Key Points
Next-generation sequencing revealed variants in cancer-associated genes at diagnosis of CML more frequently in patients with poor outcomes.
All patients at BC had mutated cancer genes, including fusions, that predated BCR-ABL1 kinase domain mutations in a majority.
Abstract
Genomic events associated with poor outcome in chronic myeloid leukemia (CML) are poorly understood. We performed whole-exome sequencing, copy-number variation, and/or RNA sequencing for 65 patients to discover mutations at diagnosis and blast crisis (BC). Forty-six patients with chronic-phase disease with the extremes of outcome were studied at diagnosis. Cancer gene variants were detected in 15 (56%) of 27 patients with subsequent BC or poor outcome and in 3 (16%) of 19 optimal responders (P = .007). Frequently mutated genes at diagnosis were ASXL1, IKZF1, and RUNX1. The methyltransferase SETD1B was a novel recurrently mutated gene. A novel class of variant associated with the Philadelphia (Ph) translocation was detected at diagnosis in 11 (24%) of 46 patients comprising fusions and/or rearrangement of genes on the translocated chromosomes, with evidence of fragmentation, inversion, and imperfect sequence reassembly. These were more frequent at diagnosis in patients with poor outcome: 9 (33%) of 27 vs 2 (11%) of 19 optimal responders (P = .07). Thirty-nine patients were tested at BC, and all had cancer gene variants, including ABL1 kinase domain mutations in 58%. However, ABL1 mutations cooccurred with other mutated cancer genes in 89% of cases, and these predated ABL1 mutations in 62% of evaluable patients. Gene fusions not associated with the Ph translocation occurred in 42% of patients at BC and commonly involved fusion partners that were known cancer genes (78%). Genomic analysis revealed numerous relevant variants at diagnosis in patients with poor outcome and all patients at BC. Future refined biomarker testing of specific variants will likely provide prognostic information to facilitate a risk-adapted therapeutic approach.
Introduction
Chronic myeloid leukemia (CML) represents the prototype of genetically based diagnosis and management, and tyrosine kinase inhibitors (TKIs) exemplify the success of molecularly targeted therapy, with average life expectancy approaching that of the general population.1 However, therapy still fails in a proportion of patients. At 10 years of first-line imatinib, 47% of patients were alive and still receiving imatinib.2 Up to 26% of all imatinib discontinuation was for drug resistance.2-4 More-potent inhibitors rescued response for a proportion of patients with resistant chronic-phase (CP) disease; however, the risk of transformation and death for imatinib-resistant patients was 34% to 43%.5,6
BCR-ABL1 kinase domain (KD) mutations remain the major known mechanism of acquired TKI resistance,7-12 but they are rarely responsible for primary resistance. The events initiating BCR-ABL1–independent resistance remain poorly understood.13 However, studies have demonstrated that transformation to blast crisis (BC) is due to the accumulation of genetic abnormalities detectable as additional cytogenetic changes and mutations in individual genes identified by deep sequencing.14-22 Furthermore, gene expression changes may distinguish CP from BC and predict response to TKI therapy.23-27 The increasingly mainstream use of next-generation sequencing for studying cancer genomes is advancing our understanding of disease pathogenesis and prognosis.28-32 In CML, this technology may reveal the most pathologically relevant additional genetic events to identify high-risk patients and direct trials to test new combinational treatment approaches.
Here, we used whole-exome (WES) and whole-transcriptome sequencing (RNA-Seq) to determine somatic point or small insertion/deletion (indel) variants, copy-number variation (CNV), and fusion genes that underlie disease transformation. Patients with outlier responses have been studied to identify biomarkers of drug response,33-35 which demonstrates the potential of such cases to advance understanding of the genetic basis of treatment outcome. We therefore included diagnosis samples of patients at the extremes of TKI response: achievement of a major molecular response (MMR) vs BC transformation. Our study revealed clinically relevant variants in addition to BCR-ABL1 at CP diagnosis in patients with poor outcome and in all patients at BC. The types of variants were diverse and included gene fusions and CNVs. Many of the mutated genes have been identified in CML and other hematologic diseases.15-22,29,36 However, some of the recurrent variants, mutated genes, fusions, and splice variants are novel in CML. We highlight the importance of incorporating multiple sequencing platforms to fully characterize the various mutation classes.
Patients and methods
Patient samples and type of sequencing
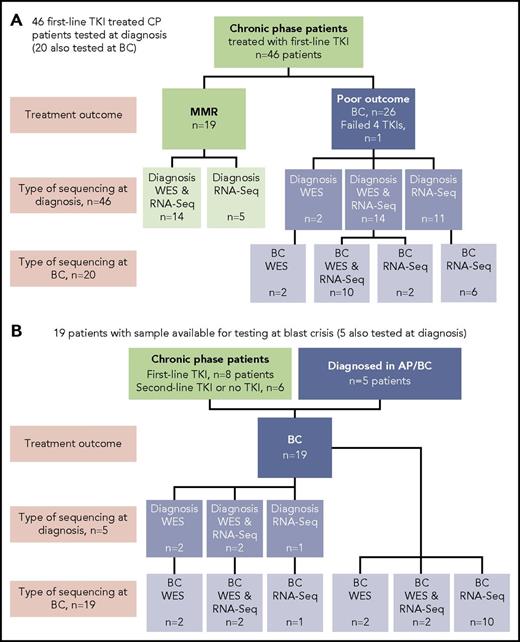
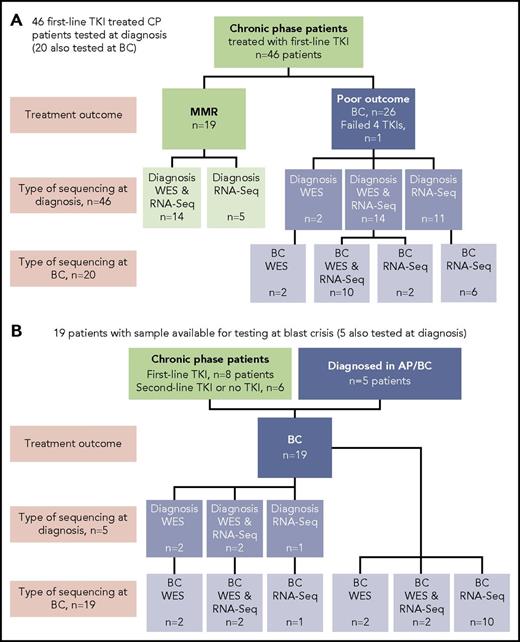
Sixty-five patients were studied, and the type of sequencing technique performed was dependent on the sample type available: DNA and/or RNA (Figure 1). DNA was available for WES for 38 of 65 patients. The somatic status of variants was assessed with a matching nonleukemic control sample in 35 of these 38 patients. Two sources of control were used: mesenchymal stromal cells or remission samples. RNA-Seq was possible for 59 of 65 patients.
Patients tested and type of sequencing. Of the total cohort of 65 patients, 38 had samples tested by WES and 59 had samples tested by RNA-Seq. (A) Forty-six CP patients treated with first-line TKIs who were all sequenced at diagnosis. Twenty of the 46 were also sequenced at BC. (B) Nineteen patients selected for sequencing on the basis of available samples at BC. Five of these patients also had samples available for sequencing at diagnosis: 2 patients diagnosed in accelerated phase (AP), 2 patients who received no TKI therapy, and 1 patient treated with a second-line TKI.
Patients tested and type of sequencing. Of the total cohort of 65 patients, 38 had samples tested by WES and 59 had samples tested by RNA-Seq. (A) Forty-six CP patients treated with first-line TKIs who were all sequenced at diagnosis. Twenty of the 46 were also sequenced at BC. (B) Nineteen patients selected for sequencing on the basis of available samples at BC. Five of these patients also had samples available for sequencing at diagnosis: 2 patients diagnosed in accelerated phase (AP), 2 patients who received no TKI therapy, and 1 patient treated with a second-line TKI.
