Key Points
IL-3/GM-CSF stimulation is required for MYC-transduced human hematopoietic cells to transition from normalcy to AML.
Early and late granulopoietic progenitor types generate AML rapidly and efficiently after MYC transduction and IL-3/GM-CSF exposure.
Abstract
Hematopoietic clones with leukemogenic mutations arise in healthy people as they age, but progression to acute myeloid leukemia (AML) is rare. Recent evidence suggests that the microenvironment may play an important role in modulating human AML population dynamics. To investigate this concept further, we examined the combined and separate effects of an oncogene (c-MYC) and exposure to interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), and stem cell factor (SCF) on the experimental genesis of a human AML in xenografted immunodeficient mice. Initial experiments showed that normal human CD34+ blood cells transduced with a lentiviral MYC vector and then transplanted into immunodeficient mice produced a hierarchically organized, rapidly fatal, and serially transplantable blast population, phenotypically and transcriptionally similar to human AML cells, but only in mice producing IL-3, GM-CSF, and SCF transgenically or in regular mice in which the cells were exposed to IL-3 or GM-CSF delivered using a cotransduction strategy. In their absence, the MYC+ human cells produced a normal repertoire of lymphoid and myeloid progeny in transplanted mice for many months, but, on transfer to secondary mice producing the human cytokines, the MYC+ cells rapidly generated AML. Indistinguishable diseases were also obtained efficiently from both primitive (CD34+CD38−) and late granulocyte-macrophage progenitor (GMP) cells. These findings underscore the critical role that these cytokines can play in activating a malignant state in normally differentiating human hematopoietic cells in which MYC expression has been deregulated. They also introduce a robust experimental model of human leukemogenesis to further elucidate key mechanisms involved and test strategies to suppress them.
Introduction
Healthy adult humans commonly develop expanded clones of normal blood cells bearing mutations associated with acute myeloid leukemia (AML) as they age, but progression to AML is rare.1-3 This finding has raised interest in the likelihood that other factors, both extrinsic and intrinsic, are important contributors to the eventual genesis of a malignant disease. Evidence of microenvironmental regulation of human AML cell growth has been demonstrated in vitro,4,5 in patient-derived xenografts in immunodeficient mice,6-9 and is inferred from studies of de novo models produced in transgenic mice.10,11 However, because experimental protocols for generating human AML de novo have been difficult to develop, the role of the cytokine milieu in supporting or sustaining a leukemic state in human cells remains poorly understood.
To investigate this question, we designed an experimental strategy to examine the separate and combined effects in freshly isolated normal human CD34+ hematopoietic cells of a deregulated oncogene (c-MYC) and 1 or both of 2 cytokines typically released in inflammatory states (ie, interleukin-3 [IL-3] and granulocyte-macrophage colony-stimulating factor [GM-CSF]).12,13 MYC was chosen because its increased expression is commonly seen in many AMLs with different mutational profiles.14-17 IL-3 and GM-CSF were also of primary interest because many studies have shown that the leukemic cells in most AML patients exhibit IL-3 and/or GM-CSF responsiveness in vivo (in patient-derived xenografts)6,7 and in vitro,18,19 and acquisition of an autocrine GM-CSF phenotype by AML blasts has also been documented.20,21
Methods
Human cells
Discarded normal cord blood (CB) and adult bone marrow (BM) transplant collections were obtained with consent, anonymized, and used in accordance with procedures approved by the Research Ethics Board of the University of British Columbia. Populations enriched in viable CD34+ cells (>70%) were isolated using the EasySep protocol (STEMCELL Technologies) from low-density (<1.077 g/mL) cells isolated by centrifugation on Lymphoprep (STEMCELL Technologies). For many experiments, variably CD34+ cell-enriched CB cells were combined to make large pools of frozen cells. For transduction, thawed cells were incubated in a serum-free medium supplemented with 100 ng/mL stem cell factor (SCF) and Fms-like tyrosine kinase 3 ligand (FLT3L) and 20 ng/mL IL-6, IL-3, and granulocyte colony-stimulating factor (G-CSF) for 16 hours at 37°C before a 6-hour exposure to 1 or more lentiviral vectors (supplemental Methods available on the Blood Web site). Cells were then washed in Hanks balanced salt solution plus 2% fetal bovine serum (FBS) and transplanted into mice. Aliquots cultured in the same pretransduction medium for another 2 to 4 days were used for gene transfer efficiency assessments or fluorescence-activated cell sorting (FACS) of transduced cells for in vitro experiments. The numbers of transduced cells present in experiments initiated with unselected cells were calculated using the gene transfer efficiency values obtained in the same experiment. These values were 10% to 40% for the SFFV-MYC-PGK-GFP lentiviral vector containing the human MYC cDNA,22 30% to 40% for the SFFV-PGK-GFP or -YFP control lentiviruses, and 20% to 50% for the lentiviruses containing human IL-3, GM-CSF, and SCF sequence-verified cDNAs (MNDU3-PGK-mCherry lentiviral vector, from a construct originally obtained from Don Kohn, University of California in Los Angeles).23 Lentiviruses (supplemental Figure 1A) were produced as previously described.24
