with PTCL relapse after first-line chemotherapy treatment. Consequently, 5-year survival rates are as low as 10% to 30%, which underscores the urgent medical need for new therapeutic options.1-3
The MYC family of transcription factors has been widely studied regarding its oncogenic properties and has been implicated in many different cancer types. MYC amplification was previously described in PTCL and was found to be enriched in the GATA3+ PTCL, not otherwise specified (PTCL-NOS) subgroup.4 In accordance with these findings, MYC overexpression was associated with a worse prognosis in nodal PTCL.5MYCN, another member of the MYC oncogene family, was previously found to be amplified and/or aberrantly overexpressed in several solid tumors, including neuroblastoma, prostate cancer, and breast cancer.6 However, it was not yet described as an oncogene in PTCL, although it had been found in rare cases of MYC-negative Burkitt lymphoma.7 The oncogenic role of MYCN has already been extensively studied in neuroblastoma, in which amplification of MYCN is an initiating event present in 50% of high-risk patients.8 Deregulated MYCN has been shown to invade tissue-specific promoter and enhancers, leading to the activation of oncogenic transcriptional programs,9 and this MYCN-induced cell state was shown to be regulated by a transcriptional core regulatory circuitry consisting of a small number of transcription factors.10 Furthermore, a genome-wide CRISPR screen identified a specific dependency of MYCN-driven neuroblastoma on EZH2.11 Indeed, EZH2 was found to directly interact with MYCN, thereby inducing its stabilization by protecting it from ubiquitination, and EZH2 depletion leads to downregulation of the MYCN-driven transcriptional program independent of its methyltransferase activity.12 The oncogenic function of EZH2 has been mostly associated with its canonical function as the enzymatic component of the PRC2 complex driving gene repression. Remarkably, in multiple cancer types, EZH2 was found to have a noncanonical function outside of the PRC2 complex as a transcriptional activator via its partially disordered transactivation domain that can be unlocked through cancer-specific phosphorylation.13,14 This was found to be mostly independent of the methyltransferase activity.
In this study, we uncovered MYCN as an oncogenic driver in PTCL, and we identified EZH2 as an important cofactor for transcriptional activation of MYCN target genes.
Methods
Patient material
Samples from patients with PTCL were collected retrospectively from the tumor banks of UZ Leuven and the CHU Mont-Godinne and prospectively in UZ Leuven. For prospectively obtained samples, we obtained informed consent from all patients. All cases were reviewed by two hematopathologists.
Flow cytometry
Single-cell suspensions were prepared from infiltrated organs and incubated in red blood cell lysis buffer (150 mM NH4Cl, 0.1 mM EDTA, 10 mM KHCO3) for 10 minutes prior to staining. Cells were then washed with phosphate-buffered saline (PBS) and stained in the dark for 20 minutes at 4°C. Next, stained cells were washed with PBS and then analyzed on a MACSQuant VYB (Miltenyi) or on a LSRFortessa X-20 (BD Biosciences).
Annexin V stainings were performed using the APC Annexin V Apoptosis Detection Kit (BioLegend) according to the manufacturer’s instructions.
Intracellular stainings were performed using the Foxp3/Transcription Factor Staining Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Data were analyzed with the FlowJo software (Tree Star). Antibodies are listed in supplemental Table 1, available on the Blood website.
ChIPmentation ChIP-sequencing
ChIPmentation chromatin immunoprecipitation followed by sequencing (ChIP-seq) was performed as described previously.15-17 Primary spleen cells (20 to 50 million) were washed with PBS and cross-linked with 1% formaldehyde for 10 minutes and then quenched by addition of glycine. For nuclei isolation, cells were resuspended in 1× resuspension buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2) and left on ice for 10 min. Cells were collected and resuspended in RSBG40 buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 10% glycerol, 0.5% NP40) with 1:10 volume-to-volume ratio of 10% detergent (3.3% weight-to-volume ratio sodium deoxycholate, 6.6% volume-to-volume ratio Tween-40). Nuclei were resuspended in L3B buffer (10 mM Tris-Cl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-Lauroylsarcosine, 0.2% sodium dodecyl sulfate). Chromatin was fragmented to 200 to 400 bp using a Bioruptor (Diagenode) for 20 to 25 cycles (30 seconds on, 30 seconds off, high setting). The chromatin was supplemented with 1% Triton-X100 after fragmentation. Antibodies (listed in supplemental Table 4) were preconjugated to magnetic protein A/G beads (Millipore). Chromatin immunoprecipitation was carried out overnight. Tagmentation and library preparation was performed using the Nextera DNA library prep kit (Illumina). DNA was purified using triple-sided solid-phase reversible immobilization bead cleanup (Agencourt AMPure Beads, Beckman Coulter).
Dual luciferase assay
HEK293T cells at 80% to 90% confluency in a 12-well plate were transfected using 6 μL GeneJuice Transfection Reagent (Sigma-Aldrich, 70967) and 100 μL RPMI 1640 medium (Thermo Fisher) per well. Each well was transfected with 500 ng of pGL4.27 (Promega), in which we cloned a fragment of the murine Aurka promoter (supplemental Figure 3D) or no response element, 25 ng of renilla luciferase control reporter vector (RL-TK, Promega), and the respective plasmids for the expression of each transcription factor (MYCN, EZH2, and variants) or empty vector in triplicates. Cells were lysed after 24 hours with Passive Lysis Buffer and the luciferase signal was measured on a Victor multilabel plate reader (PerkinElmer) using the Dual-Luciferase Reporter Assay System (Promega). The signal was normalized to the Renilla luciferase activity.
In vitro and ex vivo inhibitor treatments
RPMI-8402 cells were seeded into 96-well plates at a concentration of 5.105 cells per mL in RPMI 1640 supplemented with 20% fetal calf serum. Cells harvested from the spleen of mice that developed MYCN-driven T-cell lymphoma (TCL) or TAL1 AKT leukemia18 were seeded into 96-well plates at a concentration of 1.106 cells per mL in RPMI 1640 supplemented with 20% fetal calf serum, 2 mM l-glutamine, nonessential amino acids solution (Gibco), 50 μM β-mercapto-ethanol, primocin (InVivoGen), 100 ng/mL interleukin-2, and 50 ng/mL interleukin-7.
The compounds (all dissolved in dimethyl sulfoxide [DMSO]) were added in a randomized fashion using a D300e digital dispenser (Tecan) and the DMSO concentration was normalized. All compounds are listed in supplemental Table 6. Plates were incubated overnight and cell proliferation was measured after 24 hours using the ATPlite luminescence system (PerkinElmer) using a Victor multilabel plate reader. For combination treatments with multiple compounds, data were analyzed using the SynergyFinder software.19
Data analysis
All statistical analyses were performed using Prism software (GraphPad Software).