At diagnosis, patients were selected from among >500 patients for sequencing in CP on the basis of their subsequent response to first-line TKIs and availability of appropriate samples. Nineteen patients had an optimal response, defined as durable MMR (median time to MMR, 3 months; range, 3-55 months). MMR was durable, with a median follow-up of 34 months (range, 18-174 months). Twenty-seven patients had a poor outcome, where 26 developed BC after a median of 6 months (range, 1-60 months), and for 1 patient, 4 TKIs had failed by 18 months (best response, complete hematologic response). These 27 patients were collectively termed poor-outcome patients. At BC, samples of 39 patients were sequenced, including 20 of the first-line TKI–treated patients who were also sequenced at diagnosis. The study was approved by the Human Research Ethics Committee of the Royal Adelaide Hospital, and patients provided written informed consent.
Next-generation sequencing
For WES, we used the Roche SeqCap EZ Exome v3.0 kit for 100-bp paired-end sequencing. Somatic variants were called using published algorithms (supplemental Methods, available on the Blood Web site). CNV detection in normal and tumor samples was performed using a method developed in house (supplemental Methods). Structural variants and genomic fusions were additionally assessed using Manta.37
RNA-Seq libraries were prepared using the Illumina TruSeq Stranded Total RNA protocol for 100-bp or 150-bp paired-end sequencing. The protocol proved capable of identifying sense and antisense spliced fusion transcripts as well as genomic breakpoints from intron-retaining precursor messenger RNA that are usually absent in standard poly(A)-enriched RNA-Seq. Gene fusions and other genomic rearrangements were analyzed using in-house tools based on output provided by the STAR RNA-Seq mapper38 (supplemental Methods), and single-nucleotide variants (SNVs) and indels in the RNA-Seq data were detected using FreeBayes.39 Differential gene expression analysis is described in the supplemental Methods. Sequencing was performed on Illumina HiSeq2500 and NextSeq 500 instruments.
The sequence data have been deposited at the European Genome-phenome Archive, which is hosted by the European Bioinformatics Institute, under accession #EGAS00001003071.
Variant validation
Variants were selected for validation (>3500) using independent methods (supplemental Methods and Results), including Sanger sequencing, reverse transcription polymerase chain reaction, and single-nucleotide polymorphism array. A comparison was performed between the somatic variant calls when either mesenchymal stromal cells or remission samples were used as the source of nonleukemic control, and the minimum detectable somatic variant allele frequency was established (supplemental Results).
Filtering strategy for potentially pathogenic variants
Annotation of variants is described in the supplemental Methods. Indels in protein-coding regions and nonsynonymous and essential splice-site SNVs predicted to be deleterious using in silico functional prediction models were retained and classified according to clinical relevance, as were fusions and CNVs (supplemental Methods). Missense-coding variants designated as deleterious were predicted to be damaging by at least 3 of the 4 functional prediction algorithms, which indicates that they are likely to modify protein function. These variants also met minimum evolutionary conservation scores (supplemental Methods).
Classification systems for clinical relevance were adapted to enrich for potentially pathogenic variants.40-42 Retained deleterious variants were classified as: 1) clinically relevant, 2) possibly relevant, or 3) unknown relevance (supplemental Methods). Briefly, clinically relevant variants were associated with hematologic malignancy and occurred in genes listed in the Catalogue of Somatic Mutations in Cancer Cancer Gene Census.43 These genes have high-level evidence as drivers of hematologic malignancy and are hereafter termed cancer genes. Possibly relevant variants occurred in genes recurrently mutated in our cohort. Novel somatic fusions and recurrent CNVs were also considered possibly relevant.
Statistics
Groups were compared using the Mann-Whitney rank sum test. Fisher’s exact test and the χ2 test were used to compare frequencies.
Results
Relevant variants at diagnosis in CP
Patient characteristics for the 46 patients diagnosed in CP and treated with first-line TKIs are listed in Table 1, including the best response to first-line TKI therapy. The type of sequencing performed for each patient is outlined in supplemental Table 1 and summarized in Figure 1.
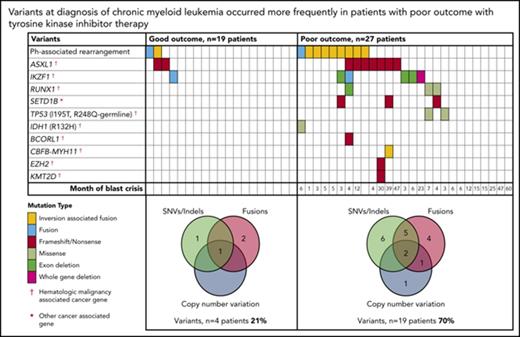
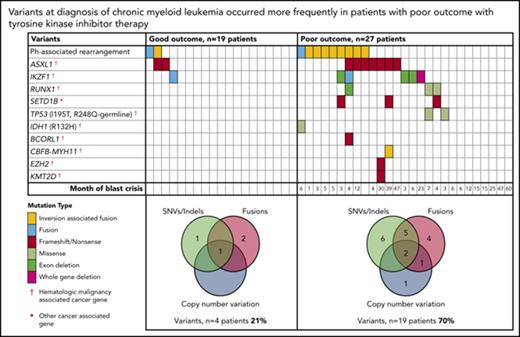
At diagnosis, variants that met our criteria for clinically relevant or possibly relevant were detected in 23 (50%) of 46 patients (Figure 2; supplemental Table 1). A majority occurred in cancer genes and were detected in patients with a poor outcome: 19 (70%) of 27 patients compared with 4 (21%) of 19 patients with subsequent MMR (P = .002). All coding variants are listed in supplemental Tables 6 and 7. When available, RNA-Seq data established whether somatic variants were detected in the transcripts. The coding variants classified as relevant were expressed, whereas some of the others were not. These were likely to be passengers that do not play a role in treatment response.
Relevant variants at diagnosis in CP for patients treated with first-line TKIs. (A) Variants at diagnosis and overlap of variant type per patient at diagnosis. The average Combined Annotation Dependent Depletion score of the SNVs and indels was 32 (range [r], 22-44), indicating they were predicted to be deleterious. All relevant SNVs were predicted to be damaging by at least 3 of 4 in silico prediction tools. For the Venn diagrams, patients with >1 variant within a category (eg, 2 SNVs) were only counted once for that category. SNVs and indels were detected by WES and/or RNA-Seq. Fusions represent fusion transcripts detected by RNA-Seq and genomic fusions where breakpoints were detected by RNA-Seq or WES, including focal deletions. CNVs were detected from the WES data. All variants that met the criteria for classification for clinical relevance or possible relevance were included. (B) Graphs show characteristics of variants that occurred in ≥3 patients. The median month and r of subsequent BC was calculated for patients with any 1 of the variants. Ph, Philadelphia.
Relevant variants at diagnosis in CP for patients treated with first-line TKIs. (A) Variants at diagnosis and overlap of variant type per patient at diagnosis. The average Combined Annotation Dependent Depletion score of the SNVs and indels was 32 (range [r], 22-44), indicating they were predicted to be deleterious. All relevant SNVs were predicted to be damaging by at least 3 of 4 in silico prediction tools. For the Venn diagrams, patients with >1 variant within a category (eg, 2 SNVs) were only counted once for that category. SNVs and indels were detected by WES and/or RNA-Seq. Fusions represent fusion transcripts detected by RNA-Seq and genomic fusions where breakpoints were detected by RNA-Seq or WES, including focal deletions. CNVs were detected from the WES data. All variants that met the criteria for classification for clinical relevance or possible relevance were included. (B) Graphs show characteristics of variants that occurred in ≥3 patients. The median month and r of subsequent BC was calculated for patients with any 1 of the variants. Ph, Philadelphia.
