Key Points
Almost one-third of myeloma patients have genetic alterations in CRBN (IMiD binding protein) by the time they are refractory to POM.
Genetic changes to CRBN are associated with inferior outcomes to POM-based regimes in those already refractory to LEN.
Abstract
Emergence of drug resistance to all available therapies is the major challenge to improving survival in myeloma. Cereblon (CRBN) is the essential binding protein of the widely used immunomodulatory drugs (IMiDs) and novel CRBN E3 ligase modulator drugs (CELMoDs) in myeloma, as well as certain proteolysis targeting chimeras (PROTACs), in development for a range of diseases. Using whole-genome sequencing (WGS) data from 455 patients and RNA sequencing (RNASeq) data from 655 patients, including newly diagnosed (WGS, n = 198; RNASeq, n = 437), lenalidomide (LEN)-refractory (WGS, n = 203; RNASeq, n = 176), and pomalidomide (POM)-refractory cohorts (WGS, n = 54; RNASeq, n = 42), we found incremental increases in the frequency of 3 CRBN aberrations, namely point mutations, copy losses/structural variations, and a specific variant transcript (exon 10 spliced), with progressive IMiD exposure, until almost one-third of patients had CBRN alterations by the time they were POM refractory. We found all 3 CRBN aberrations were associated with inferior outcomes to POM in those already refractory to LEN, including those with gene copy losses and structural variations, a finding not previously described. This represents the first comprehensive analysis and largest data set of CBRN alterations in myeloma patients as they progress through therapy. It will help inform patient selection for sequential therapies with CRBN-targeting drugs.
Introduction
Myeloma remains incurable because of the universal emergence of drug resistance. Identification of resistance mechanisms in relapsed/refractory myeloma would facilitate appropriate targeting of novel therapeutics to resistant myeloma cells.
Antimyeloma therapies immunomodulatory drugs (IMiDs) and their newer derivatives CRBN E3 ligase modulators (CELMoDs) bind the E3 ligase substrate-recognition adapter protein cereblon (CRBN). CRBN is essential for the therapeutic effect of these drugs in myeloma.1,2 CRBN mutations have been described as rare events (<1%) in newly diagnosed (ND) patients,3,4 but they have been reported at a higher frequency (12%) in IMiD-treated patients, although the numbers reported have been low.5 Beyond mutation, CRBN function may be impaired by other genomic (structural variation, copy loss) or transcriptomic aberrations (epigenetic, RNA splicing/stability). Gene expression loss and transcript splice variation have been associated with acquired resistance to IMiDs lenalidomide (LEN) and pomalidomide (POM).6-8 One-third of LEN-refractory patients go on to respond to POM.9 However, a systematic analysis of the diversity of CRBN changes associated with acquired resistance to IMiD-based therapies and their clinical significance has not been undertaken. We report CRBN changes identified in 454 patients by whole-genome sequencing (WGS) and 655 patients by RNA sequencing (RNASeq), including ND, LEN-refractory, and POM-refractory cases.
Methods
DNA and RNA extracted from bead-enriched CD138+ myeloma cells and peripheral blood germ line DNA were collected at baseline and relapse from patients recruited to trials CC4074-MM010 (STRATUS; registered at www.clinicaltrials.gov as #NCT01712789; entry criteria included LEN-refractory status),9 CC220-MM001 (registered at www.clinicaltrials.gov as #NCT02773030),10 and CC122-ST-001MM2 (registered at www.clinicaltrials.gov as #NCT01421524) (entry criteria to both included LEN- and POM-refractory status). In all cases, clinical annotation data were collected. Progression-free survival (PFS) data were not available for the latter 2 phase 1/dose-finding trials. Illumina WGS (depth coverage 60/30x tumor/germline) and RNASeq were performed. Newly diagnosed (ND) patient data from the IFM/DFCI-2009 trial (registered at www.clinicaltrials.gov as #NCT01191060; WGS)11 and the Myeloma Genome Project12 (RNASeq) were also used. Computational methods are described in supplementary Methods (supplemental Figure 1, available on the Blood Web site). This analysis is complementary to a landscape analysis of relapsed/refractory myeloma using the same cohorts, the subject of a separate publication (in preparation).