Paired-end RNA-seq data were collected from the patient samples, and all other RNA-seq data are single 3′ end transcriptome data. Libraries were sequenced on a HiSeq 4000 with 125-bp single-end reads (Illumina). All data were first cleaned with fastq-mcf from ea-utils, after which a quality control was performed with FastQC. Subsequently the reads were mapped to the respective reference genomes (GRCh38/hg38 or GRCm38/mm10) with HISAT2. Further processing of the reads was executed with the SAMtools package and the number of reads per transcript was determined with HTSeq-count. DESeq2 was used to determine the significantly differentially expressed genes. Heat maps and expression plots were constructed with R. Gene set enrichment analyses were performed with the gene set enrichment analysis software from the Broad Institute. Possible regulatory transcription factor motifs were determined with i-CisTarget.20
ChIP-seq data with either specific antibodies against transcription factor or histone marks were preprocessed similarly as the RNA-seq data, ie, cleaning with fastq-mcf and quality control with FastQC. Subsequently, these cleaned reads were mapped to the respective reference genome (GRCm38/mm10) with the Bowtie2 software, after which the reads were further processed with SAMtools. The ChIP signals were normalized with the deepTools package (bamCoverage) and the peaks were called with MACS2. These peaks were then annotated with a custom script. The centered heat maps were constructed with the deepTools package (computeMatrix, plotHeatmap).
The RNA-seq and ChIP-seq data were deposited in the Gene Expression Omnibus database with accession number GSE198126.
Results
MYCN is aberrantly expressed in PTCL and drives a MYC target gene expression signature
To gain a deeper understanding of the mechanisms that drive PTCL development, we carried out RNA-seq on 28 PTCL samples, including 23 PTCL-NOS cases, 2 ALCL cases, and 3 follicular T-cell lymphoma (FTCL) cases.21 In 5 of these 28 PTCL cases (18%), we identified aberrant overexpression of MYCN (Figure 1A). In addition, we were able to identify additional cases with MYCN overexpression in publicly available PTCL data sets22–24 (Figure 1B), although MYCN overexpression was extremely rare in diffuse large B-cell lymphoma (DLBCL) (supplemental Figure 1A).
MYCN is aberrantly overexpressed in PTCL. (A) Normalized counts for MYCN in the Leuven PTCL cohort of 28 cases. (B) Normalized counts for MYCN in three published data sets (RNA-seq24 and microarray22,23). AITL, angioimmunoblastic T-cell lymphoma. (C) Heat map showing expression of MYC target genes from the hallmark gene sets in PTCL cases from the Leuven and Kyoto cohorts. Samples indicated in green harbor overexpression of MYCN. (D) Gene set enrichment analysis (GSEA) showing enrichment of different MYC target gene sets in the differentially expressed genes of PTCL with a high MYC signature compared with PTCL with a low MYC signature. NES, normalized enrichment score. (E) Normalized enrichment scores for different hallmark gene sets in MYC high vs low PTCL. (F) GSEA showing enrichment of an embryonic stem-cell-like signature as described by Wong et al.25 (G) Lugano staging for the patients with PTCL in the Leuven cohort. (H) Pie chart representing the sex of the patients with PTCL in the MYC low (left) and MYC high (right) subgroups. The difference between the female presence in the 2 groups was calculated using a hypergeometrical distribution (P = .016). See also supplemental Figure 1.
MYCN is aberrantly overexpressed in PTCL. (A) Normalized counts for MYCN in the Leuven PTCL cohort of 28 cases. (B) Normalized counts for MYCN in three published data sets (RNA-seq24 and microarray22,23). AITL, angioimmunoblastic T-cell lymphoma. (C) Heat map showing expression of MYC target genes from the hallmark gene sets in PTCL cases from the Leuven and Kyoto cohorts. Samples indicated in green harbor overexpression of MYCN. (D) Gene set enrichment analysis (GSEA) showing enrichment of different MYC target gene sets in the differentially expressed genes of PTCL with a high MYC signature compared with PTCL with a low MYC signature. NES, normalized enrichment score. (E) Normalized enrichment scores for different hallmark gene sets in MYC high vs low PTCL. (F) GSEA showing enrichment of an embryonic stem-cell-like signature as described by Wong et al.25 (G) Lugano staging for the patients with PTCL in the Leuven cohort. (H) Pie chart representing the sex of the patients with PTCL in the MYC low (left) and MYC high (right) subgroups. The difference between the female presence in the 2 groups was calculated using a hypergeometrical distribution (P = .016). See also supplemental Figure 1.
As MYC amplification was previously described in PTCL-NOS,4 and we now identified aberrant overexpression of MYCN, we investigated the expression of known MYC transcription family target genes in our PTCL cohort. Unsupervised clustering showed that approximately half of the PTCL cases are characterized by a high MYC signature (Figure 1C-D), and these cases showed the highest expression levels of MYCN, MYC, MYB, MYBL1, and/or MYBL2 (supplemental Figure 1B). Remarkably, the presence of a MYC signature was mutually exclusive with a diagnosis of Lennert lymphoma, a lymphoepithelioid variant of PTCL-NOS (Figure 1C; supplemental Figure 1C). PTCL cases with a high MYC signature showed strong positive enrichment of gene sets related to E2F targets, cell cycle checkpoints, and DNA repair (Figure 1E; supplemental Figure 1D), indicative of a proliferative state, which is in line with previous findings that describe a proliferative signature in PTCL that predicts a poorer survival.26 We also observed strong enrichment of a gene set representing an embryonic stem cell–like state that was previously associated with MYC-induced cancers25 (Figure 1F). Our MYC signature classification did not replicate the classification based on TBX21 or GATA3 expression22 (supplemental Figure 1E-H). Clinically, cases with a high MYC signature presented with more advanced disease, as reflected by the Lugano staging at diagnosis (Figure 1G), and were more often female (P = .016, Figure 1H). No difference in age at diagnosis was observed (supplemental Figure 1I).
Taken together, we identified MYCN as a novel recurrent oncogene in PTCL and uncovered the presence of a high MYC signature in approximately half of the PTCL cases, which correlates with a more aggressive and proliferative disease.
MYCN overexpression drives the development of TCLs in a bone marrow transplant mouse model
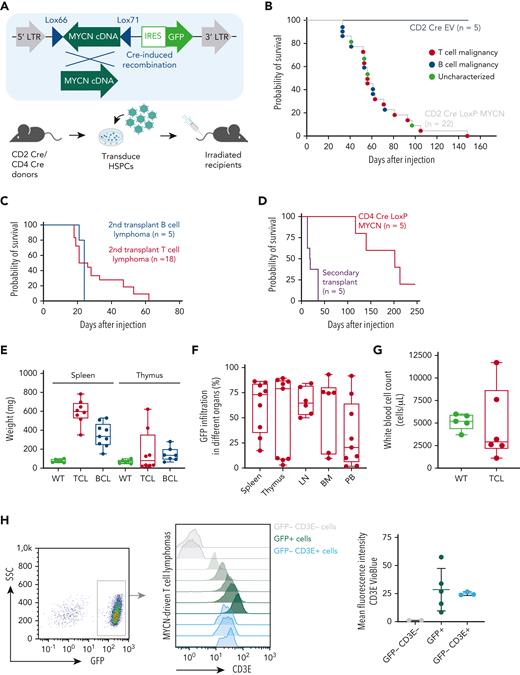
As MYCN overexpression had not been described in PTCL before, we aimed to further explore the oncogenic potential of MYCN overexpression in driving T-cell malignancies in vivo. We used a bone marrow transplant assay to generate mice expressing MYCN in a subset of hematopoietic stem and progenitor cells (HSPCs). However, constitutive expression of MYCN in HSPCs led to a rapid development of myeloid leukemia, as previously described27 (supplemental Figure 2A-B). To model TCL, we restricted MYCN expression to the lymphoid lineage by using a Cre-inducible retroviral vector16 in combination with CD2 Cre or CD4 Cre transgenic mice as donors (Figure 2A). Recipient mice transplanted with CD2 Cre HSPCs transduced with the inducible MYCN vector (also expressing green fluorescent protein [GFP]) developed lymphomas with a B- or T-cell phenotype with an incidence of 100% and a median survival of 56 days (Figure 2B; supplemental Figure 2C-E). Both T-cell and B-cell lymphomas were transplantable into secondary recipients with a short latency (Figure 2C). Recipient mice transplanted with CD4 Cre HSPCs transduced with the inducible MYCN vector uniquely developed TCLs with an incidence of 80% and a longer median latency (202 days) (Figure 2D; supplemental Figure 2G).
