Abstract
Since a report of some 50 years ago describing refractory anemia associated with group C monosomy, monosomy 7 (−7) and interstitial deletions of chromosome 7 (del(7q)) have been established as one of the most frequent chromosomal aberrations found in essentially all types of myeloid tumors regardless of patient age and disease etiology. In the last century, researchers sought recessive myeloid tumor-suppressor genes by attempting to determine commonly deleted regions (CDRs) in del(7q) patients. However, these efforts were not successful. Today, tumor suppressors located in 7q are believed to act in a haploinsufficient fashion, and powerful new technologies such as microarray comparative genomic hybridization and high-throughput sequencing allow comprehensive searches throughout the genes encoded on 7q. Among those proposed as promising candidates, 4 have been validated by gene targeting in mouse models. SAMD9 (sterile α motif domain 9) and SAMD9L (SAMD9-like) encode related endosomal proteins, mutations of which cause hereditary diseases with strong propensity to infantile myelodysplastic syndrome (MDS) harboring monosomy 7. Because MDS develops in SAMD9L-deficient mice over their lifetime, SAMD9/SAMD9L are likely responsible for sporadic MDS with −7/del(7q) as the sole anomaly. EZH2 (enhancer of zeste homolog 2) and MLL3 (mixed lineage leukemia 3) encode histone-modifying enzymes; loss-of-function mutations of these are detected in some myeloid tumors at high frequencies. In contrast to SAMD9/SAMD9L, loss of EZH2 or MLL3 likely contributes to myeloid tumorigenesis in cooperation with additional specific gene alterations such as of TET2 or genes involved in the p53/Ras pathway, respectively. Distinctive roles with different significance of the loss of multiple responsible genes render the complex nature of myeloid tumors carrying −7/del(7q).
Monosomy 7 in a wide variety of myeloid tumors
The monosomy 7 story started when a report was published in 1964.1 This short paper of ∼200 words proposed a new clinical syndrome, namely, refractory anemia missing one of the group 6 to 12 chromosomes in marrow cells and terminating as acute myelomonocytic leukemia. According to today’s criteria, the proposed disease would be “sporadic adult myelodysplastic syndrome (MDS) carrying monosomy 7 as the sole chromosomal anomaly that frequently develops into overt leukemia.” This clinical entity, constituting ∼10% of sporadic adult MDS, seems to represent the core hematological disease caused by monosomy 7.
Monosomy 7 as the sole anomaly was later identified in children with preleukemia,2 which includes MDS and juvenile myelomonocytic leukemia (JMML) according to the current criteria. In addition, familial monosomy 7, defined as bone marrow monosomy 7 (hereafter referred to as −7) occurring as the sole anomaly affecting >2 siblings, has been reported in 14 families (see references in Gaitonde et al3 ), in which most patients are children or adolescents.
In addition to cases for which −7 is the sole anomaly, it has been found together with chromosomal translocations in acute myelogenous leukemia (AML) and in chronic myelogenous leukemia in blast crisis, as well as in congenital diseases with a propensity to evolve into myeloid diseases, such as those caused by germ line GATA2 (3q21.3) mutations,4 neurofibromatosis,5 and severe congenital neutropenia (SCN).6 The frequency of −7 is particularly high among therapy-related and/or radiation-induced MDS or AML.7
Effects of monosomy 7
Because −7 is detected in a broad range of myeloid tumors regardless of age and disease etiology, its contribution to the development and/or promotion of each myeloid malignancy clearly varies from being the major driving force to having a supportive role. For instance, adult sporadic MDS associated with −7 manifests at more advanced disease stages with increased blast counts, so that the revised International Prognostic Scoring System for MDS assigns −7 patients to a poor risk group.8 In contrast, there are no significant differences in clinical features or prognosis of JMML or childhood MDS patients whether or not they carry −7.9-11 Nevertheless, childhood MDS provides many opportunities for speculation as to the roles of −7.