Results and discussion
We included WGS data from 198 ND, 203 LEN-refractory, and 54 POM-refractory patients and RNASeq data from 437 ND, 176 LEN-refractory, and 42 POM-refractory patients (summary patient characteristics listed in supplemental Tables 1 and 2). Only those with confirmed drug-refractory status were included in the LEN- and POM-refractory cohorts, defined as myeloma patients with either nonresponsive or progressive disease while taking the named drug or progressive disease within 60 days of stopping it.
CRBN genetic variation and copy loss increase in incidence at sequential IMiD-refractory states
CRBN mutations were detected in 1 (0.5%) of 198 ND myeloma patients, 5 (2.2%) of 203 LEN-refractory patients, and 5 (9%; 8 mutations detected) of 54 POM-refractory patients (Figure 1A), significant increases (P < .05) from baseline. Mutation clonal fraction distribution (Figure 1A) remained variable at LEN- and POM-refractory states, and mutations displayed no clustering along the CRBN gene (Figure 1B), but structural modeling predicted a majority to have a deleterious effect on protein stability or function (supplemental Figure 2).
Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN increases with sequential IMiD refractoriness. (A) Incidence of CRBN mutations (left-handed system [LH] y-axis) and their cancer clonal fractions (CCFs; right-handed system [RH] y-axis) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (B) Amino acid changes resulting from the mutations in CRBN gene found at each state. CCF of each mutation shown. (C) Incidence of copy loss or structural variation (LH y-axis) and their CCFs (RH y-axis; copy loss only) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (D) Diagrammatic representation of site of structural variant breakpoints identified at LEN refractoriness. (E) Change in ratio of exon 10–spliced/full-length transcripts between ND, LEN-, and POM-refractory states; ratio expressed as log2(exon 10–spliced/full-length transcript count) × tumor sample purity. Significant difference detected by Kruskal-Wallis 1-way analysis of variance (nonparametric). (F) Landscape of total and coincidence of 4 types of CRBN aberrations in LEN- and POM-refractory cases analyzed. *Mutations from same patient.
Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN increases with sequential IMiD refractoriness. (A) Incidence of CRBN mutations (left-handed system [LH] y-axis) and their cancer clonal fractions (CCFs; right-handed system [RH] y-axis) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (B) Amino acid changes resulting from the mutations in CRBN gene found at each state. CCF of each mutation shown. (C) Incidence of copy loss or structural variation (LH y-axis) and their CCFs (RH y-axis; copy loss only) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (D) Diagrammatic representation of site of structural variant breakpoints identified at LEN refractoriness. (E) Change in ratio of exon 10–spliced/full-length transcripts between ND, LEN-, and POM-refractory states; ratio expressed as log2(exon 10–spliced/full-length transcript count) × tumor sample purity. Significant difference detected by Kruskal-Wallis 1-way analysis of variance (nonparametric). (F) Landscape of total and coincidence of 4 types of CRBN aberrations in LEN- and POM-refractory cases analyzed. *Mutations from same patient.
CRBN gene copy loss occurred in 3 (1.5%) of 198 ND patients, 16 (7.9%) of 203 LEN-refractory patients, and 13 (24%) of 54 POM-refractory patients (Figure 1C), again significant increases (P < .05) from baseline. CRBN copy loss was compared with background loss of heterozygosity (LOH) rate using a permutation test with false discovery rate correction. In LEN-refractory patients, CRBN loss was not enriched beyond background LOH. However, in POM-refractory patients, CRBN LOH was significantly enriched (P = .006). The size of the lost region varied widely, but neither whole-chromosome nor homozygous loss was seen (supplemental Figure 3). CRBN gene expression was no different in cases with gene copy loss (supplemental Figure 4B), so how copy loss of CRBN could affect IMiD resistance remains unclear. Additional structural variants (translocations, inversions) of the CRBN gene or promoter region were found only in the LEN-refractory cohort (Figure 1C); all but 1 of those identified were highly likely to impair gene function (Figure 1D).