MYCN drives PTCL in a bone marrow transplant model. (A) Schematic overview of the bone marrow transplant assay strategy used to restrict MYCN expression to the lymphoid lineage. (B) Kaplan-Meier overall survival curve for mice transplanted with CD2 Cre HSPCs transduced with empty vector (EV) (n = 5) or the LoxP MYCN retroviral vector (n = 22). (C) Kaplan-Meier overall survival curve for secondary transplants of MYCN-driven B-cell lymphoma (n = 5) or MYCN-driven T-cell lymphoma (n = 18). (D) Kaplan-Meier overall survival curve for mice transplanted with CD4 Cre HSPCs transduced with the LoxP MYCN retroviral vector (n = 5) and secondary transplants from the resulting T-cell lymphomas (n = 5). (E) Spleen and thymus weights in mice that develop MYCN-driven B-cell lymphomas (BCL) or TCL or wild-type (WT) mice. (F) Infiltration of GFP-positive cells in different organs from mice that develop MYCN-driven TCL. LN, lymph node; BM, bone marrow; PB, peripheral blood. (G) White blood cell counts of WT mice or recipient mice transplanted with CD2 Cre cells transduced with the LoxP MYCN retroviral vector at end-stage disease (TCL). (H) Representative fluorescence-activated cell sorter (FACS) analysis showing CD3e expression in GFP-positive spleen cells of mice that developed MYCN-driven TCL (left and middle). The mean fluorescence intensity is shown on the right. SSC, side scatter. (I) Representative FACS analysis showing CD4 expression in GFP-positive spleen cells of mice that developed MYCN-driven TCL (left). The mean fluorescence intensity is shown on the right. (J) Hematoxylin and eosin stainings on a spleen from a mouse transplanted with HSPCs expressing EV (GFP) and a spleen from the murine MYCN-driven TCL. Scale bars indicate 200 μm (black) or 20 μm (white). (K) Immunofluorescent staining showing GFP or Cd3e in a spleen from a mouse transplanted with HSPCs expressing EV and a spleen from a mouse that developed MYCN-driven TCL. Scale bars indicate 200 μm. See also supplemental Figure 2. DAPI, 4′,6-diamidino-2-phenylindole.
MYCN drives PTCL in a bone marrow transplant model. (A) Schematic overview of the bone marrow transplant assay strategy used to restrict MYCN expression to the lymphoid lineage. (B) Kaplan-Meier overall survival curve for mice transplanted with CD2 Cre HSPCs transduced with empty vector (EV) (n = 5) or the LoxP MYCN retroviral vector (n = 22). (C) Kaplan-Meier overall survival curve for secondary transplants of MYCN-driven B-cell lymphoma (n = 5) or MYCN-driven T-cell lymphoma (n = 18). (D) Kaplan-Meier overall survival curve for mice transplanted with CD4 Cre HSPCs transduced with the LoxP MYCN retroviral vector (n = 5) and secondary transplants from the resulting T-cell lymphomas (n = 5). (E) Spleen and thymus weights in mice that develop MYCN-driven B-cell lymphomas (BCL) or TCL or wild-type (WT) mice. (F) Infiltration of GFP-positive cells in different organs from mice that develop MYCN-driven TCL. LN, lymph node; BM, bone marrow; PB, peripheral blood. (G) White blood cell counts of WT mice or recipient mice transplanted with CD2 Cre cells transduced with the LoxP MYCN retroviral vector at end-stage disease (TCL). (H) Representative fluorescence-activated cell sorter (FACS) analysis showing CD3e expression in GFP-positive spleen cells of mice that developed MYCN-driven TCL (left and middle). The mean fluorescence intensity is shown on the right. SSC, side scatter. (I) Representative FACS analysis showing CD4 expression in GFP-positive spleen cells of mice that developed MYCN-driven TCL (left). The mean fluorescence intensity is shown on the right. (J) Hematoxylin and eosin stainings on a spleen from a mouse transplanted with HSPCs expressing EV (GFP) and a spleen from the murine MYCN-driven TCL. Scale bars indicate 200 μm (black) or 20 μm (white). (K) Immunofluorescent staining showing GFP or Cd3e in a spleen from a mouse transplanted with HSPCs expressing EV and a spleen from a mouse that developed MYCN-driven TCL. Scale bars indicate 200 μm. See also supplemental Figure 2. DAPI, 4′,6-diamidino-2-phenylindole.
The mice that developed TCLs presented with splenomegaly (Figure 2E), and some animals developed paraplegia. We typically observed high infiltration of GFP-positive cells in spleen and lymph nodes and more variable levels of infiltration in thymus, bone marrow, and peripheral blood (Figure 2F), but no strong increase in white blood cell count (Figure 2G). Phenotypic analysis revealed that the malignant T cells expressed surface CD3E (Figure 2H) and showed variable levels of CD4 expression (Figure 2I) but low or absent CD8 expression (supplemental Figure 2G) and low or absent terminal deoxynucleotidyltransferase (TdT) expression (supplemental Figure 2H). Histopathological analysis showed a profound loss of spleen architecture and infiltration of mitotic cells and confirmed high levels of CD3E and GFP expression in the affected organs (Figure 2J-K).
To further investigate the transcriptional programs underlying MYCN-driven TCL, we performed a global gene expression analysis using RNA-seq on six murine MYCN-driven TCL samples and on CD4 T cells isolated from the spleens of three wild type mice (supplemental Figure 3A-B). Motif analysis revealed a strong enrichment of E2F and MYC motifs in genes that are upregulated in murine MYCN-driven TCL (Figure 3A). In line with this, gene set enrichment analysis revealed strong enrichment of gene sets containing E2F target genes, MYC target genes, gene sets related to cell cycle progression, DNA repair, and MYC-driven stemness25 (Figure 3B-C), as was the case in the human PTCL cases characterized by a high MYC signature. Malignant cells expressed intermediate levels of Pdcd1 but low levels of the T follicular helper cell markers Icos, Cxcr5, and Bcl6 and the ALCL markers Batf3 and Irf4 (supplemental Figure 3C-E), ruling out an angioimmunoblastic T-cell lymphoma or ALCL phenotype. Furthermore, we did not observe significant changes in the B-cell compartment (supplemental Figure 3F). Expression analysis of T-cell receptor variable genes confirmed the oligoclonal nature of the lymphomas (supplemental Figure 3G).