Among the 27 poor-outcome patients, 26 developed BC. The median time to BC was earlier for those with relevant variants at diagnosis compared with those without: 5 months (range, 1-47 months; n = 18 patients) vs 15 months (range, 6-60 months; n = 8 patients; P = .018). The most frequently mutated gene at diagnosis was ASXL1 in 9 patients. Six of these developed BC. Patients with mutated ASXL1 at diagnosis had a significantly longer time to BC than patients with other mutated genes at diagnosis that did not include ASXL1: 21 months (range, 4-47 months) vs 4.5 months (range, 1-23 months; P = .037). Furthermore, 2 patients with ASXL1 variants (allele frequencies, 22% and 40%) achieved an MMR at 3 months of first-line imatinib. The variants were not detected at remission, suggesting they were present in a leukemic clone and were reduced or eradicated with effective imatinib treatment.
Other recurrently mutated genes at CP diagnosis included cancer genes RUNX1, TP53, and IKZF1. IKZF1 variants occurred in 6 patients and comprised whole-gene (n = 1) or exon deletions (n = 3) and novel fusions (n = 2). Five of these 6 patients developed lymphoid BC (LBC) after 3 to 23 months of imatinib. A novel recurrently mutated gene was the histone methyltransferase SETD1B, in which somatic frameshift/nonsense variants occurred in 3 patients with subsequent BC. This gene, although categorized as a cancer gene in the Catalogue of Somatic Mutations in Cancer, is currently not associated with hematologic malignancy.
Thirty of the 46 patients sequenced at diagnosis had WES analysis performed and were evaluable for somatic CNV analysis. We used an algorithm developed in house, which demonstrated a high concordance (97.6%) with a gold-standard technique: Affymetrix CytoScan HD array (supplemental Results; 123 somatic CNVs were evaluable from the total cohort of patients). At diagnosis, CNVs were detected in 7 (23%) of 30 patients (supplemental Table 8). These included deletions associated with the Ph translocation in 4 of 7 patients. Six of the 7 patients with CNVs at diagnosis developed BC (n = 5) or had no response to 4 TKIs (n = 1).
Among 23 patients with poor outcome where the best response to first-line TKIs was known, 8 (30%) had primary resistance, defined as failure to achieve a major cytogenetic response. All of these patients had relevant variants at diagnosis (supplemental Table 1), compared with 6 of 9 with a major cytogenetic response (67%) and 2 of 6 with a complete cytogenetic response/MMR (33%; P = .027). The 1 patient who achieved but did not sustain an MMR transformed rapidly at 25 months of imatinib and acquired an MLL fusion.
The Sokal risk score was known for 37 of 46 CP patients. There was an association between Sokal score and relevant variants at diagnosis: high, 86% (6 of 7 patients); intermediate, 42% (5 of 12 patients); and low, 28% (5 of 18 patients; P = .03). We also tested for differentially expressed genes between good and poor responders at diagnosis to determine if an expression signature could be identified for use as a classifier for outcome. However, the expression analysis did not result in any genes differentially expressed at the level of P < .05 (Benjamini-Hochberg adjusted; supplemental Figure 18).
Relevant variants at BC
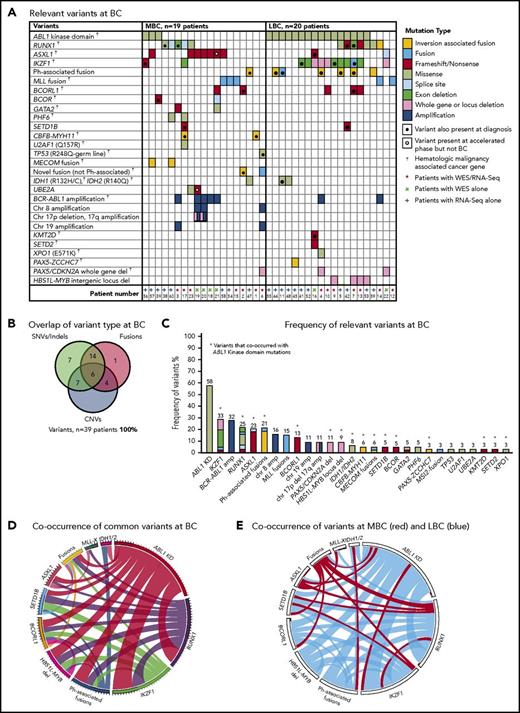
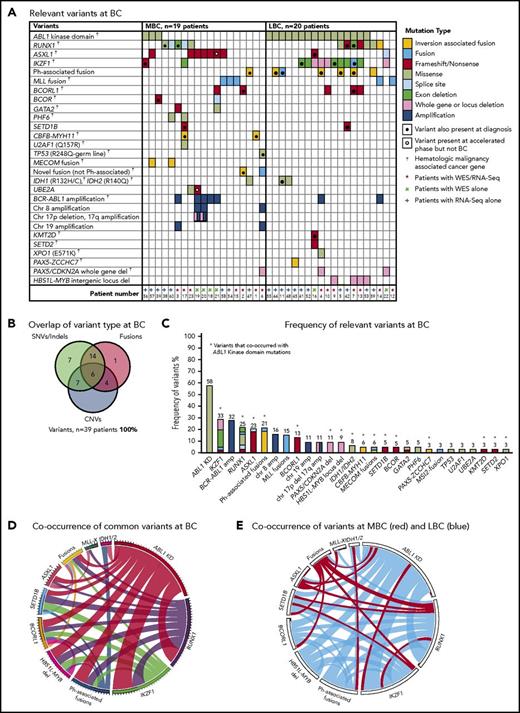
Thirty-nine patients had sequencing performed at BC, including 20 of the first-line TKI–treated patients who were sequenced at CP diagnosis described in “Patient samples and type of sequencing” (Figure 1). All 39 patients had 1 to 6 variants in cancer genes at BC (supplemental Table 1; Figure 3). Variants were acquired in genes that were also somatically mutated in other patients at CP diagnosis: IKZF1, RUNX1, ASXL1, BCORL1, and IDH1.
Relevant variants at BC for 39 patients. (A) Variants at BC. Recurrent CNVs are shown except for immunoglobulin and T-cell receptor deletions. (B) Overlap of variant type per patient at BC. (C) The percentage frequency of each mutated gene or locus is indicated above each column and calculated relative to the number of patients who underwent a particular form of sequencing or treatment (eg, the frequency of ABL1 mutations is calculated for the 33 patients who had received TKIs at the time of sequencing). Recurrent CNVs are graphed, except immunoglobulin and T-cell receptor deletions. IKZF1 and RUNX1 fusions and deletions are grouped with the other variants in these genes. The non–Ph-associated fusions are shown if 1 of the fusion partners was a known cancer gene. (D) Pairwise cooccurrence of variant types and genes. All genes that were mutated in ≥3 patients are included. The width of the ribbon correlates with the relative frequency of cooccurring mutated genes/variant type. Variants are arranged in clockwise order of frequency. Fusions where 1 of the partners was a cancer gene are grouped (fusions), except for MLL fusions (MLL-X). MLL fusions were the most frequent fusions and had few cooccurring variants, whereas ABL1 KD mutations and RUNX1 and IKZF1 variants had multiple cooccurring variants. The novel recurrently mutated gene SETD1B is shown for all patients with variants in this gene to show cooccurrence, although the variant was below the level of detection in 1 patient at BC. (E) The same plot is shown where the colors indicate cooccurring variants associated with myeloid BC (MBC) in red and LBC in blue, which reveals that cancer gene–associated fusions were rare in LBC, as were ASXL1 variants. The Ph-associated fusions were more frequently linked with LBC.