Xenotransplants
Parental NOD-Rag1−/−-IL2Rγc−/− (NRG) mice and derived NRG mice producing human IL-3, GM-CSF, and SCF constitutively (NRG-3GS mice)7,25 or NRG-W41/41-3GS26 immunodeficient mice were bred and maintained under sterile conditions in the Animal Resource Centre of the BC Cancer Research Institute. Adult NRG and NRG-3GS mice were sublethally irradiated with 850 cGy 137Cs ɣ-rays delivered over a period of 3 hours and NRG-W41/41-3GS mice with a biologically equivalent dose of 320 cGy given in a few minutes,26 and cells were injected intravenously within 24 hours. For primary xenografts, 102 to 2 × 105 total CD34+ CB or BM cells or indicated numbers of FACS-sorted CB subsets were injected immediately after transduction. For secondary and tertiary transplants of primary AML cells, 2 × 105 to 1.1 × 107 unsorted bulk cells from the BM and/or spleen of primary recipients were injected. For secondary transplants of cells from NRG to NRG-3GS or NRG recipients, matched aliquots of 1 to 2 × 106 bulk BM and spleen cells obtained from a primary NRG mouse euthanized 32 weeks after transplant in 1 experiment or from a femoral aspirate obtained from another primary NRG mouse 13 weeks after transplant in a second experiment were injected into 5 NRG and 5 NRG-3GS secondary recipients. For secondary transplants of cells from NRG-3GS into NRG mice, 10 NRG mice were transplanted with bulk leukemic cells from 4 primary NRG-3GS recipients in 2 separate experiments. Cells from 2 of these primary NRG-3GS recipients had been shown previously to regenerate leukemias in secondary NRG-3GS recipients; cells from the other 2 had not been previously tested.
All post-transplant BM chimerism assessments were performed on at least 2 × 105 live propidium iodide (PI)-negative cells from aspirates on alternating femurs or BM and/or spleen harvests from mice euthanized when moribund as determined by weight loss and lethargy or from unaffected control or other test mice to generate comparative data. All animal procedures were carried out in accordance with the Canadian Council on Animal Care guidelines using protocols approved by the Animal Care Committee of the University of British Columbia.
Tissue processing and histology
Animal tissues were mechanically dissociated, and the material was passed through a 40-μm filter (Falcon). Spleen and BM cells harvested from each animal were checked, at a minimum, for the presence of live GFP+ cells by flow cytometry. Mice were considered leukemic when they displayed enlarged spleens weighing ≥0.1 g and/or GFP+ cells made up ≥20% of the BM cells and ≥10% the spleen cells, or a predominantly human CD33+123+ population was present in their BM or spleen cells. Cells were then viably cryopreserved in 10% dimethylsulfoxide with 90% FBS and stored at −190°C for further analyses. Spleen touch-preps were generated by lightly touching dissected spleens onto a slide then staining the slide with the Wright-Giemsa stain using a Hematek automatic stainer. Photomicrographs were taken using a Leica microscope and LAS EZ v.2.1.0 software (Leica Biosystems).
Flow cytometry and cell sorting
For surface phenotype analyses or sorting, cells were suspended in blocking buffer (Hanks balanced salt solution plus 5% human serum and 0.5% anti-human CD32 antibody Clone IV.3 [0.5 mg/mL, STEMCELL Technologies]) and 0.7% anti-mouse CD32 antibody Clone 2.4 G2 (0.5 mg/mL, BD Pharmingen) and then incubated for 1 hour on ice in the dark with selected fluorochrome-conjugated antibodies, washed in phosphate-buffered saline (STEMCELL Technologies) with 2% FBS, and resuspended in 0.1% PI (1 mg/mL, Sigma-Aldrich) or a 0.1-µg/mL 4′,6-diamidine-2′-phenylindole solution (Sigma-Aldrich) to detect viable cells. Compensation was performed using UltraComp eBeads Compensation beads (ThermoFisher Scientific) on the FACS Diva v.8.0 (BD) or FloJo v.10 (TreeStar) software. For phenotyping cells, cells were analyzed on an LSR Fortessa or LSR Fortessa II cytometer (BD), and for isolating phenotyped cells, a FACSAria Fusion or FACSARIA III sorter (BD) was used. CD34+38− CB cells were isolated as CD34+38low/− cells. GMPs were isolated as CD34+38+CD10−CD7−CD45RA+CD135+ cells (supplemental Figure 4A). The fluorochrome-labeled antibodies used to isolate these are listed in supplemental Table 1. The purity of sorted cells was assessed by immediate reanalysis and was 80% to 100%. Flow cytometry data were analyzed using FloJo v.10 (TreeStar).