Unlike adult MDS, −7 is present in nearly half of all children with refractory anemia as diagnosed by French-American-British criteria.12 This suggests that loss of chromosome 7 could be an early event. Although refractory anemia patients with −7 progress to advanced MDS at high frequency,12 spontaneous regression has been reported in childhood MDS with −7 (see references in Mantadakis et al13 ). In familial monosomy 7 syndrome, a puzzling finding is the different parental origin of the retained chromosome 7 in some sibling pairs.14-16 Although difficult to understand, this suggests the possibility that the gene responsible for monosomy 7 is located on another chromosome. However, because cytopenias are often associated with MDS or even non-MDS/AML in siblings, the essence of familial monosomy 7 syndrome could be a bone marrow environment conducive to the expansion of a −7 clone. These findings suggest that cells with −7 have a relative survival advantage over the surrounding bone marrow cells. Enhanced responsiveness to cytokines, rather than cytokine-independent growth potential, which leukemia cells usually develop, may be the reason for a temporary growth advantage. It may be that this is augmented by the long-term use of high doses of granulocyte colony-stimulating factor (G-CSF) in patients with aplastic anemia and SCN,17-20 although some investigators reported a similar incidence of AML/MDS with −7 in patients who have not been treated with G-CSF.21-23 G-CSF is not likely to induce −7 clones directly24 but may stimulate and expand a preexisting −7 clone, as suggested by the disappearance of such clones coincidental with the discontinuation of G-CSF therapy.25,26 Preferential stimulation of cells carrying −7 by cytokines has also been demonstrated in in vitro assays.20
Interstitial deletion of chromosome 7
By the mid-1970s, cytogeneticists had established chromosome banding techniques and were able to determine that the chromosome reported as lost in the original paper of 19641 was chromosome 7.27 Subsequently, using the same techniques, they identified patients harboring an interstitial deletion of the long arm of chromosome 7 (hereafter del(7q)) in myeloid diseases. Just like −7, del(7q) is detected either as the sole chromosomal anomaly or as an aberration additional to chromosomal translocations in essentially all myeloid tumors at all ages, albeit generally at frequencies less than −7.
A difficult question is whether the effects of del(7q) are identical to −7 or whether there are clinically relevant differences. Needless to say, −7 and del(7q) are caused by totally different mechanisms. The former is a result of chromosome dissegregation in mitosis, whereas the latter requires a process involving chromosome rearrangement. Many studies have reported that the prognosis of AML/MDS with −7 is worse than for the same malignancies with del(7q). This could be due in part to additional chromosome and/or gene alterations, because −7 tends to be associated with other unfavorable chromosome translocations such as those involving the EVI1 (3q26.2) gene. Indeed, −7 is not only the most frequent (>50%) chromosomal abnormality in tumors overexpressing the EVI1 transcription factor28-30 but also significantly worsens the prognosis,30 whereas high EVI1 expression worsens the prognosis of AML with −7 as the sole aberration.31 A bona fide cooperation of −7 and EVI1 overexpression in the progression of the disease was further supported by results from 2 patients with chronic granulomatous disease treated by gene therapy who thereafter developed MDS with −7 as a result of insertional activation of EVI1.32 In contrast, more favorable karyotypes in AML, such as t(8;21), inv(16), t(15;17), and t(9;11), are associated with del(7q).31 Moreover, the prognosis of del(7q) AML with a more favorable karyotype is better than in patients with del(7q) alone,31 suggesting a limited role of del(7q) by itself in these cases.
Comparing −7 and del(7q) as the sole anomaly indicated in one study that prognosis of adult sporadic MDS with −7 is better than with del(7q),33 but another report failed to confirm this.34 These disparate findings may have resulted from grouping all del(7q) patients, who have a wide variety of deleted regions, into one “del(7q)” category. A recent paper reported the adverse effects of del(7p) as the sole anomaly to the prognosis of myeloid neoplasms.35 This could be a cause of possible worse prognosis of −7 than del(7q). The revised International Prognostic Scoring System assigns del(7q) patients to an intermediate risk group.8
Broad nature of commonly deleted regions in del(7q) patients
Investigating del(7q) cases provided researchers with an opportunity to identify the responsible myeloid tumor-suppressor gene(s). Indeed, del(7q) cases were almost the only clue in this regard in the last century, but this was the real start of confusion. In the 1970s and 1980s, recessive tumor suppressors were the focus of intensive research. These genes typically lose their function completely by the loss of 1 allele together with a loss-of-function mutation on the remaining gene. Thus, a commonly deleted region (CDR) is an ideal guide to lead researchers to the place where the tumor suppressor is located. Once a CDR is identified, candidate genes within the CDR can be sequenced to find mutations. The identification of the retinoblastoma (Rb) gene (13q14.2) in 1982 encouraged hematologists to seek CDR(s) in chromosome 7. However, G-banding chromosome spreads of unstimulated myeloid tumor cells did not have as high a resolution as could be obtained from the lectin-stimulated peripheral blood T cells that had been employed for the identification of the Rb gene. Nonetheless, 2 regions, 7q22 and 7q34-36, seemed promising, but these bands were too large to be sequenced by the methods available at that time. Researchers tried to detail CDRs using then-current techniques such as restriction fragment–length polymorphism assessments, fluorescence in situ hybridization, and microsatellite surveys. Results from multiple laboratories were published around 1990, but the CDRs identified by each laboratory did not overlap (reviewed in Honda et al36 and Todd et al37 ) with the result that instead of narrowing down the location of the relevant CDRs, they were reported to be spread over the whole 7q region. Of note, at least these results showed that the conclusion that 7q22 and 7q34-36 bands were promising CDRs based on chromosomal analysis was incorrect. The wide distribution of CDRs among del(7q) myeloid malignancies was later confirmed by comparative genomic hybridization microarraying.38