Relevant variants at BC for 39 patients. (A) Variants at BC. Recurrent CNVs are shown except for immunoglobulin and T-cell receptor deletions. (B) Overlap of variant type per patient at BC. (C) The percentage frequency of each mutated gene or locus is indicated above each column and calculated relative to the number of patients who underwent a particular form of sequencing or treatment (eg, the frequency of ABL1 mutations is calculated for the 33 patients who had received TKIs at the time of sequencing). Recurrent CNVs are graphed, except immunoglobulin and T-cell receptor deletions. IKZF1 and RUNX1 fusions and deletions are grouped with the other variants in these genes. The non–Ph-associated fusions are shown if 1 of the fusion partners was a known cancer gene. (D) Pairwise cooccurrence of variant types and genes. All genes that were mutated in ≥3 patients are included. The width of the ribbon correlates with the relative frequency of cooccurring mutated genes/variant type. Variants are arranged in clockwise order of frequency. Fusions where 1 of the partners was a cancer gene are grouped (fusions), except for MLL fusions (MLL-X). MLL fusions were the most frequent fusions and had few cooccurring variants, whereas ABL1 KD mutations and RUNX1 and IKZF1 variants had multiple cooccurring variants. The novel recurrently mutated gene SETD1B is shown for all patients with variants in this gene to show cooccurrence, although the variant was below the level of detection in 1 patient at BC. (E) The same plot is shown where the colors indicate cooccurring variants associated with myeloid BC (MBC) in red and LBC in blue, which reveals that cancer gene–associated fusions were rare in LBC, as were ASXL1 variants. The Ph-associated fusions were more frequently linked with LBC.
The most frequently mutated gene was ABL1, where KD mutations were detected in 19 (58%) of 33 TKI-treated patients. These mutations were more frequently detected at LBC (16 [84%] of 19 patients) compared with MBC (3 [21%] of 14 patients; supplemental Results). The average allele frequency of the KD mutations was 31% detected by WES (range, 13%-65%) and 47% by RNA-Seq (range, 10%-88%). All of these mutations were known imatinib-resistant mutations,44,45 and all were confirmed to be present in the BCR-ABL1 transcript by Sanger sequencing.8 Only 2 of 19 patients had ABL1 KD mutations as the sole mutated cancer gene. Pairwise interactions between mutated genes were assessed and potential interactions identified (supplemental Results; supplemental Table 10). The number of mutants was low, but no patient with an MLL fusion had an ABL1 KD mutation. Conversely, ABL1 KD mutations were frequently observed in patients with IKZF1 variants.
CNVs at BC
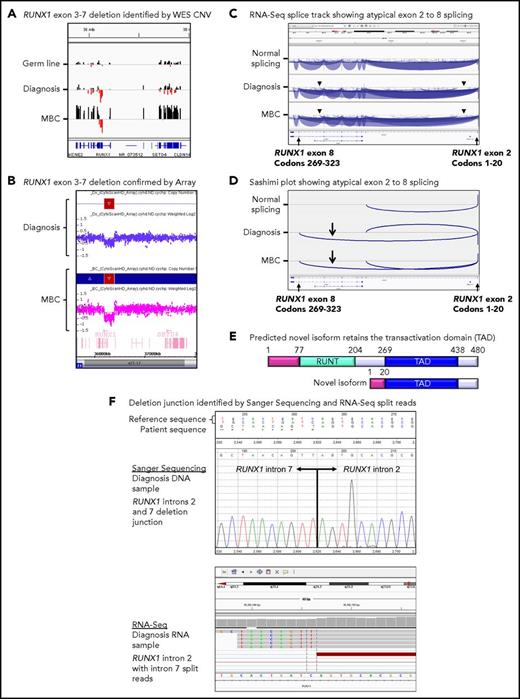
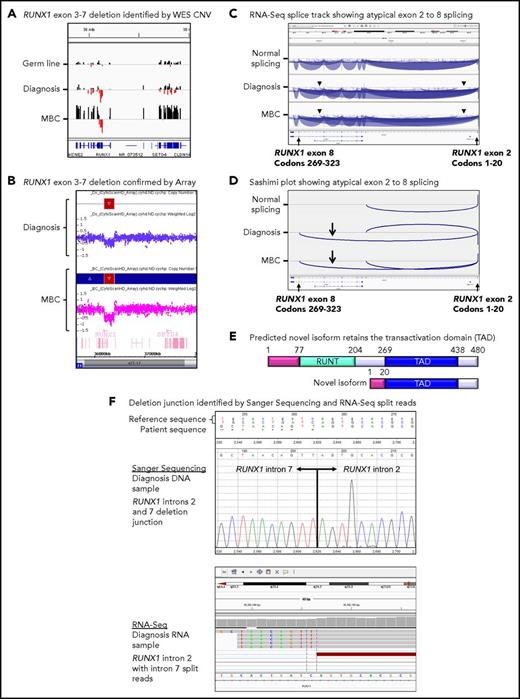
Twenty patients had WES analysis performed at BC and were evaluable for somatic CNV analysis. CNVs were detected in 18 (90%) of 20 patients (supplemental Table 8; supplemental Figures 4 and 5). Recurrent CNVs involved large regions of chromosomes 8, 17, and 19 and the Ph chromosome [der(22)]. Patients with LBC had a high frequency of submicroscopic deletions of immunoglobulin and T-cell receptor loci, consistent with clonal populations of lymphoid progenitors. A novel recurrent ∼60-Kb deletion comprising exons 1 to 2 of HBS1L and an intergenic sequence between HBS1L and MYB (termed HBS1L-MYB locus deletion) was detected in 3 of 10 patients with LBC (supplemental Figure 6). Genes with recurrent submicroscopic deletions included RUNX1 (Figure 4) and IKZF1 (supplemental Figure 9), and atypical splice junctions were frequently detectable by RNA-Seq in patients with exon deletions (Figure 4; supplemental Figures 8 and 9).
Somatic RUNX1 deletion and novel transcript. (A) CNV BedGraphs were generated by an in-house algorithm from the WES data for the germ line, diagnosis, and MBC samples of patient 3. The germ line gray central horizontal line indicates 2 copies of each gene. Extended red bars below the central line indicate a deletion of RUNX1 exons 3 to 7 at diagnosis and MBC. The extended black bars above the central line at MBC indicate chromosome 21 gain. (B) The RUNX1 deletion was confirmed by the CytoScan HD Array, where additional intronic markers revealed the full extent of the deletion. The genomic region is shown using the Affymetrix Chromosome Analysis Suite for the diagnosis and MBC samples. Red boxes represent the deletion, and blue boxes at MBC confirm the gain. (C) The deletion generated a novel in-frame RUNX1 exon 2 to 8 transcript. The splice junction tracks visualized in the Integrative Genomics Viewer are shown for a representative sample analyzed by RNA-Seq with normal RUNX1 splicing and the diagnosis and MBC samples of patient 3 with atypical RUNX1 exon 2 to 8 splicing. Arcs represent splice junctions that connect exons. Junctions from the minus strand are colored blue and extend below the center line. The arrowheads indicate the location of the genomic deletion breakpoints. RefSeq transcripts are shown at the bottom. (D) Sashimi plots of the diagnosis and MBC samples generated from the RUNX1 RNA-Seq splice junction track, which allows visualization of the novel splice generated by the deletion. Only the junctions that overlap RUNX1 RefSeq transcript ID NM_001754 exon 2 are shown. (E) The predicted novel isoform excludes the Runt DNA binding domain. (F) The stranded RNA-Seq protocol enabled sequencing of intron-retaining precursor RNA and genomic breakpoint detection. The intronic breakpoints were identified by RNA-Seq of the corresponding RNA samples at chr21:36392146 in intron 2 and chr21:36201259 in intron 7 (hg19). The genomic deletion junction was polymerase chain reaction amplified and Sanger sequenced, which confirmed a 190 888 base deletion.