Murine MYCN-driven T-cell lymphoma recapitulates human PTCL with a high MYC signature. (A) Heat map showing the top differentially expressed genes in MYCN-driven TCL compared with normal CD4 T cells and depicting the transcription factor binding motifs of the top transcription factors identified by i-CisTarget in the up- and downregulated genes. NES = normalized enrichment score. (B) NESs for different hallmark gene sets in MYCN-driven TCL compared with normal CD4 T cells. (C) GSEA showing enrichment of an embryonic stem cell–like signature as described by Wong et al.25 (D) Centered read-density heat maps showing binding locations of MYCN, H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. Heat maps centered and ranked on MYCN signal strength. (E) ChIP-seq peak occurrence of MYCN in relation to promoter regions (H3K4me3+) and enhancer regions (H3K27ac+ and H3K4me3−). (F) Venn diagram showing the total amount of ChIP-seq peaks of MYCN in promoters of genes that are differentially (Diff.) expressed in MYCN-driven TCL compared with normal CD4 T cells. (G) Volcano plot showing the log2 fold change of direct MYCN target genes in MYCN-driven TCL compared with CD4 T cells. (H) GSEA to show negative enrichment of the top 500 downregulated direct MYCN target genes and positive enrichment of the top 500 upregulated direct MYCN target genes in the differentially expressed genes in PTCL with a high vs low MYC signature. See also supplemental Figure 3.
Murine MYCN-driven T-cell lymphoma recapitulates human PTCL with a high MYC signature. (A) Heat map showing the top differentially expressed genes in MYCN-driven TCL compared with normal CD4 T cells and depicting the transcription factor binding motifs of the top transcription factors identified by i-CisTarget in the up- and downregulated genes. NES = normalized enrichment score. (B) NESs for different hallmark gene sets in MYCN-driven TCL compared with normal CD4 T cells. (C) GSEA showing enrichment of an embryonic stem cell–like signature as described by Wong et al.25 (D) Centered read-density heat maps showing binding locations of MYCN, H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. Heat maps centered and ranked on MYCN signal strength. (E) ChIP-seq peak occurrence of MYCN in relation to promoter regions (H3K4me3+) and enhancer regions (H3K27ac+ and H3K4me3−). (F) Venn diagram showing the total amount of ChIP-seq peaks of MYCN in promoters of genes that are differentially (Diff.) expressed in MYCN-driven TCL compared with normal CD4 T cells. (G) Volcano plot showing the log2 fold change of direct MYCN target genes in MYCN-driven TCL compared with CD4 T cells. (H) GSEA to show negative enrichment of the top 500 downregulated direct MYCN target genes and positive enrichment of the top 500 upregulated direct MYCN target genes in the differentially expressed genes in PTCL with a high vs low MYC signature. See also supplemental Figure 3.
Subsequently, we performed ChIP-seq for MYCN and various histone marks. MYCN was found to bind most often in promoter regions, and MYCN binding locations were enriched in H3K4me3 and H3K27ac (Figure 3D-E). We integrated our ChIP-seq and RNA-seq data to determine which genes are directly regulated by MYCN (eg, genes that are bound by MYCN in their promoter region and that are differentially expressed in MYCN-driven TCL compared with nonmalignant CD4 T cells). We identified 3754 direct MYCN target genes, from which 2429 genes were upregulated and 1325 genes were downregulated (Figure 3F-G). These direct MYCN target genes were also found to be enriched in differentially expressed genes in PTCL with a high vs a low MYC signature (Figure 3H).
Altogether, these results show that MYCN acts as a strong oncogene in the lymphoid lineage, and that this mouse model of MYCN-driven TCL reflects human PTCL with a high MYC signature.
Ezh2 is implicated in transcriptional activation of direct MYCN target genes
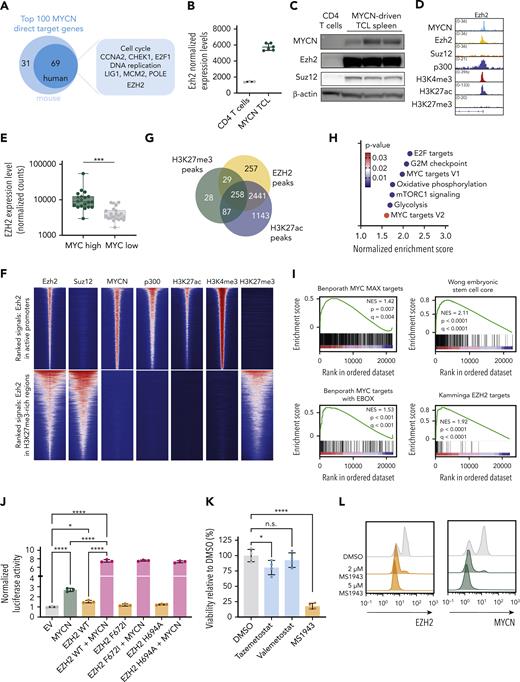
Within the most significantly upregulated direct MYCN target genes that were also differentially expressed in human PTCL with a high MYC signature, we identified genes related to cell cycle and DNA replication (Figure 4A). Surprisingly, Ezh2, but not Suz12, Eed, or Ezh1, was found among the most significantly upregulated direct MYCN target genes (Figure 4B-D; supplemental Figure 4A-B). Similarly, we observed increased EZH2 expression levels in human PTCL with a high MYC signature (Figure 4E). Interestingly, Ezh2 peaks were found both in repressed regions (defined by H3K27me3 marks) where cobinding with Suz12 was observed, as well as in active promoter regions (defined by H3K4me3 and H3K27ac marks, p300 binding, and active gene expression). Co-occupancy of Ezh2 with MYCN and p300 in active promoter regions was not accompanied by Suz12 or the repressive H3K27me3 mark, indicating that Ezh2 binds these regions independent of the PRC2 complex (Figure 4F). Remarkably and unexpectedly, when looking at direct MYCN target genes, Ezh2 binding was mostly associated with H3K27ac rather than H3K27me3 (Figure 4G) and these activating Ezh2 peaks were enriched for the same gene sets as MYCN-driven TCL and for a gene set containing EZH2 targets that preserve stem cell potential in hematopoietic stem cells28 (Figure 4H-I). Together, these findings suggest that Ezh2 acts as a transcriptional cofactor for MYCN.