In vitro experiments
For liquid cultures of single cells, transduced single GFP+ cells were sorted 48 hours after transduction into the individual wells of a 96-well plate preloaded with 100 µL Iscove modified Dulbecco medium supplemented with 30% FBS and human cytokines (50 ng/mL SCF, 20 ng/mL GM-CSF, IL-3, IL6, and G-SCF and 3 U/mL erythropoietin). After 12 days at 37°C, the number of live (PI−) cells in each well containing > 128 PI− cells was enumerated by flow cytometry using fluorescent AccuCheck counting beads (Invitrogen). Cloning efficiency was 69% to 71% for the MYC-transduced cells and 77% to 81% for control cells (total of 384 cells for each group in 2 experiments). Similar cultures with bulk preselected transduced cells and individual and combinations of cytokines were similarly assessed after 8 days of incubation. Stromal cell-containing cultures were initiated by plating 4.5 × 103MYC- and/or control-transduced CB CD34+ cells (sorted for GFP+ cells 48 hours after transduction) in 1 mL Myelocult H5100 media/well (STEMCELL Technologies) plus 10−6 M hydrocortisone (Sigma-Aldrich) onto irradiated murine M2-10B4 and Sl/Sl mouse fibroblasts engineered to produce human FLT3L, SCF, IL-3, and G-CSF. Matched numbers of MYC-GFP–transduced or control-YFP–transduced CD34+ cells (4.5 × 103 cells each) were either cultured in separate wells or cocultured together in the same well. These cultures were maintained for 12 weeks with weekly half-media changes and addition of freshly prepared hydrocortisone. Live (PI−) nonadherent cells removed weekly were enumerated by flow cytometry using AccuCheck counting beads and assessed biweekly in Wright-Giemsa–stained cytospin preparations. For colony-forming cell assays, 150 GFP+PI−MYC-transduced cells sorted 48 hours after transduction or 0.2 to 1 × 105 FACS-sorted PI−GFP+ (CD33+) cells from the BM or spleen of leukemic mice were plated into replicate 1-mL cultures of Methocult H4230 (STEMCELL Technologies) supplemented with the same human cytokine cocktail used in the liquid suspension cultures or no cytokines, and colonies of >20 cells were scored after 12 to 14 days.
RNA sequencing
MYC+ (GFP+) human CD33+ BM or spleen cells from moribund leukemic mice were ribodepleted, RNA was extracted, libraries were generated and sequenced, and reads were aligned and sorted. Gene expression was characterized by calculating reads per kilobase of transcript per million mapped reads values that were then compared with similarly generated and analyzed RNA-seq data for normal cells. Hierarchical clustering of the data was generated from 1 − the Spearman correlation coefficients of all protein-coding genes with reads per kilobase of transcript per million mapped reads values >0.01 in ≥1 sample. For single-cell RNA sequencing, cells were loaded onto a 10X Genomics single-cell controller. Libraries were generated and sequenced on an Illumina NextSeq 500 machine. For additional details, see the supplemental Methods.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assays for human IL-3, GM-CSF, and SCF were performed on supernatants harvested from cultures of CD34+ CB cells 6 days after being transduced with vectors encoding the corresponding human cytokine cDNAs and then frozen. Test and control cells were cultured in serum-free medium (STEMSpan containing 20% custom bovine serum albumin, insulin, and transferrin) supplemented with low-density lipoproteins, 10−4 M β-mercaptoethanol, 1% glutamine, 100 ng/mL each of SCF and FLT3L, and 20 ng/mL of each of IL6, IL-3, and G-CSF. Enzyme-linked immunosorbent assays were performed using the Quantikine kits (R&D Systems), and the analysis of absorbance was conducted on a Sunrise Absorbance Microplate Reader using Magellan v.7.1 software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism or R. The L-Calc software (STEMCELL Technologies) was used for limiting dilution analysis. Two-tailed Student t test, 2-way analysis of variance, or 2-tailed Mann-Whitney tests, as appropriate, were used to determine statistical significance. Sidak's or Holm-Sidak's tests were used to adjust for multiple comparisons when appropriate. Log-rank (Mantel-Cox) tests were used to determine statistical significance for survival curves. An α of 0.05 was used as cutoff for significance. Reported n values refer to different experiments with different human cells and/or separate mice.
RNA-seq data obtained from the model AML cells are available from Gene Expression Omnibus (GSE130931). RNA-seq data obtained for normal CD34+ CB cell subsets are available from the European Genome-phenome Archive (EGAD00001004950). RNA-seq data (level 3 access) and clinical data of childhood27 and adult28 AML samples were downloaded from ocg.cancer.gov. All code for the RNA-seq data analysis is available upon request.
Results
MYC-transduced human CD34+ cells generate a lethal AML in immunodeficient mice producing human IL-3, GM-CSF, and SCF
CD34+ CB cells exposed to MYC-GFP or -YFP vectors (supplemental Figure 1A) showed gene transfer efficiencies that ranged from 10% to 35% in individual experiments. Analysis of their progeny isolated by FACS and then incubated for several days in vitro in the presence of multiple cytokines revealed a stable two- to fivefold increase in MYC transcripts and protein levels compared with control-transduced cells (supplemental Figure 1B-C). This was accompanied by an expanded, but still cytokine-dependent, output of their clonal progeny in short-term, single-cell suspension cultures (Figure 1A-B; supplemental Figure 1D-E). MYC-transduced CD34+ CB cells also produced greater numbers of nonadherent progeny than controls when these were cultured separately for 12 weeks in wells containing a feeder layer of irradiated mouse fibroblasts engineered to produce human IL-3, G-CSF, SCF, and FLT3L29,30 (Figure 1A,Ci). Examination of the nonadherent cells produced from the MYC-transduced cells as of 2 weeks later showed these displayed an abnormal immature morphology, in contrast to the expected mature neutrophils found in the matching cultures initiated with control vector-transduced cells (Figure 1C, inset). The MYC-transduced (GFP+) cells showed a similar output of cells when they were cultured together with the control-YFP–transduced CD34+ CB cells in the same wells. However, in this case, the output of nonadherent cells generated from the control (YFP+) cells was markedly suppressed (Figure 1Cii), thus mimicking the suppression of normal hematopoiesis also seen in many AML patients at diagnosis.