Somatic RUNX1 deletion and novel transcript. (A) CNV BedGraphs were generated by an in-house algorithm from the WES data for the germ line, diagnosis, and MBC samples of patient 3. The germ line gray central horizontal line indicates 2 copies of each gene. Extended red bars below the central line indicate a deletion of RUNX1 exons 3 to 7 at diagnosis and MBC. The extended black bars above the central line at MBC indicate chromosome 21 gain. (B) The RUNX1 deletion was confirmed by the CytoScan HD Array, where additional intronic markers revealed the full extent of the deletion. The genomic region is shown using the Affymetrix Chromosome Analysis Suite for the diagnosis and MBC samples. Red boxes represent the deletion, and blue boxes at MBC confirm the gain. (C) The deletion generated a novel in-frame RUNX1 exon 2 to 8 transcript. The splice junction tracks visualized in the Integrative Genomics Viewer are shown for a representative sample analyzed by RNA-Seq with normal RUNX1 splicing and the diagnosis and MBC samples of patient 3 with atypical RUNX1 exon 2 to 8 splicing. Arcs represent splice junctions that connect exons. Junctions from the minus strand are colored blue and extend below the center line. The arrowheads indicate the location of the genomic deletion breakpoints. RefSeq transcripts are shown at the bottom. (D) Sashimi plots of the diagnosis and MBC samples generated from the RUNX1 RNA-Seq splice junction track, which allows visualization of the novel splice generated by the deletion. Only the junctions that overlap RUNX1 RefSeq transcript ID NM_001754 exon 2 are shown. (E) The predicted novel isoform excludes the Runt DNA binding domain. (F) The stranded RNA-Seq protocol enabled sequencing of intron-retaining precursor RNA and genomic breakpoint detection. The intronic breakpoints were identified by RNA-Seq of the corresponding RNA samples at chr21:36392146 in intron 2 and chr21:36201259 in intron 7 (hg19). The genomic deletion junction was polymerase chain reaction amplified and Sanger sequenced, which confirmed a 190 888 base deletion.
Gene fusions and expression analysis at BC
Thirty-three patients had RNA-Seq performed at BC and were assessed for gene fusions. Fourteen patients (42%) had 18 known or novel fusions that were not associated with the Ph translocation (see “Novel class of variant associated with the Ph translocation”). MLL (KMT2A) fusions were the most frequent (n = 5), and 2 of these were cytogenetically cryptic. The other known fusions were CBFB-MYH11 and PAX5-ZCCHC7. Novel fusions in 6 patients involved cancer genes implicated in hematologic malignancy: RUNX1, IKZF1, MECOM, and MSI2 (supplemental Table 11).
In total, 14 (78%) of 18 fusions at BC were known leukemia-associated fusions (n = 7) or were fusions in which a fusion partner was a known leukemia-associated gene (n = 7). A number of the novel fusions were cytogenetically cryptic, including an intrachromosomal RUNX1-MX1 out-of-frame fusion. A submicroscopic 6.6-Mb inversion brought the RUNX1 gene into the same transcriptional orientation as MX1 (supplemental Figure 10). A similar inversion generated a novel intrachromosomal MBNL1-MECOM fusion. A complex series of events generated a cryptic MSI2-PRMT2 fusion, including a 3-way exchange of sequence from chromosomes 17, 19, and 21 (supplemental Figure 11). In total, 151 of the fusions that met our criteria for relevance were evaluated using reverse transcription polymerase chain reaction, and 150 (99%) were validated (supplemental Results).
Gene expression analysis among patients with specific mutated genes was confounded by multiple mutations in different cancer genes in most patients. These multiple variants included SNVs, indels, and fusions, plus CNVs in some patients. However, expression analysis between disease phases revealed differentially expressed genes (supplemental Results). Samples of TKI-treated patients at MBC (n = 11) or LBC (n = 18) were compared with the diagnosis samples of 43 patients who were subsequently treated with first-line TKIs. Supplemental Figure 18A shows a heat map for hierarchical clustering of 1156 differentially expressed genes with an adjusted P value <.001. Two clusters were dominated by the BC samples, whereas there was no discernable clustering of the diagnosis samples according to treatment outcome. One BC cluster only included patients with LBC (n = 15), and the predominant genes mutated were ABL1 and IKZF1 in 93% and 53% of patients, respectively (supplemental Figure 18B). Most patients in the other BC cluster had MBC (10 [63%] of 16 patients), and the predominant variants were fusions (not Ph-associated) in 56% of patients. All 5 patients with MLL fusions at BC clustered in this group, including 2 patients with LBC.
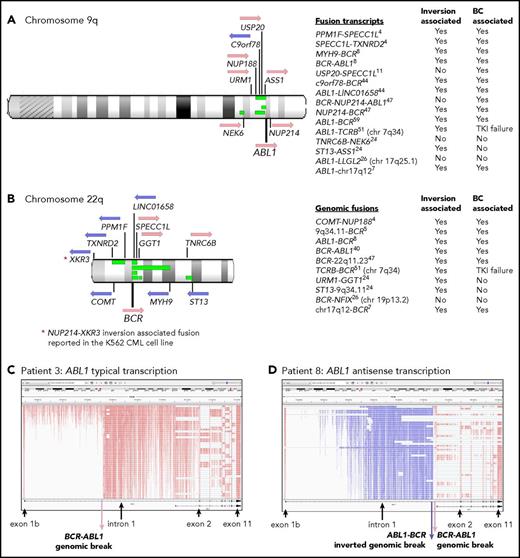
Novel class of variant associated with the Ph translocation
Twelve of 59 patients with RNA-Seq performed had gene rearrangement and novel fusions on the chromosome arms involved in the Ph translocation (supplemental Table 11; Figure 5). These are termed Ph-associated rearrangements. They were detected in 11 patients at diagnosis and in 1 patient tested only at BC. The somatic status of the rearrangements was confirmed in 5 of 5 patients with available nonleukemic control. No newly emerging Ph-associated rearrangements were detected at BC, suggesting they occurred with the initiating translocation.
Ph translocation–associated fusions and rearrangement. Chromosomes 9q (A) and 22q (B) are shown along with the location of genes involved in the fusions. Pink arrows indicate transcription from the plus strand and blue arrows from the minus strand. Green bars indicate the relative location of deletions detected in 4 patients: patients 4, 8, 11, and 26. These align with the fusion partners in some cases. Sequence inversions brought some genes into the same transcriptional orientation. Fusion partners were located upstream and downstream of ABL1 and BCR, indicating the complex nature of the rearrangements. Fusion partners were also located on other chromosomes in 3 patients with variant Ph translocations. Listed are fusion transcripts and the genomic fusions where no corresponding fusion transcript was identified. Superscripts indicate the patient number. Two fusions involved intragenic sequence inversions: BCR sequence inversion (NUP214-BCR) and GGT1 sequence inversion (URM1-GGT1). Some fusions involving an intergenic sequence also contained an inverted sequence. (C) RNA-Seq aligned sequencing reads that mapped to the ABL1 gene for a representative patient at diagnosis (patient 3) viewed in the Integrative Genomics Viewer. Reads are colored by first-of-pair strand. A large number of reads were evident in intronic regions because the protocol sequenced total RNA, including intron-retaining primary RNA transcripts. ABL1 is transcribed on the plus strand, and reads are colored pink. (D) Patient 8 had a large ∼93-Kb inversion within ABL1 intron 1, as indicated by the reads colored blue, which specify antisense transcription. A genomic break was evident at the distal end of the antisense transcripts, which was an ABL1-BCR genomic fusion inversion. The BCR-ABL1 genomic breakpoint was located 224 bp upstream in the correct transcriptional orientation. The patient also had a 13-Mb deletion between BCR and the MYH9 gene on chromosome 22 (shown in panel B), plus an inversion that brought MYH9 into the same transcriptional orientation as BCR and generated an MYH9-BCR fusion transcript.