New approaches
After great efforts over decades, researchers accepted that the identification of CDRs as the first step of the orthodox approach to determining classical-type recessive tumor suppressors was not effective. No rational explanation for this has been forthcoming so far. Many researchers now consider that there are several responsible myeloid tumor suppressors that act in a haploinsufficient manner, but this cannot be the reason for the broad nature of CDRs on chromosome 7. Indeed, in del(5q), another chromosomal deletion frequently detected in myeloid tumors, 2 CDRs within the length of a chromosome subband were identified, each containing several genes where the loss of one allele contributes to the development of MDS.39,40
In this century, advances in technology have enabled researchers to apply new approaches for seeking the responsible genes. For instance, high-throughput sequencing allows a strategy where the whole of 7q is seen as one large CDR and a list of nonsilent mutations in genes located in this area can be made.38,41,42 Not surprisingly, many mutated genes have been identified. To select those truly responsible, the effects of single allele loss on gene expression were determined by comparing AML carrying −7 to AML with a normal karyotype using quantitative real-time polymerase chain reaction.43 The median expression level of 1088 probe sets was 0.57-fold, indicating a strong gene dosage effect on messenger RNA expression. This was confirmed in single hematopoietic stem/progenitor cells (HSPCs) using single-cell RNA sequencing.44 Thus, significant reduction of expression levels is not a useful indicator in searching for or evaluating them.
Several attempts were made to directly analyze the biological effects of large segment loss. For instance, HSPCs differentiated in vitro from isogenic induced pluripotent stem cells established from patients with del(7q) recapitulated disease-associated phenotypes, including impaired hematopoietic differentiation.45 Shannon et al attempted a straightforward but technically very difficult approach in which mice with a heterozygous germ line deletion of a 2-Mb portion of chromosome band 5A3 syntenic to a segment of human 7q22 were generated.46,47 Although these mice did not develop myeloid diseases, cell-autonomous abnormalities in HSPCs were found that were exacerbated by physiological aging and upon serial transplantation.46,47
Another approach is to use microarray comparative genomic hybridization to search for microdeletions in patients with myeloid malignancies that might be present in an apparently normal chromosome 7. Because there are many insignificant alterations in the genome of tumor cells in general, careful selection of patients to avoid “passenger deletions” is necessary. Asou et al selected JMML and JMML-like diseases for this, because it was expected that insignificant deletions are much less frequent in leukemia cells of children than adults. A common microdeletion spanning approximately 100 kb was identified in the 7q21.3 subband that contains 3 genes: SAMD9, SAMD9L, and MIKI (LOC253012).48,49 Similarly, from EVI1-dysregulated AML cell lines, 2 small microdeletions (0.39 and 1.33 Mb) in subband 7q36.1 were detected, the latter of which included the EZH2 gene.50,51 In addition, a focal deletion (8.8 Mb) at 7q35-36 encompassing the MLL3 gene (7q36.1) was isolated from a relapsed AML with a normal karyotype.52 A contribution of these 4 genes to the development of myeloid diseases has now been validated by gene targeting in mouse models.
Current candidates for responsible genes
SAMD9 and SAMD9L
Human SAMD9 and SAMD9L encode related endosomal proteins (60% amino acid identity) that facilitate homotypic fusion of primary/early endosomes critical for endosomal trafficking, including metabolism of cytokine receptors.53 Only vertebrates have SAMD9/SAMD9L, which have no similarity to any other genes. Fish, frogs, and birds all possess the common ancestral genes of SAMD9 and SAMD9L. Mammals fall into 3 groups regarding these genes. For example, (1) humans (and other higher primates), horses, and rats have both SAMD9 and SAMD9L; (2) cows, sheep, and primitive primates (such as galagos) possess only SAMD9; and (3) cats, dogs, and mice have only SAMD9L.54 This odd distribution implies that the 2 gene products have common functions. Nonetheless, because of differences in gene regulation in tissues and organs, the 2 genes do not fully compensate each other, because loss-of-function mutations of SAMD9, despite intact SAMD9L, cause the life-threatening autosomal-recessive disease normophosphatemic familial tumoral carcinosis in humans.55
Heterozygous (SAMD9L+/−) as well as homozygous (SAMD9L−/−) mice were found to develop MDS and die after 1.5 years.53 Most SAMD9L+/− as well as SAMD9L−/− mice exhibited leukocytopenia and anemia with dysplasia in multiple hematopoietic lineages in their normal-to-hypercellular bone marrow. Almost all mice of both genotypes developed AML together with EVI1 overexpression at younger ages, indicating cooperation between the haploinsufficiency of SAMD9L and EVI1 overfunction in disease progression. Colony-replating and competitive repopulation assays revealed that SAMD9L deficiency confers a proliferative advantage on HSPCs. Indeed, SAMD9L-deficient HSPCs possessed an enhanced sensitivity to cytokines, most likely due to disturbed metabolism of ligand-bound cytokine receptors. However, it is not fully understood why such a growth advantage of HSPCs mainly induces MDS rather than MPD or AML.