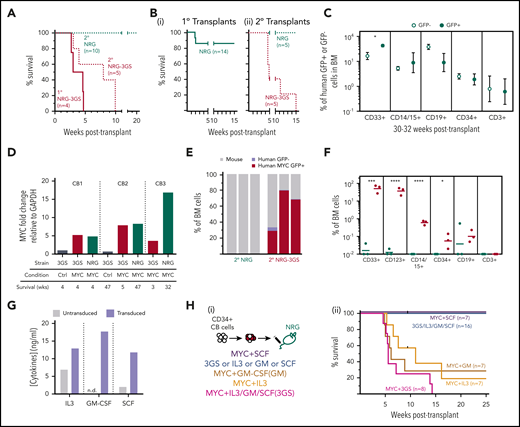
Rapid generation of a lethal human AML in NRG-3GS mice transplanted with MYC-transduced human CD34+ CB cells. (A) General experimental design. (B) Distributions of 12-day clone sizes from 238 MYC-GFP and 256 control-YFP–transduced cells sorted and plated 2 days after transduction in single-cell, cytokine-containing suspension cultures; 2 experiments, 2-tailed Mann-Whitney test, P < .0001. (C) Weekly nonadherent cell outputs in cultures containing human cytokine-producing stromal cells, normalized to the cell input numbers (4.5 × 103MYC-GFP– or control-YFP–transduced cells). (i) Results when these were cultured separately. Insets are Giemsa-stained cytospins of week 8 cells. (ii) Results when the same numbers of MYC-GFP– and control-YFP–transduced cells were cultured together in the same wells. Mean ± standard error of the mean of replicate cultures from 1 of 2 experiments with different CB cells; 2-way analysis of variance. (D) Survival of NRG-3GS recipients of 0.6 to 2 × 104MYC-GFP–transduced cells or 1 to 3 × 104 empty-vector GFP-transduced (control) cells; log-rank test, P < .0001. Inset shows Giemsa-stained touch preps of spleen cells at 3 weeks after transplant. (E) GFP chimerism in the BM of recipients of MYC (n = 6) or control (n = 4) cells 3 to 5 weeks after transplant. (F) Phenotype of GFP+ cells shown in panel E; unpaired 2-tailed Student t test. MYC, red; control, steel blue. Scale bars in photomicrograph insets are 10 μm.
Rapid generation of a lethal human AML in NRG-3GS mice transplanted with MYC-transduced human CD34+ CB cells. (A) General experimental design. (B) Distributions of 12-day clone sizes from 238 MYC-GFP and 256 control-YFP–transduced cells sorted and plated 2 days after transduction in single-cell, cytokine-containing suspension cultures; 2 experiments, 2-tailed Mann-Whitney test, P < .0001. (C) Weekly nonadherent cell outputs in cultures containing human cytokine-producing stromal cells, normalized to the cell input numbers (4.5 × 103MYC-GFP– or control-YFP–transduced cells). (i) Results when these were cultured separately. Insets are Giemsa-stained cytospins of week 8 cells. (ii) Results when the same numbers of MYC-GFP– and control-YFP–transduced cells were cultured together in the same wells. Mean ± standard error of the mean of replicate cultures from 1 of 2 experiments with different CB cells; 2-way analysis of variance. (D) Survival of NRG-3GS recipients of 0.6 to 2 × 104MYC-GFP–transduced cells or 1 to 3 × 104 empty-vector GFP-transduced (control) cells; log-rank test, P < .0001. Inset shows Giemsa-stained touch preps of spleen cells at 3 weeks after transplant. (E) GFP chimerism in the BM of recipients of MYC (n = 6) or control (n = 4) cells 3 to 5 weeks after transplant. (F) Phenotype of GFP+ cells shown in panel E; unpaired 2-tailed Student t test. MYC, red; control, steel blue. Scale bars in photomicrograph insets are 10 μm.
Transplantation of MYC and control-transduced CD34+ CB cells into sublethally irradiated immunodeficient NRG-3GS mice (≤2 × 104 transduced cells/mouse; Figure 1A) caused 21 of 22 mice injected with the MYC-transduced cells to become moribund within 7 weeks (Figure 1D). When autopsied, these showed widespread indications of a human AML characterized by the presence in the BM and frequently enlarged spleens of a prominent MYC+GFP+ population of human cells that showed a blast morphology and a CD123+CD33+CD15±CD34−CD14−CD19−CD3− phenotype (Figure 1D, inset, 1E-F; supplemental Figure 2A-C) typical of patients and other models of human AML cells.31,32 Transfer of these cells to secondary NRG-3GS mice produced phenotypically indistinguishable fatal human AML populations, demonstrating the presence in the primary recipients of transplantable leukemia-initiating cells (Figure 2A, dotted red survival curve). In contrast, no evidence of disease was noted in any of the primary NRG-3GS recipients of control cells at any of the time points when these were killed, in some cases to provide matching repopulation data (Figure 1D).