EZH2 is a cofactor for transcriptional activation together with MYCN. (A) Schematic representation of the top 100 MYCN direct target genes. (B) Normalized expression levels of Ezh2 in CD4 T cells and in MYCN-driven lymphoma cells. (C) Western blot showing expression levels of MYCN, Ezh2, and Suz12 in normal CD4 T cells and in MYCN-driven TCL spleen cells. (D) Representative ChIP-seq tracks for the Ezh2 promoter showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (E) Normalized counts for EZH2 from the PTCL-NOS and FTCL cases from the Leuven cohort and the Kyoto cohort. Patients with overexpression of MYCN are indicated with stars. (F) Centered read-density heat maps showing binding locations of Ezh2, Suz12, MYCN, p300, H3K27ac, H3K4me3, and H3K27me3 in MYCN-driven TCL. Heat maps centered and ranked on Ezh2 signal strength in active promoters (top) or in regions enriched for H3K27me3 (bottom). (G) Venn diagram of EZH2 peaks in direct MYCN target genes and their overlap with H3K27ac or H3K27me3 peaks. (H) Normalized enrichment scores for different hallmark gene sets in the differentially expressed direct EZH2+MYCN target genes. (I) GSEA showing enrichment of different MYC target gene sets and stemness-associated gene sets in the differentially expressed direct EZH2+MYCN target genes. (J) Luciferase assay in HEK293T cells showing transcriptional activation of Firefly luciferase by MYCN and/or EZH2 (wild type or methyltransferase-inactivating mutants). (K) Forty-eight–hour ex vivo treatment of MYCN-driven TCL spleen cells with 2 μM of the EZH2 methyltransferase inhibitors tazemetostat and valemetostat, or the EZH2 degrader MS1943. n.s., not significant. (L) Fluorescence-activated cell sorter (FACS) analysis of EZH2 and MYCN in MYCN-driven TCL spleen cells treated ex vivo with DMSO or MS1943 for 12 hours. (M) In vitro treatment of RPMI-8402 cells with 5 μM MS1943, tazemetostat (Taz), or valemetostat (Val). (N) FACS analysis of EZH2 and MYCN in RPMI-8402 cells after treatment with 7 μM MS1943 or DMSO. (O) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 36 hours with 2.5 μM MS1943 or DMSO. See also supplemental Figures 4 and 5. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
EZH2 is a cofactor for transcriptional activation together with MYCN. (A) Schematic representation of the top 100 MYCN direct target genes. (B) Normalized expression levels of Ezh2 in CD4 T cells and in MYCN-driven lymphoma cells. (C) Western blot showing expression levels of MYCN, Ezh2, and Suz12 in normal CD4 T cells and in MYCN-driven TCL spleen cells. (D) Representative ChIP-seq tracks for the Ezh2 promoter showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (E) Normalized counts for EZH2 from the PTCL-NOS and FTCL cases from the Leuven cohort and the Kyoto cohort. Patients with overexpression of MYCN are indicated with stars. (F) Centered read-density heat maps showing binding locations of Ezh2, Suz12, MYCN, p300, H3K27ac, H3K4me3, and H3K27me3 in MYCN-driven TCL. Heat maps centered and ranked on Ezh2 signal strength in active promoters (top) or in regions enriched for H3K27me3 (bottom). (G) Venn diagram of EZH2 peaks in direct MYCN target genes and their overlap with H3K27ac or H3K27me3 peaks. (H) Normalized enrichment scores for different hallmark gene sets in the differentially expressed direct EZH2+MYCN target genes. (I) GSEA showing enrichment of different MYC target gene sets and stemness-associated gene sets in the differentially expressed direct EZH2+MYCN target genes. (J) Luciferase assay in HEK293T cells showing transcriptional activation of Firefly luciferase by MYCN and/or EZH2 (wild type or methyltransferase-inactivating mutants). (K) Forty-eight–hour ex vivo treatment of MYCN-driven TCL spleen cells with 2 μM of the EZH2 methyltransferase inhibitors tazemetostat and valemetostat, or the EZH2 degrader MS1943. n.s., not significant. (L) Fluorescence-activated cell sorter (FACS) analysis of EZH2 and MYCN in MYCN-driven TCL spleen cells treated ex vivo with DMSO or MS1943 for 12 hours. (M) In vitro treatment of RPMI-8402 cells with 5 μM MS1943, tazemetostat (Taz), or valemetostat (Val). (N) FACS analysis of EZH2 and MYCN in RPMI-8402 cells after treatment with 7 μM MS1943 or DMSO. (O) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 36 hours with 2.5 μM MS1943 or DMSO. See also supplemental Figures 4 and 5. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
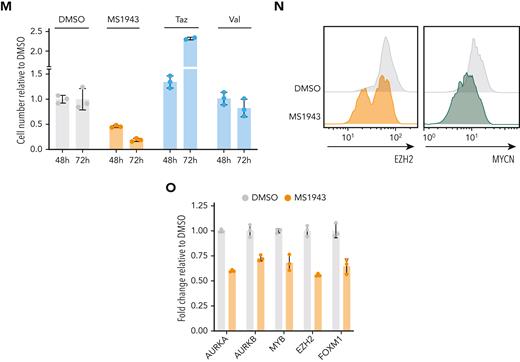
To further investigate the role of EZH2 in MYCN transcriptional activity, we performed a luciferase reporter experiment with a luciferase expression construct under control of the Aurka promoter, a region that we identified as strongly bound by MYCN and EZH2 (supplemental Figure 4C). Expression of MYCN alone caused increased luciferase activity, which was further enhanced when MYCN was coexpressed with EZH2. Mutations in EZH2 that inactivate the methyltransferase activity had no effect on the transcriptional activity in the luciferase reporter assay, indicating that the enzymatic activity of EZH2 is not required (Figure 4J; supplemental Figure 4D-F). In line with this, MYCN-driven TCL cells showed only a slight sensitivity to inhibitors that block the methyltransferase activity of Ezh2, yet they were strongly sensitive to targeted Ezh2 degradation by MS1943 (Figure 4K; supplemental Figure 4G-H). Furthermore, Ezh2 depletion induced MYCN destabilization and subsequent degradation (Figure 4L), as previously reported in neuroblastoma.12 We confirmed these findings in the human T-cell line RPMI-8402, which is also characterized by high MYCN expression and a strong MYC signature (Figure 4M-O; supplemental Figure 5A-D).
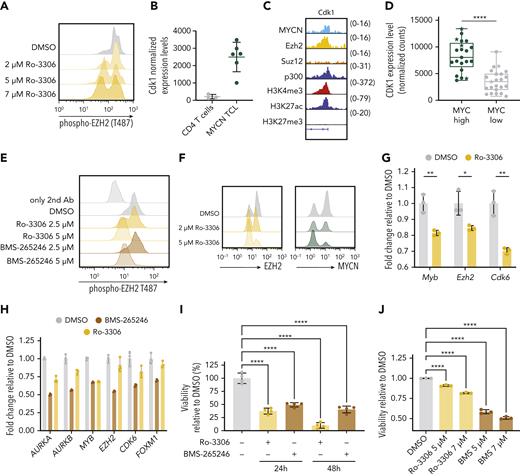
Subsequently, we performed immunoprecipitation of Ezh2 in MYCN-driven TCL cells, followed by mass spectrometry. We uncovered phosphorylation of Ezh2 at Threonine 487 (Figure 5A; supplemental Figure 4I), a posttranslational modification catalyzed by CDK1, which was previously associated with decreased interaction with PRC2 complex members SUZ12 and EED.29 Interestingly, Cdk1 was found to be a direct target gene of MYCN and Ezh2 in MYCN-driven TCL (Figure 5B-C). Moreover, CDK1 was significantly upregulated in human PTCL with a high MYC signature (Figure 5D). Treatment with a CDK1 inhibitor reduced phosphorylation of EZH2 (Figure 5A,E), leading to degradation of both EZH2 and MYCN (Figure 5F; supplemental Figure 5E), and reduced expression of genes directly regulated by MYCN and EZH2 (Figure 5G-H). Consequently, MYCN-driven TCL cells and RPMI-8402 cells were sensitive to CDK1 inhibition (Figure 5I-J).
