Although acute graft-versus-host-disease (AGVHD) is one of the major causes of non-relapse mortality after hematopoietic stem cell transplant (HCT), we are still unable to predict which patients will develop the most severe form of this disease, or which molecular pathways are dysregulated in the T cells that cause disease. Thus, understanding the molecular features of AGVHD is a critical unmet need. To address this, we have performed a companion mechanistic study as a part of our completed Phase 2 trial of abatacept, a CD28:CD80/86 costimulation blockade agent, for severe AGVHD prevention (Clinicaltrials.gov # NCT01743131, 'ABA2'). ABA2 has demonstrated significant improvement in AGVHD in patients prophylaxed with abatacept in addition to calcineurin inhibition (CNI) + Methotrexate (MTX) compared to controls receiving CNI/MTX alone. To begin to uncover mechanisms responsible for the control of AGVHD with abatacept, and given that CD4+ T cells have been consistently documented to be the main therapeutic target of this drug, we interrogated the transcriptome of CD4+ T cells reconstituting in patients prophylaxed with abatacept compared to CNI/MTX.
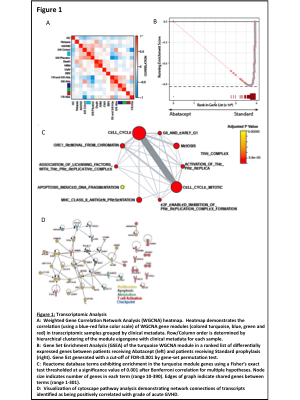
To perform this analysis, we flow cytometrically sorted CD4+ T cells on Days 21-28 post-transplant from all patients on ABA2, as well as a cohort of 12 untransplanted healthy controls, and subsequently performed mRNA-sequencing on these cells. Weighted Gene Correlation Network Analysis (WGCNA) was performed on the top 6000 most variant transcripts from the resulting sequencing data. Hierarchical clustering of the WGCNA co-expression matrix enabled the identification of self-assembling modules (SAMs) that met a threshold of coexpression (Figure 1A). For the ABA2 dataset, we considered the following variables in the WGCNA model: patient cohort (7/8 patients, 8/8 patients, healthy controls), +/- prophylaxis with abatacept, CMV reactivation, EBV reactivation, Grade of GVHD (0-4), relapse, non-relapse mortality, and all-cause mortality.
The WGCNA clustering analysis resulted in the identification of 4 discrete SAMs, which were highly correlated with clinical variable metamodules. This analysis revealed a strong positive correlation of a 476-gene SAM (the Turquoise module) in patients prophylaxed with CNI/MTX + placebo and anti-correlation of this module in patients prophylaxed with CNI/MTX + abatacept, as demonstrated in both the WGCNA heatmap and through Gene Set Enrichment Analysis (Figure 1 A-B). These opposing correlations suggested that interrogation of this module would reveal mechanistic correlates with standard prophylaxis that were decoupled by abatacept. Pathway analysis using the Reactome database (Figure 1C) revealed the turquoise SAM to be dominated by four types of pathways: (1) Those that define canonical cell-cycle pathways (2) Those involved in T cell metabolism (3) Those involved in apoptosis and (4) Those involved in T cell activation, consistent with upregulation of these transcripts in placebo versus abatacept patients.
In addition to being highly correlated with patients receiving placebo, the expression of a subset of the transcripts in the Turquoise module were also directly correlated with the severity of AGVHD in these patients. Thus, linear regression analysis of the 476 transcripts in this module identified a subset of 93 genes for which transcript expression level was increased both in placebo compared to abatacept, and for which expression level also positively correlated with Grade of AGVHD. As with the Turquoise module as a whole, this subset of genes also formed a highly correlated network, linking transcripts involved in T cell proliferation, apoptosis, activation, metabolism as well as the T cell checkpoint (Figure 1D).
This analysis represents the first comprehensive interrogation of the transcriptomic correlates of AGVHD. It identifies a novel set of transcripts which positively associate with the severity of AGVHD, and which costimulation blockade with abatacept down-regulates and de-couples from AGVHD severity. These results suggest a profound reprograming of T cell activation with abatacept that is correlated with the control of AGVHD.
Qayed:Bristol-Myers Squibb: Honoraria. Langston:Astellas Pharma: Other: Research Support; Incyte: Other: Research Support; Jazz Pharmaceuticals: Other: Research Support; Chimerix: Other: Research Support; Takeda: Other: Research Support; Kadmon Corporation: Other: Research Support; Novartis: Other: Research Support; Bristol Myers Squibb: Other: Research Support. Blazar:Fate Therapeutics, Inc.: Research Funding; RXi Pharmaceuticals: Research Funding; Alpine Immune Sciences, Inc.: Research Funding; Abbvie Inc: Research Funding; Leukemia and Lymphoma Society: Research Funding; Childrens' Cancer Research Fund: Research Funding; KidsFirst Fund: Research Funding; Tmunity: Other: Co-Founder; BlueRock Therapeutics: Membership on an entity's Board of Directors or advisory committees; Kamon Pharmaceuticals, Inc: Membership on an entity's Board of Directors or advisory committees; Five Prime Therapeutics Inc: Co-Founder, Membership on an entity's Board of Directors or advisory committees; Regeneron Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees; Magenta Therapeutics and BlueRock Therapeuetics: Membership on an entity's Board of Directors or advisory committees. Kean:HiFiBio: Consultancy; BlueBirdBio: Research Funding; Gilead: Research Funding; Regeneron: Research Funding; EMDSerono: Consultancy; FortySeven: Consultancy; Magenta: Research Funding; Kymab: Consultancy; Jazz: Research Funding; Bristol Meyers Squibb: Patents & Royalties, Research Funding.
Abatacept: Approved for Rheumatoid Arthritis; used in this trial for prevention of GVHD.
Author notes
Asterisk with author names denotes non-ASH members.