Ph translocation–associated fusions and rearrangement. Chromosomes 9q (A) and 22q (B) are shown along with the location of genes involved in the fusions. Pink arrows indicate transcription from the plus strand and blue arrows from the minus strand. Green bars indicate the relative location of deletions detected in 4 patients: patients 4, 8, 11, and 26. These align with the fusion partners in some cases. Sequence inversions brought some genes into the same transcriptional orientation. Fusion partners were located upstream and downstream of ABL1 and BCR, indicating the complex nature of the rearrangements. Fusion partners were also located on other chromosomes in 3 patients with variant Ph translocations. Listed are fusion transcripts and the genomic fusions where no corresponding fusion transcript was identified. Superscripts indicate the patient number. Two fusions involved intragenic sequence inversions: BCR sequence inversion (NUP214-BCR) and GGT1 sequence inversion (URM1-GGT1). Some fusions involving an intergenic sequence also contained an inverted sequence. (C) RNA-Seq aligned sequencing reads that mapped to the ABL1 gene for a representative patient at diagnosis (patient 3) viewed in the Integrative Genomics Viewer. Reads are colored by first-of-pair strand. A large number of reads were evident in intronic regions because the protocol sequenced total RNA, including intron-retaining primary RNA transcripts. ABL1 is transcribed on the plus strand, and reads are colored pink. (D) Patient 8 had a large ∼93-Kb inversion within ABL1 intron 1, as indicated by the reads colored blue, which specify antisense transcription. A genomic break was evident at the distal end of the antisense transcripts, which was an ABL1-BCR genomic fusion inversion. The BCR-ABL1 genomic breakpoint was located 224 bp upstream in the correct transcriptional orientation. The patient also had a 13-Mb deletion between BCR and the MYH9 gene on chromosome 22 (shown in panel B), plus an inversion that brought MYH9 into the same transcriptional orientation as BCR and generated an MYH9-BCR fusion transcript.
The Ph-associated rearrangements were more frequently detected at diagnosis in patients with poor outcome: 9 (33%) of 27 first-line TKI–treated CP patients who developed BC (n = 8) or for whom 4 TKIs failed (n = 1) vs 2 (11%) of 19 with sustained MMR (P = .07). The events generated 15 novel fusion transcripts, plus 10 genomic fusions where no corresponding fusion transcript was detected. The fusions frequently involved inversions that brought genes into the same transcriptional orientation. In addition, 4 patients had intragenic inversions of ABL1 or BCR. Figure 5D shows a 93-Kb intragenic inversion of ABL1 intron 1. In total, sequence inversions were detected in 10 (83%) of 12 patients.
Interestingly, fusion partner genes were located both upstream and downstream of BCR and ABL1 (Figure 5), and some fusions were associated with deletions on chromosomes 9 and 22. These were detected in 4 of 7 patients with CNV analysis performed. An insertion of NUP214 exon 23 occurred in 1 patient between BCR exon 19 and ABL1 exon 2, which generated an in-frame BCR-NUP214-ABL1 transcript (supplemental Results). NUP214 is normally located downstream of ABL1. The additional exon could potentially alter the conformation of a generated protein and modify sensitivity to imatinib. The observed gene fusions in some patients and their transcriptional orientation provide evidence of a complex series of events, including chromosome fragmentation, sequence inversion, and imperfect reassembly generating random fusion (supplemental Figures 14-16).
Timeline of mutation acquisition in cancer genes
Various patterns of mutation acquisition with disease stage were evident. First, 5 patients diagnosed in accelerated phase/BC had 1 to 2 cancer gene variants at diagnosis: CBFB-MYH11, RUNX1, GATA2, BCOR, BCORL1, and an MSI2 fusion.
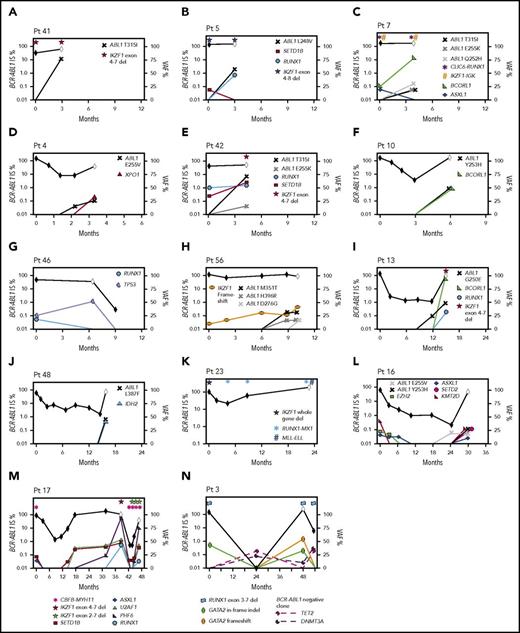
Second, 18 CP patients had 1 to 4 cancer gene variants at diagnosis, including recurrently mutated genes TP53, RUNX1, IKZF1, and ASXL1. Fifteen of these 18 developed BC (n = 14; median, 6 months; range, 3-47 months) or had no response to 4 TKIs (n = 1). Eleven of 12 patients who were tested during therapy gained 1 to 5 additional variants in cancer genes (Figure 6). The other 3 of 18 patients achieved and maintained MMR: 2 had somatic ASXL1 variants, and 1 had a novel somatic IKZF1-TCF4 fusion at diagnosis.
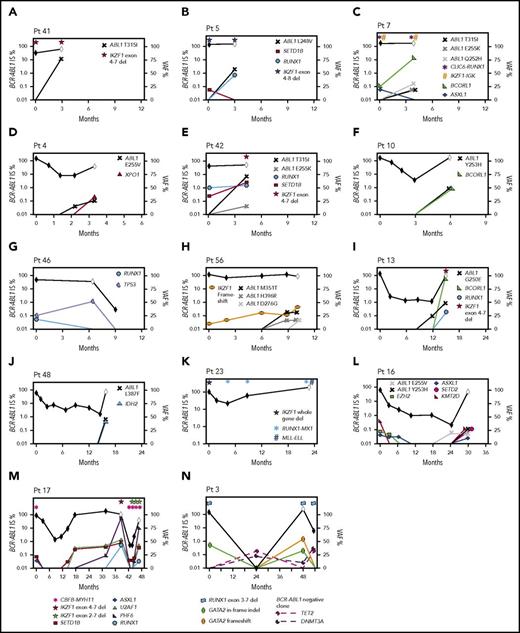
Timeline of mutation acquisition. Multiple patients had additional analyses after diagnosis by next-generation or Sanger sequencing. Shown are the BCR-ABL1 transcript levels over time (black diamond symbol; left y-axis). White diamonds indicate BC. Variants are plotted according to their time point of detection and percentage variant allele frequency (VAF; right y-axis). Lines that extend from 0 VAF indicate the time point before the first detection of the variant. BCORL1 and PHF6 are located on the X chromosome and represent a single-gene copy (all male patients). The VAFs of the ABL1 KD mutations were derived from Sanger sequencing by isolating the BCR-ABL1 allele, but the VAFs are normalized for diploid number. Fusions and intra- and intergenic deletions were not quantified and are plotted with symbols above their time point of detection. (A-H) Eight patients with BC before 12 months. In 6 of these patients, variants were present at diagnosis. ABL1 KD mutations were acquired in 7 of the 8 patients. (I-N) Six patients with BC after 12 months. Variants were present at diagnosis in 4 and ABL1 KD mutations were acquired in 3 of the 6 patients. The graphs demonstrate that the VAF decreased or increased over time for some variants, which corresponded with BCR-ABL1 levels. In some cases, the VAF changes suggest clonal competition. (N) Patient 3 had DNMT3A and TET2 mutations detected in a remission sample when BCR-ABL1 transcripts were undetectable, and the pattern of clonal selection is consistent with their presence in a BCR-ABL1− clone. IS, International Scale.