Cytokine-dependent activation of a latent leukemic state present in the in vivo progeny of MYC-transduced human CB cells. (A) Survival of secondary (dotted, NRG-3GS, red; NRG, green) recipients transplanted with bulk leukemic cells from primary NRG-3GS recipients (solid red); log-rank test, P < .005. (B) Survival of primary NRG recipients (i, solid green), and secondary NRG and NRG-3GS recipients of matched doses of cells from 2 primary NRG recipients of MYC-transduced cells (ii: NRG-3GS, dotted red; NRG, dotted green); n = 2 experiments, log-rank test, P < .01. (C) Mean ± standard error of the mean chimerism of human cell types within the MYC+GFP+ (solid green) and nontransduced populations (hollow green) detected in the BM of individual NRG recipients of unselected CD34+ or prepurified CD34+38− cells analyzed 30 to 32 weeks after transplant; paired 2-tailed Student t test, n = 4 to 7, grafts initiated from 3 different CB pools. (D) MYC transcripts in bulk GFP+ cells from primary grafts of MYC- or control-transduced cells in NRG-3GS or NRG mice. Grafts were initiated from 3 different CB pools. (E-F) Four-week BM chimerism (E) and phenotype (F) of cells from 3 secondary NRG (green) and NRG-3GS (red) recipients from (Bii); unpaired 2-tailed Student t test. (G) Cytokine concentrations in the supernatants of cytokine-transduced CD34+ CB cells cultured for 6 days. n.d., below the level of detection. (H) Survival of primary NRG recipients of CD34+ cells transduced with cytokines ± MYC cDNAs; log-rank test, P < .05. Experimental design (i) and results (ii).
Cytokine-dependent activation of a latent leukemic state present in the in vivo progeny of MYC-transduced human CB cells. (A) Survival of secondary (dotted, NRG-3GS, red; NRG, green) recipients transplanted with bulk leukemic cells from primary NRG-3GS recipients (solid red); log-rank test, P < .005. (B) Survival of primary NRG recipients (i, solid green), and secondary NRG and NRG-3GS recipients of matched doses of cells from 2 primary NRG recipients of MYC-transduced cells (ii: NRG-3GS, dotted red; NRG, dotted green); n = 2 experiments, log-rank test, P < .01. (C) Mean ± standard error of the mean chimerism of human cell types within the MYC+GFP+ (solid green) and nontransduced populations (hollow green) detected in the BM of individual NRG recipients of unselected CD34+ or prepurified CD34+38− cells analyzed 30 to 32 weeks after transplant; paired 2-tailed Student t test, n = 4 to 7, grafts initiated from 3 different CB pools. (D) MYC transcripts in bulk GFP+ cells from primary grafts of MYC- or control-transduced cells in NRG-3GS or NRG mice. Grafts were initiated from 3 different CB pools. (E-F) Four-week BM chimerism (E) and phenotype (F) of cells from 3 secondary NRG (green) and NRG-3GS (red) recipients from (Bii); unpaired 2-tailed Student t test. (G) Cytokine concentrations in the supernatants of cytokine-transduced CD34+ CB cells cultured for 6 days. n.d., below the level of detection. (H) Survival of primary NRG recipients of CD34+ cells transduced with cytokines ± MYC cDNAs; log-rank test, P < .05. Experimental design (i) and results (ii).
Interestingly, transplantation of MYC-transduced CB cells into sublethally irradiated primary NRG mice (not producing human IL-3, GM-CSF, or SCF constitutively) produced no evidence of disease throughout a follow-up period of up to 47 weeks, despite a few radiation-induced deaths that occurred in the first 2 weeks after transplant (Figure 2Bi). Similarly, paired secondary NRG recipients of the AML cells produced in primary NRG-3GS mice showed no sign of disease development (Figure 2A, dotted green survival curve).
A rapidly fatal and phenotypically similar human AML was also produced from MYC-transduced CD34+ CB cells in NRG-3GS hosts that received no prior conditioning and from MYC-transduced adult human CD34+ BM cells transplanted into sublethally irradiated NRG-3GS or NRG-W41-3GS hosts (supplemental Figure 2D-F).
MYC induces a latent AML program that requires human IL-3 or GM-CSF for activation
Interestingly, analysis of the cells present in BM aspirates obtained from the NRG recipients of MYC-transduced cells showed they were producing the same spectrum of lymphoid and myeloid progeny as were being generated from the nontransduced (GFP−) cells present in the original inoculum transplanted (Figure 2C; supplemental Figure 2G). This included the parallel appearance of human CD19+ and CD3+ (B- and T-lineage) cells, CD14+ and/or CD15+ granulocytes/monocytes, and more primitive CD34+ cells, with similar kinetics and outputs. The normal cell output activity from the MYC-transduced (GFP+) cells was not attributable to a lack of increased expression of MYC in their progeny produced in the NRG mice, because these showed elevated levels of MYC transcripts similar to those measured in the human leukemic cells isolated from NRG-3GS mice (Figure 2D).