Exon 10–spliced CRBN transcripts increase in incidence at sequential IMiD-refractory states
In a subcohort of the cases described here, we previously reported that pretherapy CRBN expression, as measured by quantitative reverse transcription polymerase chain reaction, did not correlate with subsequent response to POM.13 We confirmed this lack of correlation by RNASeq (data not shown), although as previously described,7 overall CRBN expression was lower at LEN- and/or POM-refractory states (supplemental Figure 4C). However, many splice variants of CRBN have been described,14 including a transcript lacking exon 10, deleting the LEN/POM-binding region.2 This splice variant is not expected to bind or support IMiD function. We found that the ratio of exon 10–spliced/full-length CRBN transcripts, adjusted for sample purity, was significantly increased in POM-refractory compared with ND or LEN-refractory patients (P < .0001; Figure 1E). Full-length CRBN gene expression did not significantly vary between cases with high vs low exon 10–spliced ratio (supplemental Figure 4A).
The ratio of exon 10–spliced (ENST00000424814.5)/full-length transcripts (ENST00000231948.8) has previously been proposed to correlate with outcome after IMiD-based regimens.8 We determined the optimum sample purity-adjusted cutoff for a high exon 10–spliced/full-length transcript ratio to be 2.6 (supplemental Figure 5). Using this cutoff, the overall incidence of CRBN alterations in LEN-refractory cases was 20.7%. In POM-refractory cases, it was 29.6% (Figure 1F), although poorer sample purity in the POM-refractory cohort means high exon 10–spliced incidence may have been underreported, once ratio adjustment by sample purity was applied (supplemental Figures 6 and 7; supplemental Methods).
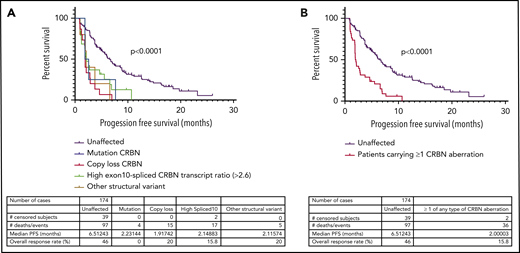
In LEN-refractory patients, the presence of CRBN genetic changes or high exon 10–spliced ratio correlates with PFS
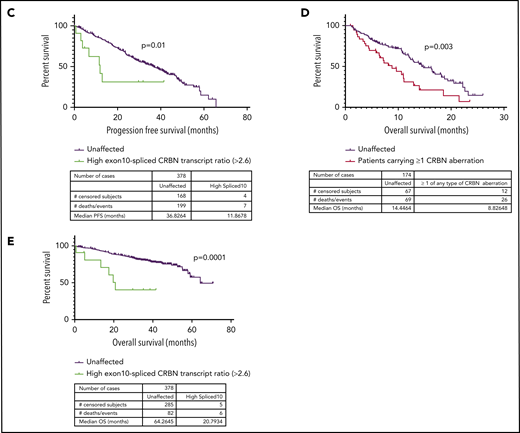
Next, we examined the relationship between the incidence of CRBN aberrations and PFS in LEN-refractory patients with both WGS and RNASeq data available (n = 174), treated with a POM-based regimen. Overall, PFS was significantly reduced (P < .0001) in LEN-refractory patients with mutations (n = 4), copy losses (n = 15), structural variations (n = 6), or high exon 10–spliced ratio (n = 17) (Figure 2A-B), compared to patients with no CRBN aberrations. We found that this association was also present in ND myeloma patients undergoing induction therapy. Only high exon 10–spliced ratio had sufficient incidence at new diagnosis to assess correlation, and it was associated with significantly reduced PFS (Figure 2C). Induction regimen was too varied to perform subgroup analysis specifically for IMiD-based induction regimen, but 62.4% (236 of 378 patients age <75 years for whom sample purity data were available) received a regimen containing an IMiD drug. In both cohorts, overall survival was also significantly reduced in those with CRBN aberrations (Figure 2D-E; aberrations by type shown in supplemental Figure 8). The incidence of high-risk cytogenetics was not different between CRBN-aberrant and unaffected groups (supplemental Table 2).
Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN correlates with outcome to POM in a LEN-refractory cohort. High exon 10–spliced transcript correlates with outcome to induction therapy in ND myeloma. (A) PFS of those who carried the 4 types of CRBN aberration vs those unaffected in the LEN-refractory cohort exposed to POM-based therapy. (B) PFS of those carrying ≥1 of any type of CRBN aberration vs those unaffected in the LEN-refractory cohort exposed to POM-based therapy. (C) Association of high exon 10–spliced CRBN transcript with PFS in the ND cohort undergoing induction therapy. (D) Overall survival (OS) of those carrying ≥1 of any type of CRBN aberration vs those unaffected in the LEN-refractory cohort exposed to POM-based therapy. (E) Association of high exon 10–spliced CRBN transcript with OS in the ND cohort undergoing induction therapy. Comparison of survival curves by log-rank testing in all cases.
Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN correlates with outcome to POM in a LEN-refractory cohort. High exon 10–spliced transcript correlates with outcome to induction therapy in ND myeloma. (A) PFS of those who carried the 4 types of CRBN aberration vs those unaffected in the LEN-refractory cohort exposed to POM-based therapy. (B) PFS of those carrying ≥1 of any type of CRBN aberration vs those unaffected in the LEN-refractory cohort exposed to POM-based therapy. (C) Association of high exon 10–spliced CRBN transcript with PFS in the ND cohort undergoing induction therapy. (D) Overall survival (OS) of those carrying ≥1 of any type of CRBN aberration vs those unaffected in the LEN-refractory cohort exposed to POM-based therapy. (E) Association of high exon 10–spliced CRBN transcript with OS in the ND cohort undergoing induction therapy. Comparison of survival curves by log-rank testing in all cases.
This is the first attempt to quantify the range of defects in CRBN, the molecular target of IMiDs and CELMoDs. Our analysis confirms the rarity of CRBN mutations in ND myeloma. As patients receive therapy, the incidence of mutations and other genetic alterations increases with the acquisition of LEN- and POM-refractory states. All alteration types are associated with adverse clinical outcomes, irrespective of CRBN gene expression. Interestingly, the drug binding region of the protein is not favored in these mutations, which occur along the length of the protein. We describe structural variants in the CRBN gene for the first time and show association with adverse outcomes.
Taken together, direct CRBN genetic alterations occur in nearly 30% of relapsed/refractory myeloma patients treated with IMiDs, making it the single most clinically significant contributor to clinical resistance. The novel CELMoDs iberdomide and CC92480 are currently in clinical trials in the LEN/POM-refractory myeloma population (registered at www.clinicaltrials.gov as #NCT02773030 and #NCT03374085). It is premature to ascertain the impact of CRBN dysregulations on the efficacy of CELMoDs at present, although analysis of data from ongoing trials may reveal this in time. This initial focused analysis excluded CRBN-independent genomic and transcriptomic features of resistance, nor did we explore the underlying mechanisms that cause such CRBN aberrations and result in drug resistance, which will be the subject of future analyses.
For original data, please e-mail the corresponding author.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by the NIHR Oxford Biomedical Research Centre (P.D., N.A.-P., K.R., P.V.) and a Celgene-Oxford Fellowship (S.G.). S.G. and P.V. work in a UK Medical Research Council–funded institute. Funding for data generation, processing, and storage was provided by Bristol-Myers-Squibb.
Authorship
Contribution: S.G. designed research questions, analyzed and interpreted the data, and wrote the manuscript; N.A.-P., F.T., P.D., and M.O.E. performed data analysis, statistical design, and data interpretation; P.P.C. generated supplemental Figure 2; K.-T.T., M.H., and D.R. curated and maintained clinical trial data sets; E.F., N.B., P.N., K.R., and P.V. assisted with study design and the manuscript; E.F. and F.T. performed oversight and management of resources (data generation, infrastructure, and processing); and A.T. designed research questions, oversaw scientific direction, and assisted with the manuscript.