Ezh2-driven transcriptional activation requires CDK1-mediated phosphorylation. (A) FACS analysis of phospho-EZH2 (T487) in MYCN-driven TCL spleen cells treated ex vivo with DMSO or Ro-3306 (CDK1 inhibitor) for 8 hours. (B) Normalized expression levels of Cdk1 in CD4 T cells and in MYCN-driven lymphoma cells. (C) Representative ChIP-seq tracks for the Cdk1 promoter showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (D) Normalized counts for CDK1 from the PTCL cases from the Leuven cohort and the Kyoto cohort. Stars indicate patients with MYCN overexpression. (E) FACS analysis of phospho-EZH2 (T487) in RPMI-8402 cells treated for 16 hours with CDK1 inhibitors Ro-3306 or BMS-265246. (F) FACS analysis of EZH2 or MYCN in MYCN-driven TCL spleen cells treated ex vivo with DMSO or Ro-3306 for 8 hours. (G) qRT-PCR analysis of MYCN and EZH2 target gene expression in MYCN-driven TCL cells treated ex vivo for 8 hours with 4 μM Ro-3306. Data are represented as mean ± standard deviation. (H) qRT-PCR analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 36 hours with Ro-3306 or BMS-265246. (I) Twenty-four- and 48-hour ex vivo treatment of MYCN-driven TCL spleen cells with 2 μM of the CDK1 inhibitors Ro-3306 or BMS-265246. (J) Sixty-hour treatment of RPMI-8402 cells with CDK1 inhibitors Ro-3306 and BMS-265246. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Ezh2-driven transcriptional activation requires CDK1-mediated phosphorylation. (A) FACS analysis of phospho-EZH2 (T487) in MYCN-driven TCL spleen cells treated ex vivo with DMSO or Ro-3306 (CDK1 inhibitor) for 8 hours. (B) Normalized expression levels of Cdk1 in CD4 T cells and in MYCN-driven lymphoma cells. (C) Representative ChIP-seq tracks for the Cdk1 promoter showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (D) Normalized counts for CDK1 from the PTCL cases from the Leuven cohort and the Kyoto cohort. Stars indicate patients with MYCN overexpression. (E) FACS analysis of phospho-EZH2 (T487) in RPMI-8402 cells treated for 16 hours with CDK1 inhibitors Ro-3306 or BMS-265246. (F) FACS analysis of EZH2 or MYCN in MYCN-driven TCL spleen cells treated ex vivo with DMSO or Ro-3306 for 8 hours. (G) qRT-PCR analysis of MYCN and EZH2 target gene expression in MYCN-driven TCL cells treated ex vivo for 8 hours with 4 μM Ro-3306. Data are represented as mean ± standard deviation. (H) qRT-PCR analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 36 hours with Ro-3306 or BMS-265246. (I) Twenty-four- and 48-hour ex vivo treatment of MYCN-driven TCL spleen cells with 2 μM of the CDK1 inhibitors Ro-3306 or BMS-265246. (J) Sixty-hour treatment of RPMI-8402 cells with CDK1 inhibitors Ro-3306 and BMS-265246. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Collectively, these data show that, in the context of PTCL, EZH2 plays an important role outside of the PRC2 complex as a transcriptional coactivator together with MYCN, thereby reinforcing the transcriptional programs established by MYCN overexpression.
MYCN-driven TCL is addicted to high levels of EZH2 and displays a strong sensitivity for histone deacetylase (HDAC) inhibition
Having established EZH2 as a strong transcriptional cofactor for MYCN, we wanted to assess EZH2 inhibition as therapeutic approach for MYCN-driven TCL. Therefore, we performed an ex vivo treatment of MYCN-driven TCL cells with the EZH2 selective degrader MS1943, and we could observe a strong dependency on Ezh2 (Figure 6A). Similarly, we inhibited Ezh2 phosphorylation on T487 using the CDK1 inhibitor Ro-3306, and this induced apoptosis in MYCN-driven TCL cells (Figure 6B-C). As expected,30 MYCN-driven TCL cells also showed sensitivity to BET inhibition (supplemental Figure 6A).
MYCN-driven TCL is sensitive to EZH2 depletion and HDAC inhibition. (A) Dose-response curve for 24-hour treatment with the EZH2 degrader MS1943 on ex vivo cultured MYCN-driven TCL cells. (B) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 on ex vivo cultured MYCN-driven TCL cells. (C) Annexin V staining of MYCN-driven TCL cells treated ex vivo for 8 hours with DMSO, 10 nM romidepsin, 5 μM Ro-3306, 5 μM MS1943, or a combination of these inhibitors. (D) Normalized counts for HDAC1, HDAC2, and HDAC3 from the PTCL cases from the Leuven cohort and the Kyoto cohort. (E) Representative ChIP-seq tracks for the Hdac1, Hdac2, and Hdac3 promoter regions showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (F) Normalized expression levels of Hdac1, Hdac2, and Hdac3 in CD4 T cells and in MYCN-driven lymphoma cells. n.s., not significant. (G) Dose response curve for 24-hour treatment with the HDAC inhibitors romidepsin and belinostat on ex vivo cultured MYCN-driven TCL cells. (H) Dose-response curve for 48-hour treatment of RPMI-8402 cells with different HDAC inhibitors. (I) qRT-PCR analysis of MYCN and EZH2 target gene expression in MYCN-driven TCL cells treated ex vivo for 4 hours with 500 nM romidepsin. Data are represented as mean ± standard deviation. (J) qRT-PCR analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 16 hours with HDAC inhibitors belinostat or romidepsin. (K) Dose response curve for 24-hour treatment with the EZH2 degrader MS1943 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (L) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + MS1943 (max zero interaction potential [ZIP] synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (M) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (N) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + Ro-3306 (max ZIP synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (O) Viability of RPMI-8402 cells treated for 40 hours with the HDAC inhibitor romidepsin (Romi) (3 nM), the EZH2 degrader MS1943 (5 μM), the CDK1 inhibitor Ro3306 (5 μM), the EZH2 methyltransferase inhibitor tazemetostat (Taz) (5 μM), or a combination. See also supplemental Figure 6. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
MYCN-driven TCL is sensitive to EZH2 depletion and HDAC inhibition. (A) Dose-response curve for 24-hour treatment with the EZH2 degrader MS1943 on ex vivo cultured MYCN-driven TCL cells. (B) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 on ex vivo cultured MYCN-driven TCL cells. (C) Annexin V staining of MYCN-driven TCL cells treated ex vivo for 8 hours with DMSO, 10 nM romidepsin, 5 μM Ro-3306, 5 μM MS1943, or a combination of these inhibitors. (D) Normalized counts for HDAC1, HDAC2, and HDAC3 from the PTCL cases from the Leuven cohort and the Kyoto cohort. (E) Representative ChIP-seq tracks for the Hdac1, Hdac2, and Hdac3 promoter regions showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (F) Normalized expression levels of Hdac1, Hdac2, and Hdac3 in CD4 T cells and in MYCN-driven lymphoma cells. n.s., not significant. (G) Dose response curve for 24-hour treatment with the HDAC inhibitors romidepsin and belinostat on ex vivo cultured MYCN-driven TCL cells. (H) Dose-response curve for 48-hour treatment of RPMI-8402 cells with different HDAC inhibitors. (I) qRT-PCR analysis of MYCN and EZH2 target gene expression in MYCN-driven TCL cells treated ex vivo for 4 hours with 500 nM romidepsin. Data are represented as mean ± standard deviation. (J) qRT-PCR analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 16 hours with HDAC inhibitors belinostat or romidepsin. (K) Dose response curve for 24-hour treatment with the EZH2 degrader MS1943 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (L) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + MS1943 (max zero interaction potential [ZIP] synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (M) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (N) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + Ro-3306 (max ZIP synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (O) Viability of RPMI-8402 cells treated for 40 hours with the HDAC inhibitor romidepsin (Romi) (3 nM), the EZH2 degrader MS1943 (5 μM), the CDK1 inhibitor Ro3306 (5 μM), the EZH2 methyltransferase inhibitor tazemetostat (Taz) (5 μM), or a combination. See also supplemental Figure 6. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
As EZH2 degraders are not yet ready to use in clinical practice, we subsequently investigated the potential of clinically available inhibitors. The class I HDAC inhibitors romidepsin and belinostat are already approved by the US Food and Drug Administration for treatment of PTCL but only induce durable response in a subset of patients, and no reliable biomarkers are available yet.1 We observed upregulation of class I HDACs in PTCL cases with a high MYC signature (Figure 6D), and in line with this, Hdac1 and Hdac2 were found to be direct MYCN targets in MYCN-driven TCL (Figure 6E-F). Therefore, we investigated the sensitivity of MYCN-driven TCL to HDAC inhibition. The class I HDAC inhibitors romidepsin and belinostat both induced strong inhibitory effects at low concentrations in MYCN-driven TCL (Figure 6G; supplemental Figure 6B) and in RPMI-8402 (Figure 6H), which resulted in downregulation of MYCN or EZH2 target genes (Figure 6I-J) and induction of apoptosis after only 8 hours of treatment (Figure 6C).