To determine whether the normal phenotypes of MYC+ (GFP+) human blood cell precursors produced in NRG mice might still be sensitive to leukemic transformation if exposed to human IL-3, GM-CSF, and SCF, we harvested the cells from these primary NRG hosts 13 to 23 weeks after initial transplantation and then transplanted them into paired groups of secondary NRG-3GS and NRG mice. This resulted in the rapid appearance of a fatal human AML population in all of the secondary NRG-3GS mice but not in any of the secondary NRG recipients (Figure 2Bii, E-F). In fact, in the secondary NRG hosts, the numbers of human cells detected after transplant was minimal and transient (Figure 2E-F), as would be expected for transplants of normal cells regenerated in primary NRG mice.33
To determine which or how many of the 3 human cytokines produced in the NRG-3GS mice are required to activate the leukemic transformation of MYC-transduced human CD34+ cells, we transplanted different groups of NRG mice with CD34+ CB cells that were exposed to the MYC-GFP vector simultaneously with separate mCherry vectors encoding IL-3, GM-CSF, or SCF alone or all 3 cytokine vectors together (Figure 2G-Hi; supplemental Figure 1A). This strategy showed that MYC plus either IL-3 or GM-CSF (or all 3 cytokines) supported the production of AML within 20 weeks, whereas the mice injected with cells transduced with MYC plus SCF, or all 3 cytokines but without MYC, failed to develop any evidence of AML (Figure 2Hii). Interestingly, analysis of the ultimately fatal AML populations produced showed in all cases that these included cells expressing only MYC-GFP, plus variable numbers of cells coexpressing MYC and 1 or more of the cytokine-encoding (mCherry) vectors (supplemental Figure 3A-B). Thus, exogenous delivery of either IL-3 or GM-CSF using this strategy was sufficient to induce AML in MYC-transduced cells.
Phenotypically and transcriptionally similar AML populations are generated efficiently from MYC-transduced CD34+38− and GMP cells in NRG-3GS hosts
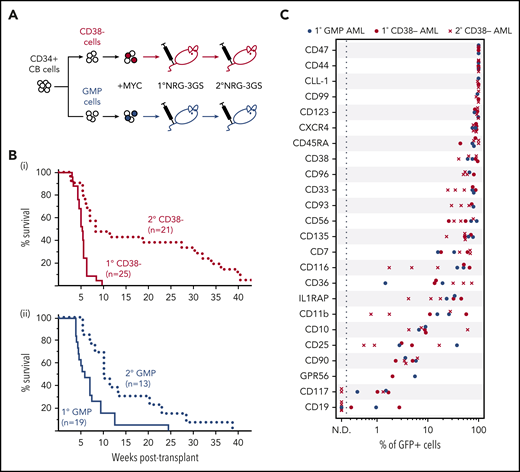
To examine the range of human CD34+ CB cell types from which AML populations could be generated, we isolated the very primitive CD34+38− cells (selectively enriched in hematopoietic stem cells) and CD34+38+45RA+135+7−10− GMPs (enriched in late-stage progenitors restricted to granulocyte-macrophage differentiation34,35 ) by FACS (supplemental Figure 4A). We then transduced each subset using the same MYC transduction transplant protocol used for the total CD34+ CB cell transplants (Figure 3A). Varying numbers of each of these transduced subsets were injected into NRG-3GS recipients, and the frequency of cells capable of producing a fatal human AML was then determined by limiting dilution analysis for both subsets. The results showed that both the CD34+38− cells and the GMPs were highly susceptible to MYC-induced leukemic transformation (frequencies of 1 in 14 and 1 in 46 cells, respectively; Figure 3B; supplemental Figure 4B-E).
Phenotypically similar AMLs are rapidly generated from MYC-transduced CD34+ 38− and GMP CB cells. (A) Experimental design. (B) Survival of mice transplanted with MYC-transduced CD34+38− (i, red) or GMP (ii, blue) CB cells (solid, primary transplants; dotted, secondary transplants). (C) Percent of GFP+ (MYC+) cells positive for the indicated surface markers in primary AMLs produced from MYC-transduced CD34+38− cells (n = 2) or GMPs (n = 2) or secondary AMLs originally initiated from MYC-transduced CD34+38− cells (n = 3), all in NRG-3GS recipients.
Phenotypically similar AMLs are rapidly generated from MYC-transduced CD34+ 38− and GMP CB cells. (A) Experimental design. (B) Survival of mice transplanted with MYC-transduced CD34+38− (i, red) or GMP (ii, blue) CB cells (solid, primary transplants; dotted, secondary transplants). (C) Percent of GFP+ (MYC+) cells positive for the indicated surface markers in primary AMLs produced from MYC-transduced CD34+38− cells (n = 2) or GMPs (n = 2) or secondary AMLs originally initiated from MYC-transduced CD34+38− cells (n = 3), all in NRG-3GS recipients.