Conflict-of-interest disclosure: S.G., N.A.-P., N.B., and P.V. receive research funding from Bristol-Myers Squibb. F.T., M.O.E., E.F., A.T., and K.-T.T. are employees and equity shareholders of Bristol-Myers Squibb. P.P.C. is an equity shareholder of Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Anjan Thakurta, Bristol-Myers Squibb, 556 Morris Ave, Summit, NJ 07901; e-mail: anjan.thakurta@bms.com.
REFERENCES
Author notes
N.A.-P. and F.T. contributed equally to this work.


![Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN increases with sequential IMiD refractoriness. (A) Incidence of CRBN mutations (left-handed system [LH] y-axis) and their cancer clonal fractions (CCFs; right-handed system [RH] y-axis) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (B) Amino acid changes resulting from the mutations in CRBN gene found at each state. CCF of each mutation shown. (C) Incidence of copy loss or structural variation (LH y-axis) and their CCFs (RH y-axis; copy loss only) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (D) Diagrammatic representation of site of structural variant breakpoints identified at LEN refractoriness. (E) Change in ratio of exon 10–spliced/full-length transcripts between ND, LEN-, and POM-refractory states; ratio expressed as log2(exon 10–spliced/full-length transcript count) × tumor sample purity. Significant difference detected by Kruskal-Wallis 1-way analysis of variance (nonparametric). (F) Landscape of total and coincidence of 4 types of CRBN aberrations in LEN- and POM-refractory cases analyzed. *Mutations from same patient.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/137/2/10.1182_blood.2020007081/3/m_bloodbld2020007081f1.png?Expires=1767836159&Signature=CpKOlcf7JD2Q~9C4k9X9yW4R09sem7l~8LQinBNi-iEK27l3Ki00ZoHwCl0hLfHJnynxTCwAsE7R-eMXmnkgKajUPKpqRE2ziXsnErLgnQOMp1wjfnBSFlnjWzciu68J2MyPo4yj3R6s86X2CXovjawpfVF7jF7gUXZd6qivvgOqrOA-8YYiQDluPROn5pzJd4yOrbIKbKy4geJn8cOshYkFsmsWwzvY0RTWwy5FzF9nJ635a3z3GJOZ6tUen5~3IPSFeiVgB7nBCCATnwFRLzhIuPR69seot4V8ThulkUBSEEDRD6Kkymr6NOD5IksepMYIZBoJgKsMRL9eGZMpwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN increases with sequential IMiD refractoriness. (A) Incidence of CRBN mutations (left-handed system [LH] y-axis) and their cancer clonal fractions (CCFs; right-handed system [RH] y-axis) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (B) Amino acid changes resulting from the mutations in CRBN gene found at each state. CCF of each mutation shown. (C) Incidence of copy loss or structural variation (LH y-axis) and their CCFs (RH y-axis; copy loss only) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (D) Diagrammatic representation of site of structural variant breakpoints identified at LEN refractoriness. (E) Change in ratio of exon 10–spliced/full-length transcripts between ND, LEN-, and POM-refractory states; ratio expressed as log2(exon 10–spliced/full-length transcript count) × tumor sample purity. Significant difference detected by Kruskal-Wallis 1-way analysis of variance (nonparametric). (F) Landscape of total and coincidence of 4 types of CRBN aberrations in LEN- and POM-refractory cases analyzed. *Mutations from same patient.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/137/2/10.1182_blood.2020007081/3/m_bloodbld2020007081f1.png?Expires=1767837334&Signature=Wy-CMtKwOEK0ZYRHT3i5luThpDqUwiWvKmIrjSX0NOD72quMdzAi5c57Ja3qH8clI7MfAx3EUoB1VQl74Flk02JgwthB9rd4RczzO2t6ozUu2XIXzsFTKU9JFlcyEEm7EiRm~8LQFkyom7lFDFIi3NyYv0tVNz8ETXUObC~b4mXLIxz2ZT6Gd0e-7VgeF236pgsdFNpgoyfjvAyfNMIVVfzbDGxUbQJXshb4zTyR1XKWlglIun-4hgd1gWoy9-7BdKy9uMR~vmRnymrLBWJ-XY3PrJZ~DQiu-1e6F8ricK4FQA0l-HhGFY2B3-uS3m8h92qJPhob-CDHMpHX5QkUSQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)