Finally, we combined HDAC inhibition with EZH2 degradation, CDK1 inhibition, or BET inhibition. We calculated synergy using the zero interaction potential method19 and observed synergistic effects between EZH2 degradation or dephosphorylation and HDAC inhibition (Figure 6K-O; supplemental Figure 6C-F).
Discussion
PTCL is a heterogeneous and poorly characterized malignancy, for which very few treatment options other than chemotherapy are currently available. Despite recent advances in our understanding of the genetic and transcriptomic landscape of PTCL, no novel targeted treatment strategies have been developed, as it became apparent that PTCL tumors are characterized by many different aberrations that occur at a low frequency. For many of these genetic or transcriptomic alterations, their exact contribution to PTCL development is unknown.
In this study, we uncovered MYCN as a novel recurrent oncogenic driver in 18% of PTCL cases. The oncogenic role of MYCN was already extensively studied in other tumor types, such as neuroblastoma and neuroendocrine prostate cancer, but its implication in PTCL was not previously described. Similar to neuroblastoma mouse models, in which MYCN overexpression is restricted to noradrenergic neurons,31 overexpression of MYCN in the lymphoid lineage was sufficient to drive development of both B- and T-cell malignancies. Although MYCN overexpression is activated during early lymphoid development in our bone marrow transplant models, recipient mice only develop mature T- or B-cell malignancies. This suggests that, although MYCN has been described as an oncogenic driver in T-cell acute lymphoblastic leukemia,32 additional factors are needed to block T-cell differentiation, and that MYCN preferentially transforms mature lymphoid cells.
Interestingly, ∼50% of all PTCL cases was characterized by a MYC signature, indicating that MYC, whether or not accompanied by MYCN overexpression, is likely to play a significant role in PTCL development. MYC amplification has indeed been described before in PTCL-NOS and was significantly associated with GATA3+ PTCL-NOS.4 In addition, MYC expression was shown to predict a poorer survival in PTCL.5 However, the molecular mechanism of MYC-driven T-cell transformation remains elusive.
Intriguingly, we identified EZH2 as an essential cofactor for MYCN-driven transcriptional activation in TCLs. The dependency of MYCN-overexpressing tumors on EZH2 already became apparent from a genome-wide CRISPR screen in MYCN-amplified neuroblastoma, which revealed a specific dependency on PRC2 complex members EZH2, EED, and SUZ12.11 Moreover, MYCN was shown to directly activate EZH2 expression. Nevertheless, EZH2 binding in these tumors was strongly correlated with repressive histone marks, and the methyltransferase activity of EZH2 was found to be essential in MYCN-driven neuroblastoma.11 Similarly, in neuroendocrine prostate cancer, EZH2 was found to act as a corepressor of MYCN target genes, and the cancer cells showed sensitivity to EZH2 methyltransferase inhibition.33 Thus, the mechanism of cooperation between MYCN and EZH2 in these tumors seems more reliant on the canonical function of EZH2 to induce gene repression within the PRC2 complex. In sharp contrast, our data from MYCN-driven TCL illustrate that EZH2 functions in PTCL as a transcriptional activator independent from its methyltransferase function in the PRC2 complex. Furthermore, a recent study showed that EZH2 can protect MYCN from degradation by inhibiting interaction with the ubiquitin ligase FBXW7 and that this is mostly independent of its methyltransferase activity.12 Interestingly, the authors suggest that other cancers with high expression levels of MYCN or MYC could also be dependent on high levels of EZH2.12 Indeed, in mixed-lineage leukemia (MLL)-rearranged leukemia, EZH2 was found to bind Myc via its transactivation domain, and EZH2 and Myc were found to colocalize on promoters, resulting in transcriptional activation of non-PRC2 target genes. These leukemias were also strongly sensitive to EZH2 degradation.34 In patients with PTCL characterized with a high MYC signature but without overexpression of MYCN, we also observed high expression levels of EZH2 and activation of similar gene expression patterns, indicating that EZH2 could play an analogous role as transcriptional activator together with MYC family transcription factors in this broader subgroup of PTCL.
Our findings on the oncogenic mechanisms of MYCN and EZH2 provide inspiration for improved targeted treatment strategies for patients with PTCL with a high MYC expression signature. We found a striking sensitivity for HDAC inhibition in our MYCN-driven TCL mouse model, which showed synergistic effects with EZH2 degradation or dephosphorylation via CDK1 inhibition. In line with our findings, high levels of EZH2 were found to be correlated with high levels of HDAC1/2 in human PTCL tumors, and high levels of EZH2 and/or HDAC2 were associated with advanced clinical stage and with a significantly worse prognosis.35 As the US Food and Drug Administration–approved HDAC inhibitors induce significant toxicities and are only effective in a limited group of patients, combination therapy of HDAC inhibition with EZH2-targeting compounds could improve their efficacy while decreasing their toxicity, as combination therapy will allow for lower doses. Finally, our findings imply that treatment strategies beyond EZH2 methyltransferase inhibitors, such as EZH2 degraders,34,36 need to be considered for EZH2-overexpressing tumors.
In conclusion, we have identified MYCN as a novel oncogenic driver in PTCL, and we demonstrate its direct cooperation with EZH2 as a transcriptional coactivator of the MYCN-driven gene expression program, which can be effectively targeted by EZH2 degradation or dephosphorylation combined with HDAC inhibition.
Acknowledgments
The authors thank the KU Leuven Genomics Core, the VIB proteomics core (Sara Dufour), the VIB Histopathology Expertise Center, and the VIB FACS expertise center (Jochen Lamote) for their technical services.
This study was supported by the Fund Tom Debackere for lymphoma research and by Stichting Tegen Kanker (2018/1272). M.V.B. and S.D. are supported by postdoctoral mandates for fundamental research from Stichting tegen Kanker. K.D. and Q.V.T. are supported by a PhD fellowship from the Research foundation Flanders. C.P. is supported by a postdoctoral fellowship from the Research Foundation Flanders. T.T. holds a Mandate for Fundamental and Translational Research from Stichting tegen Kanker (2019-091) and is a cofounder of the Fund “Me to You” supporting research in lymphoma and leukemia. D.D. is supported by a postdoctoral mandate for translational research from Kom op Tegen Kanker.