The populations of AML cells produced from both subsets of CD34+ CB cells displayed the same human CD33+CD123+ phenotype as those generated from MYC-transduced bulk CD34+ cells (supplemental Figure 4E). Both were also serially transplantable in NRG-3GS recipients on transfer of similarly high AML cell numbers and a similarly prolonged latency (Figure 3B; supplemental Figure 4F). These features of the blast populations obtained from moribund primary mice with AML suggested the establishment of a hierarchical system similar to that characteristic of AML populations that arise spontaneously in patients, in which the bulk of the blasts are incapable of further division and the frequency of cells with proliferative ability decreases with their innate proliferative potential.19 To test this possibility, we isolated MYC+ (GFP+) cells by FACS from the same primary recipients’ AML cells and assayed them for colony-forming cell activity in methylcellulose cultures with and without added growth factors. The results showed that such cells could be detected, but at highly variable and generally low (10−4 to 10−3) frequencies, regardless of the phenotype from which the AML had been generated, and only in cultures that contained the growth factors (supplemental Figure 4G).
Further characterization of the AML populations produced from MYC-transduced CD34+38− cells and GMPs in NRG-3GS mice by multiparameter flow cytometry showed that the leukemic cells generated from either initial source expressed many of the cell surface markers variably found on AML patients’ blasts. These included a consistently high expression of CD123, variably elevated expression of CD116 (the respective ligand-specific binding receptors for IL-3 and GM-CSF), and a lack of detectable expression of CD117, the receptor for SCF (Figure 3C; supplemental Figure 4E,H).
Comparison of the transcriptomes of MYC+CD33+ cells isolated from AMLs derived from CD34+38− cells or GMPs showed these to be indistinguishable (Spearman correlation coefficient ≥ 0.95 in all comparisons; Figure 4A; supplemental Figure 5A). Compared with RNA-seq data for normal human CB GMPs, both sources of leukemic cells contained similarly higher levels of transcripts of genes associated with cell cycle activation and metabolism and reduced levels of transcripts for signaling and adhesion-related genes (Figure 4B-C; supplemental Figure 5B-C). Comparison of the bulk transcriptomes with published RNA-seq data for pediatric27 and adult28 AML patient cells showed the synthetic AMLs formed a relatively distinct group more similar to some AMLs than others (supplemental Figure 5D-E). Compared with published RNA-seq data for pediatric AML patient cells,27 our analysis showed that our de novo MYC-induced AML cells are most closely related transcriptionally to the leukemic cells from a subset of children diagnosed with an M5 AML or an mixed lineage leukemia (MLL)rearrangement at <3 years of age, whose diseases were classified as standard risk AML with elevated MYC expression (supplemental Figure 5F-G).
AMLs generated from CD34+ 38− and GMP CB cells are transcriptionally similar to each other and to unmanipulated CB GMPs. (A) Unsupervised clustering (complete linkage) of transcript levels in GFP+ CD33+ (leukemic) cells isolated from NRG-3GS mice and compared with cell types purified by FACS from normal low-density CB cells. (B-C) REACTOME gene sets upregulated (B) and downregulated (C) in leukemic cells compared with normal CB GMPs. Reactome pathways enriched (P < .05 hypergeometric test) in differentially expressed genes were identified using ReactomePA, and networks clustering was performed based on the similarity of genes present in each pathway. (D-F) t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis of the top 1000 variable genes of 4570 single GFP+CD33+ leukemic cells isolated from 2 AMLs generated from MYC-transduced CD34+38− and 2 from MYC-transduced GMPs, following sample normalization and integration with Seurat v3, with t-SNE distributions of these cells shown by sample (D), by identified transcriptomic clusters (E), and cell cycle gene expression profiles (F).
AMLs generated from CD34+ 38− and GMP CB cells are transcriptionally similar to each other and to unmanipulated CB GMPs. (A) Unsupervised clustering (complete linkage) of transcript levels in GFP+ CD33+ (leukemic) cells isolated from NRG-3GS mice and compared with cell types purified by FACS from normal low-density CB cells. (B-C) REACTOME gene sets upregulated (B) and downregulated (C) in leukemic cells compared with normal CB GMPs. Reactome pathways enriched (P < .05 hypergeometric test) in differentially expressed genes were identified using ReactomePA, and networks clustering was performed based on the similarity of genes present in each pathway. (D-F) t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis of the top 1000 variable genes of 4570 single GFP+CD33+ leukemic cells isolated from 2 AMLs generated from MYC-transduced CD34+38− and 2 from MYC-transduced GMPs, following sample normalization and integration with Seurat v3, with t-SNE distributions of these cells shown by sample (D), by identified transcriptomic clusters (E), and cell cycle gene expression profiles (F).
Single-cell RNA-seq analysis of 4 of the same de novo AML populations used in the bulk transcriptomic analysis (2 from each source of GMP- or CD38-derived leukemias) were highly overlapping in multidimensional space confirming their transcriptional similarity (Figure 4D). In addition, it showed that these MYC-induced AML blasts could be segregated into 4 GFP+CD33+ clusters (Figure 4E; supplemental Figure 6A-D; supplemental Tables 3 and 4). Notably, cluster 2 contained a high proportion of cells with an actively cycling RNA profile suggestive of a proliferating subset (Figure 4F).