Timeline of mutation acquisition. Multiple patients had additional analyses after diagnosis by next-generation or Sanger sequencing. Shown are the BCR-ABL1 transcript levels over time (black diamond symbol; left y-axis). White diamonds indicate BC. Variants are plotted according to their time point of detection and percentage variant allele frequency (VAF; right y-axis). Lines that extend from 0 VAF indicate the time point before the first detection of the variant. BCORL1 and PHF6 are located on the X chromosome and represent a single-gene copy (all male patients). The VAFs of the ABL1 KD mutations were derived from Sanger sequencing by isolating the BCR-ABL1 allele, but the VAFs are normalized for diploid number. Fusions and intra- and intergenic deletions were not quantified and are plotted with symbols above their time point of detection. (A-H) Eight patients with BC before 12 months. In 6 of these patients, variants were present at diagnosis. ABL1 KD mutations were acquired in 7 of the 8 patients. (I-N) Six patients with BC after 12 months. Variants were present at diagnosis in 4 and ABL1 KD mutations were acquired in 3 of the 6 patients. The graphs demonstrate that the VAF decreased or increased over time for some variants, which corresponded with BCR-ABL1 levels. In some cases, the VAF changes suggest clonal competition. (N) Patient 3 had DNMT3A and TET2 mutations detected in a remission sample when BCR-ABL1 transcripts were undetectable, and the pattern of clonal selection is consistent with their presence in a BCR-ABL1− clone. IS, International Scale.
The third pattern involved 14 patients tested at CP diagnosis who did not have variants in cancer genes but developed BC (median, 14 months; range, 1-64 months). Eleven of these 14 were tested at BC, and all gained 1 to 4 cancer gene variants. Four patients had samples available to track the time point of variant acquisition by Sanger sequencing (Figure 6).
Of 16 patients with ABL1 KD mutations at BC who were sequenced at prior time points, 10 (62%) had cancer gene variants that predated the ABL1 mutations. Clonal competition between variants was evident over time, where the allele frequency decreased after diagnosis for some variants and increased for others (Figure 6).
One patient had DNMT3A and TET2 variants detected in a remission sample (Figure 6N), and the pattern of clonal selection is consistent with their presence in a BCR-ABL1− clone, which is detailed in the supplemental Results.
Discussion
Our study has demonstrated that somatic variants in cancer genes in addition to the BCR-ABL1 fusion are present at diagnosis in a significant subset of patients, particularly in those patients who, despite TKI therapy, experience early BC transformation. The variants included gene fusions, splice anomalies, and CNVs, and this diversity of mutation type would not have been detected by DNA sequencing alone. This underscores the importance of multiple modes of variant detection to reveal relevant genomic events in addition to the BCR-ABL1 fusion.
Among the genes mutated at diagnosis were RUNX1, ASXL1, and TP53, which confer a poor prognosis for patients with acute myeloid leukemia.46 Their role in prognosis and treatment outcome for patients with CML is yet to be established. RUNX1 is a frequent and aggressive driver of hematologic malignancies15,46-48 and has been described in multiple CML studies, but it is not always associated with poor outcome.16-22 In our cohort, mutated RUNX1 was rarely detected as the sole mutated cancer gene. We speculate that in optimal responders to TKIs, certain leukemic clones with mutated genes at diagnosis may be eradicated by TKIs. This may be dependent on a number of factors, including cooccurring mutations, the variant allele frequency, and the varying degree of innate TKI resistance conferred by particular variants. In our cohort, patients with an ASXL1 variant at diagnosis had a longer median time to BC than other patients. Furthermore, 2 of 9 patients with an ASXL1 variant at diagnosis achieved an MMR. One of these variants occurred in exon 11. A vast majority of ASXL1 variants are exon 12 frameshift/nonsense and may be gain-of-function mutations,49 whereas the functional significance of variants in other regions are unknown. The significance of mutated genes at diagnosis and their association with outcome warrant further investigation.
An unexpected finding was complex rearrangements and novel fusions associated with the Ph translocation. Chromosome 9 and 22 sequences were possibly fragmented during the Ph translocation in some patients. Some fragments were deleted and others inverted. Remaining fragments were reassembled imperfectly and generated random fusions, while maintaining the leukemia-initiating BCR-ABL1 fusion. Limitations in previous technology may have precluded their discovery. These occurred more frequently in patients with a poor outcome vs optimal responders and may constitute an important additional genomic risk factor. Derivative 9 deletions, which mainly spanned the translocation breakpoint, were associated with poorer outcome in the pre-TKI era.50-52 However, some of our deletions were noncontiguous and at locations not detectable by older techniques.50-54 Interestingly, a novel NUP214-XKR3 9;22 fusion has been reported in 2 independent studies in the BCR-ABL1+ cell line K562 that would require a similar series of events for formation, including sequence inversion.55,56 The role of the Ph-associated fusions in treatment outcome is unknown. We have revealed another layer of molecular heterogeneity, leading us to speculate that there may be a subcategory of patients in whom Ph-associated gene fusions/inversions modify outcome with TKI therapy.
Our study revealed a high rate of other gene fusions (42% of patients at BC), and most were either fusions known to be associated with hematologic malignancy or fusions in which 1 of the fusion partners was a known cancer gene.46,57-60 However, it will be important to functionally characterize the novel fusions to establish whether they are transforming and whether they confer TKI resistance or reduced sensitivity, which is planned in future studies.
Many of the mutated genes may be targets for emerging therapy. IDH1/2 inhibitors are in clinical trial for the treatment of acute myeloid leukemia.61,62 ASXL1 mutants were hypersensitive to recently developed bromodomain inhibitors.49 We detected an XPO1 E571K mutation in an isolated patient at LBC. This variant may play a role in B-cell hematologic malignancies,63 and XPO1 is implicated in ABL1-independent resistance.64,65 XPO1 inhibitors have demonstrated antileukemic activity.65,66 MLL rearrangements may be targeted by inhibitors.67,68 These were the most frequent fusions at BC, and some were cytogenetically cryptic, which emphasizes the advantage of using additional genomic tools to identify potentially targetable mutations.
The mutational spectrum in cancer genes was diverse, and subcohorts of patients with specific mutations were small in number. For this reason, we generally could not find specific gene expression signatures correlated with mutations in a statistically meaningful fashion. General differences were observed between the transcriptomes of diagnosis and BC samples, and in the latter, differences were seen between MBC and LBC samples. These differences in expression were accompanied by differences in mutational patterns.
Our study is among the most comprehensive genomic analyses of patients with CML. It included matched diagnosis/BC samples in many patients, coupled with nonleukemic controls, to determine the somatic status of variants. BC is a rare event for TKI-treated patients, and whether these findings extend to a broader population of patients will require assessment of a large cohort of unselected and consecutively treated patients. Future genomic analyses may reveal prognostic risk groups, and combining genomic data with clinical parameters could improve prognostication and disease classification, as it has in other hematologic malignancies.46,69
Although we cannot generalize our findings to current clinical practice, there are already hints that we can generate clinically useful data. There was marked heterogeneity of mutated genes in individual patients, with up to 5 mutated cancer genes at BC. Furthermore, by tracking variants in prior samples, clonal competition among mutants was evident. Additional studies will add to our understanding of treatment response and clarify the clinical usefulness of genomic analysis. These analyses are becoming increasingly affordable and in the future may be routine at cancer diagnosis. It is important to prepare for this era by gaining a deeper understanding of the somatic genome and its association with outcome in CML. Furthermore, collection of a matched nonleukemic sample at diagnosis, such as hair follicles or nail clippings,70 will aid the future interpretation of data.