Authorship
Contribution: M.V.B. designed the study, collected and analyzed data, and wrote the manuscript; K.D., Q.V.T., N. Meeuws, C.P., S.P., N. Mentens, K.J., O.G., and D.N. collected and analyzed data; S.D. performed software analysis; S.O., K.K., C.G., and T.T. provided resources; and J.C. and D.D. supervised the study and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Cools, Campus Gasthuisberg, Herestr 49 B912, 3000 Leuven, Belgium; e-mail: jan.cools@kuleuven.be; and Daan Dierickx, Campus Gasthuisberg, Herestr 49 B912, 3000 Leuven, Belgium; e-mail: daan.dierickx@kuleuven.be.
References
Author notes
This study makes use of RNA-sequencing data generated by Department of Pathology and Tumor Biology, Kyoto University, as described by Watatani et al.24 The RNA-sequencing and ChIP-sequencing data were deposited in the Gene Expression Omnibus (GEO) database with accession number GSE198126. Requests for resources and reagents should be directed to and will be fulfilled by Jan Cools (jan.cools@kuleuven.be).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




![MYCN-driven TCL is sensitive to EZH2 depletion and HDAC inhibition. (A) Dose-response curve for 24-hour treatment with the EZH2 degrader MS1943 on ex vivo cultured MYCN-driven TCL cells. (B) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 on ex vivo cultured MYCN-driven TCL cells. (C) Annexin V staining of MYCN-driven TCL cells treated ex vivo for 8 hours with DMSO, 10 nM romidepsin, 5 μM Ro-3306, 5 μM MS1943, or a combination of these inhibitors. (D) Normalized counts for HDAC1, HDAC2, and HDAC3 from the PTCL cases from the Leuven cohort and the Kyoto cohort. (E) Representative ChIP-seq tracks for the Hdac1, Hdac2, and Hdac3 promoter regions showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (F) Normalized expression levels of Hdac1, Hdac2, and Hdac3 in CD4 T cells and in MYCN-driven lymphoma cells. n.s., not significant. (G) Dose response curve for 24-hour treatment with the HDAC inhibitors romidepsin and belinostat on ex vivo cultured MYCN-driven TCL cells. (H) Dose-response curve for 48-hour treatment of RPMI-8402 cells with different HDAC inhibitors. (I) qRT-PCR analysis of MYCN and EZH2 target gene expression in MYCN-driven TCL cells treated ex vivo for 4 hours with 500 nM romidepsin. Data are represented as mean ± standard deviation. (J) qRT-PCR analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 16 hours with HDAC inhibitors belinostat or romidepsin. (K) Dose response curve for 24-hour treatment with the EZH2 degrader MS1943 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (L) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + MS1943 (max zero interaction potential [ZIP] synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (M) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (N) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + Ro-3306 (max ZIP synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (O) Viability of RPMI-8402 cells treated for 40 hours with the HDAC inhibitor romidepsin (Romi) (3 nM), the EZH2 degrader MS1943 (5 μM), the CDK1 inhibitor Ro3306 (5 μM), the EZH2 methyltransferase inhibitor tazemetostat (Taz) (5 μM), or a combination. See also supplemental Figure 6. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/23/10.1182_blood.2022016428/4/m_blood_bld-2022-016428-gr6a.jpeg?Expires=1768355033&Signature=beEfoQkEuwQJreBP4wi85sP1xShIJUKZzLFrtrYDn~YL1XZ0p9kQAtf1zlXzB-boq4I89kojv~DsSnQFRTrp3iKOZt88bEMWR29aQumm4uWv5Vi87u8X0JV56FchX-uMVxaHQ1Hqwxfq2F7YqNfnyRgR92spX7qiE8mI9FLoXLnCcCDxcc0qWzwXUP0lCIVA~t977dhTrbFeCWrk0qSlEr7HXBJNi-bg9mSYdxcFPwAT0EHod8X6Pal372XnieLvI40ZJXWAd1eyls2pNXElJVfEerQTqOCSl5nw7Uf6tvV9nNna05dVI~NeG8UlkZGL6~mk7wh7-B86mFQRiQOckw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![MYCN-driven TCL is sensitive to EZH2 depletion and HDAC inhibition. (A) Dose-response curve for 24-hour treatment with the EZH2 degrader MS1943 on ex vivo cultured MYCN-driven TCL cells. (B) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 on ex vivo cultured MYCN-driven TCL cells. (C) Annexin V staining of MYCN-driven TCL cells treated ex vivo for 8 hours with DMSO, 10 nM romidepsin, 5 μM Ro-3306, 5 μM MS1943, or a combination of these inhibitors. (D) Normalized counts for HDAC1, HDAC2, and HDAC3 from the PTCL cases from the Leuven cohort and the Kyoto cohort. (E) Representative ChIP-seq tracks for the Hdac1, Hdac2, and Hdac3 promoter regions showing binding of MYCN, Ezh2, Suz12, p300, and histone marks H3K4me3, H3K27ac, and H3K27me3 in MYCN-driven TCL. (F) Normalized expression levels of Hdac1, Hdac2, and Hdac3 in CD4 T cells and in MYCN-driven lymphoma cells. n.s., not significant. (G) Dose response curve for 24-hour treatment with the HDAC inhibitors romidepsin and belinostat on ex vivo cultured MYCN-driven TCL cells. (H) Dose-response curve for 48-hour treatment of RPMI-8402 cells with different HDAC inhibitors. (I) qRT-PCR analysis of MYCN and EZH2 target gene expression in MYCN-driven TCL cells treated ex vivo for 4 hours with 500 nM romidepsin. Data are represented as mean ± standard deviation. (J) qRT-PCR analysis of MYCN and EZH2 target gene expression in RPMI-8402 cells treated for 16 hours with HDAC inhibitors belinostat or romidepsin. (K) Dose response curve for 24-hour treatment with the EZH2 degrader MS1943 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (L) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + MS1943 (max zero interaction potential [ZIP] synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (M) Dose-response curve for 24-hour treatment with the CDK1 inhibitor Ro-3306 with or without simultaneous romidepsin treatment on ex vivo cultured MYCN-driven TCL cells. (N) Synergy matrix plot showing δ scores for ex vivo cultured MYCN-driven TCL cells treated with romidepsin + Ro-3306 (max ZIP synergy score = maximal score for a specific dose combination; average ZIP synergy score = the average δ score for the whole range of concentrations shown in the synergy matrix). (O) Viability of RPMI-8402 cells treated for 40 hours with the HDAC inhibitor romidepsin (Romi) (3 nM), the EZH2 degrader MS1943 (5 μM), the CDK1 inhibitor Ro3306 (5 μM), the EZH2 methyltransferase inhibitor tazemetostat (Taz) (5 μM), or a combination. See also supplemental Figure 6. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/23/10.1182_blood.2022016428/4/m_blood_bld-2022-016428-gr6b.jpeg?Expires=1768355033&Signature=afi9xa-WNqWd52TH5anB5h-y7mChhqYq7Po1e0~xDPmOzQnC1DKAaMLYfCIOKJkmQUeYR3wa4gZqLoCv-3lk8rCY76NsDna2ULDEVsgtgh-0GpmqeYixQgY1XgUr8va~VLpmbReaQ9oo4u7xjH8XMK6qFS27515u0xlokHxykqXmMKYRhP6RKJvzfv42lh4YtuOUP4qTCbMp-CHyBhv4jg-06VLcUxFegIRsfkVTLI2Rtq1kfb3s1oAHrPf2mjZsssKCLaWxVSZCxZ~S3GUfR2CDOqP3lzMF-culKeLrVTUD4ReiqMH9PP3iH5xFLhydw9nhHcVI5FawayxTTSe9ew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