Discussion
We report here the creation of a new experimental model of de novo human AML that is obtained at high frequency, rapidity, and reproducibility from normal human hematopoietic cells isolated directly from neonates or adults and from subsets of cells at both early and late stages of progenitors with granulopoietic differentiation capabilities. This model of de novo human AML recreates a functionally defined hierarchical system of cells with decreasing self-sustaining/proliferative potential, similar to that characteristic of patients’ AML populations, but also with strong phenotypical and transcriptional similarities to AML patients’ cells in which MYC expression is similarly increased.16,17 These features now make it possible to access, identify, and manipulate in a prospective fashion the initial events required for, and involved in, establishing a leukemic program that has the biological and molecular features of many spontaneously arising human AML populations. The present model thus overcomes many of the challenges posed by retrospective analyses of patient AML cells, for which the diversity of genetic and epigenetic abnormalities is a huge confounding issue in seeking to elucidate pathway changes that determine and mediate a leukemic state. It also bypasses caveats inherent in extrapolating from models of genetically induced AML in mice or other species where many key pathways are not well conserved.
It was of particular interest to discover that normal human GMPs and more primitive CD34+38− CB cells are highly susceptible to MYC-induced transformation when transplanted into NRG-3GS mice as previously shown only in mouse models of leukemogenesis.36-42 Such a possibility has also been inferred from phenotype analysis of the leukemic blasts from certain types of AML patients32,43-46 but not yet directly from normal GMPs. The phenotypical and transcriptional similarity of the AML cells produced by progenitors at early or late stages of differentiation to each other and normal CB GMPs found here is also noteworthy, given the accompanying evidence of the creation of a hierarchy of cells with varying proliferative potential.
Also unexpected is the finding that a de novo experimental AML similar to some spontaneously arising human AMLs can be generated by a modest overexpression of MYC in concert with, and a second requirement for, exposure to IL-3 or GM-CSF in human cells. The ability of MYC overexpression to produce this outcome provides support for the concept that it serves as a critical common mediator of many upstream driver mutations in the process of human AML development. The necessary cooperative activation of a pathway elicited by the signaling receptor used by both IL-3 and GM-CSF also points to an intriguing potential role of inflammatory conditions and/or signaling events18,47,48 in the spontaneous genesis, and subsequent maintenance, of AML populations in patients that develop this malignancy.
The observation that MYC-transduced cells produce apparently normally differentiating populations but remain capable of activating an AML program upon a delayed exposure to IL-3 or GM-CSF is also intriguing because it indicates that critical leukemia-promoting effects of an immunomodulatory factor stimulus can be temporarily dissociated from a prior mutagenic event. Potentially, this could allow the testing of pre-emptive therapeutic approaches based on targeting cells with latent but not yet active leukemogenic potential. It is also tempting to speculate that the latent potential created here by forced overexpression of MYC may have similarities to the situation created in the expanded clones characteristic of individuals with CHIP.49 In these, it appears that differentiation can also proceed in an overtly normally regulated fashion for long periods of time despite the presence of mutations known to contribute to leukemic transformation.
Overall, our findings highlight the important and tractable advantages of de novo models of human leukemogenesis for identifying previously unsuspected aspects of the process that can lead to myeloid leukemia in initially normal human hematopoietic cells. This includes the evidence that a genetic change may be necessary but insufficient to achieve this outcome. In addition, the rapidity and robustly reproducible stages of myeloid leukemia induction provided by the protocols used here should prove to be a useful and robust platform for testing novel therapeutic approaches and enabling new mechanistic analyses of the process of human AML genesis.
RNA-seq data obtained from the model AML cells are available from GEO (GSE130931). RNA-seq data obtained for normal CD34+ CB cell subsets are available from EGA (EGAD00001004950). RNA-seq data (level 3 access) and clinical data of childhood and adult AML samples were downloaded from ocg.cancer.gov. All code for the RNA-seq data analysis is available upon request.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Glenn Edin and Margaret Hale for technical assistance and the British Columbia Cancer Stem Cell Assay Laboratory for assistance with the processing and cryopreservation of CB samples.
This work was supported by grants from the Leukemia and Lymphoma Society of Canada (417871), a Terry Fox Foundation New Frontiers Program Project (1074), the Canadian Cancer Society Research Institute (704257 and 705047), the Cancer Research Society (21339), the Canadian Institutes of Health Research (CIHR EP1-120589 and CEE-151619), and Genome Canada (C41EMT and C32EMT) under the Canadian Epigenetics, Environment and Health Research Consortium. E.B., C.A.H., and A.L were supported by Canadian Institutes of Health Research Frederick Banting and Charles Best Canada Doctoral Scholarships.
Authorship
Contribution: E.B., D.P., and C.J.E. designed the experiments and wrote the manuscript. E.B. and N.N. performed the experiments. E.B., D.P., and C.A.H. performed the data analysis and interpretation. P.A.B., M.H., and A.P.W. assisted with experimental designs and interpretation. C.A.H. and A.L. assisted with the collection of cells for RNA-seq and data analysis. M.M. and A.C. assisted with the bulk RNA-seq and data quality checks. M.B. assisted with RNA-seq data analysis. N.N. and A.P.W. contributed pathologic assessments. S.L. assisted with Western blotting experiments. J.S. assisted with animal studies. B.T.W. helped provide access to patient data. All authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Connie J. Eaves, BC Cancer Agency, 675 W 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: ceaves@bccrc.ca.