Although generic imatinib has become the standard of care for patients with CML in many countries because of its cost effectiveness and safety profile, it is likely that selected patients would benefit from a therapeutic approach incorporating more potent kinase inhibition or combinational therapy targeting additional pathways. If very high-risk cases could be identified with confidence, upfront allogeneic transplantation may even be considered. A detailed understanding of the prognostic implications of specific genetic variants will be critical to the achievement of a risk-adapted approach to CML therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Korea Leukemia Bank for providing biospecimens and data (NRF-2013M3A9B8031236), the SA Cancer Research Biobank, the Australasian Leukaemia and Lymphoma Group, the Centre for Cancer Biology (an alliance between SA Pathology and the University of South Australia), Rosalie Kenyan for assistance with the SNP array work, John Toubia for valuable discussion, Monique Sinton for assistance with variant validation, and Sarah Moore, Jeff Suttle, and Mario Nicola for cytogenetic analysis and interpretation.
This work was supported by the National Health and Medical Research Council of Australia (APP1027531 and APP1104425 [S.B.], APP1135949 [T.P.H.], APP1023059 [H.S.S.], and APP1146253 [D.T.Y.]), the Ray and Shirl Norman Cancer Research Trust, the Royal Adelaide Hospital Research Foundation, and a grant from The Leukemia and Lymphoma Society. T.P.H. and H.S.S. are supported by Cancer Council SA’s Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health. The Australian Cancer Research Foundation (ACRF) Cancer Genomics Facility was established with funding from Therapeutic Innovation Australia and the ACRF. I.W. was supported by a travel grant from Bloodwise United Kingdom. W.T.P. was supported by a postdoctoral fellowship from the Leukaemia Foundation of Australia/Cure Cancer Australia.
Authorship
Contribution: S.B. designed the research, analyzed and interpreted the data, and wrote the manuscript; P.W. and A.W.S. performed the bioinformatic analyses, developed in-house bioinformatic pipelines, analyzed and interpreted the data, and wrote the manuscript; D.T.Y. designed the research, interpreted the data, provided clinical insight, and contributed to manuscript preparation; T.P.H. interpreted the data, provided clinical insight, and contributed significantly to manuscript preparation; W.T.P. and J.G. designed the research, performed experiments, and reviewed the manuscript; A.P., D.T., C.W., N.H.S., J.E.M., N.N., D.S., Z.D., H.A., J.G., J.B., and I.W. performed experiments and reviewed the manuscript; C.D., S.-H.K., S.-Y.C., and S.-H.P. provided samples and data and reviewed the manuscript; J.F. developed bioinformatic pipelines and reviewed the manuscript; H.S.S., A.B., C.H., and D.L.W. provided scientific insight and performed critical review of the manuscript; and D.M.R., A.S.M.Y., N.S., M.C.M., and D.-W.K. provided clinical insight and performed critical review of the manuscript.
Conflict-of-interest disclosure: S.B. received research funding and honoraria from and served on advisory committees for Novartis and Bristol-Myers Squibb and received consultancy fees and honoraria from Qiagen and Cepheid; D.T.Y. received research funding and honoraria from and served on advisory committees for Novartis and Bristol-Myers Squibb and has received research funding from Ariad; N.S. received travel sponsorship and honoraria from Novartis and Bristol-Myers Squibb; M.C.M. received support from Novartis, Bristol-Myers Squibb, and Ariad; A.B. received honoraria from Novartis; D.L.W. received research support from Novartis and Bristol-Myers Squibb and honoraria from Bristol-Myers Squibb; A.S.M.Y. received research funding from Novartis, Bristol-Myers Squibb, and Celgene and honoraria from Novartis and Bristol-Myers Squibb; D.-W.K. received research funding and honoraria from Novartis, Bristol-Myers Squibb, and Ariad; D.M.R. received research funding from Novartis and Celgene and honoraria from Novartis and Bristol-Myers Squibb; H.S.S. received honoraria from Celgene; and T.P.H. received research funding and honoraria from and served on advisory committees for Novartis, Bristol-Myers Squibb, Celgene, and Ariad. The remaining authors declare no competing financial interests.
Correspondence: Susan Branford, Department of Genetics and Molecular Pathology, SA Pathology, PO Box 14 Rundle Mall, Adelaide, SA, 5000, Australia; e-mail: susan.branford@sa.gov.au.

![Figure 2. Relevant variants at diagnosis in CP for patients treated with first-line TKIs. (A) Variants at diagnosis and overlap of variant type per patient at diagnosis. The average Combined Annotation Dependent Depletion score of the SNVs and indels was 32 (range [r], 22-44), indicating they were predicted to be deleterious. All relevant SNVs were predicted to be damaging by at least 3 of 4 in silico prediction tools. For the Venn diagrams, patients with >1 variant within a category (eg, 2 SNVs) were only counted once for that category. SNVs and indels were detected by WES and/or RNA-Seq. Fusions represent fusion transcripts detected by RNA-Seq and genomic fusions where breakpoints were detected by RNA-Seq or WES, including focal deletions. CNVs were detected from the WES data. All variants that met the criteria for classification for clinical relevance or possible relevance were included. (B) Graphs show characteristics of variants that occurred in ≥3 patients. The median month and r of subsequent BC was calculated for patients with any 1 of the variants. Ph, Philadelphia.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/9/10.1182_blood-2018-02-832253/4/m_blood832253f2.jpeg?Expires=1765904486&Signature=WVfHdtJPY6Eid4Z6HiZpK8oUdWVVWz1hrF313N11jtTdDR6VLi-L2OsPE-7SW4MXsmU~N0EppsTvrO~m~rdaINegd3129XXWkc1Md~8t399Ca372XiVzQmeVmmqk8l1W~i~~IQ~RU~t1sshnx2T9FQen4ZVSNwFAIS05jA08w~KjGHIECMZ665D0~o1aWBQz5fu74vq8BX2WAeYzKNXpBEd6J8WKlXlPeqJMyZEzow3oMtfidxyMvPwK4wQWyssEb~ZEC37~fFMm2cYyvdmkKDI6wRPPdL6pBeqtrPS9QGhkklQkkTM4OmjiLEwV-Dt4vxEXKrVBMDKy1MYtfIlxkQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 2. Relevant variants at diagnosis in CP for patients treated with first-line TKIs. (A) Variants at diagnosis and overlap of variant type per patient at diagnosis. The average Combined Annotation Dependent Depletion score of the SNVs and indels was 32 (range [r], 22-44), indicating they were predicted to be deleterious. All relevant SNVs were predicted to be damaging by at least 3 of 4 in silico prediction tools. For the Venn diagrams, patients with >1 variant within a category (eg, 2 SNVs) were only counted once for that category. SNVs and indels were detected by WES and/or RNA-Seq. Fusions represent fusion transcripts detected by RNA-Seq and genomic fusions where breakpoints were detected by RNA-Seq or WES, including focal deletions. CNVs were detected from the WES data. All variants that met the criteria for classification for clinical relevance or possible relevance were included. (B) Graphs show characteristics of variants that occurred in ≥3 patients. The median month and r of subsequent BC was calculated for patients with any 1 of the variants. Ph, Philadelphia.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/9/10.1182_blood-2018-02-832253/4/m_blood832253f2.jpeg?Expires=1765952834&Signature=sYIdX-5v6MDlaJ1XFLThNhD19IS0JbGJbsUg0nbUbDFTejI8UWDLBNDCXoglbi5XF5j1QA2Pf9lS8O7qhHv39ReGK0C0EUkSfUxG~dDvoLpHr0lsReRewQnd4C72FP~8gpphOYWnYTwXnakouW5y1RG8HZsMQeFidotO~iE6plNM7dB9vHOF-ySUED8eeMczb9BprOu9K5aGJPZrFbHX4TKpKZdv1XzN-zBsdIK1whgzB8yFSe8NBrzv1n~jFJEbE0SLprILaKoHgVRtTUibTZs8TbFo8cHk5jW6JfB2Vja20C1LMnx3mvVwzlPmTLc0X0qAdreFFhAp9MSgxe4MXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)