Recently, gain-of-function (g/f) mutations of the SAMD9 gene were identified in patients with MIRAGE (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, and enteropathy) syndrome,56,57 while SAMD9L mutations were found in ataxia pancytopenia syndrome.58-60 The common symptom of these 2 diseases is pancytopenia with hypocellular bone marrow in infancy that often requires transfusion but gradually improves over time. Patients with both syndromes have additional nonhematological symptoms (Figure 1). More recently, germ line g/f mutations of the Samd9/Samd9L gene were identified at high frequencies in cohorts of children and adolescents with inherited bone marrow failure61 and isolated MDS62,63 at high frequencies. The latter cohorts include familial cases.
Inherited diseases caused by Samd9/Samd9L mutations with a propensity to evolve into childhood MDS. MIRAGE and ataxia pancytopenia syndromes have nonhematological symptoms shown in blue. All disease entities except isolated MDS are characterized by inherited bone marrow failure.
Inherited diseases caused by Samd9/Samd9L mutations with a propensity to evolve into childhood MDS. MIRAGE and ataxia pancytopenia syndromes have nonhematological symptoms shown in blue. All disease entities except isolated MDS are characterized by inherited bone marrow failure.
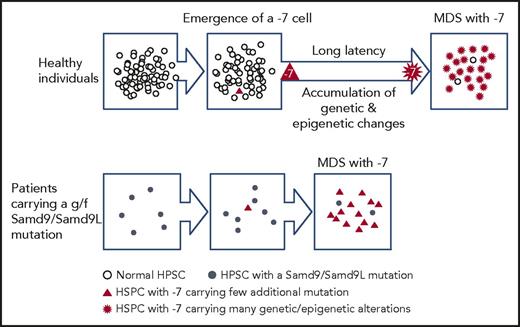
Children carrying a germ line g/f mutation of Samd9/Samd9L often develop MDS with −7. The age of onset is mostly <5 years, and, intriguingly, the mutated allele is always lost. Because g/f mutants of Samd9/Samd9L have an adverse effect on cell proliferation, loss of the mutants rescues growth potential of bone marrow cells by a mechanism proposed as “adaption by aneuploidy” and creates the condition “revertant mosaicism.”56,57 The emergence of such clones also carries a risk of developing MDS with −7. MDS cells from such children have few additional gene/chromosome alterations other than −7,63 in contrast to sporadic childhood MDS with −7, which generally carries relevant gene mutations such as GATA2 or those involved in the ras pathway.64 According to these observations, a working hypothesis is that HSPCs carrying monosomy 7 (HSPCs/−7) are susceptible to the development of MDS per se, but surrounding HSPCs suppress HSPC/−7 expansion (Figure 2). When neighboring HSPCs are “weak” and unable to suppress HSPCs/−7, as is the case for HSPCs harboring g/f mutations of Samd9/Samd9L, then HSPCs/−7 can develop into MDS.
Schematic representation of the hypothesis that the relative strength of HSPCs carrying monosomy 7 compared with the surrounding bone marrow cells affects the process of development of MDS.
Schematic representation of the hypothesis that the relative strength of HSPCs carrying monosomy 7 compared with the surrounding bone marrow cells affects the process of development of MDS.
This logic may extend to the pathogenesis of sporadic adult MDS. It is generally accepted that the accumulation of additional genetic and/or epigenetic alterations is required for HSPCs/-7 to develop MDS. This is assumed to be a reason why the great majority of MDS patients are >40 years of age. However, if the expansion potential of HSPCs/−7 would be determined by the relative strength of the surrounding HSPCs, aging may also contribute to the development of MDS by “weakening” surrounding HSPCs to allow the expansion of HSPCs/−7.
EZH2
EZH2 encodes a methyltransferase for lysine 27 of histone H3 (H3K27).65-67 EZH2 is a component of the polycomb repressive complex 2 (PRC2), which catalyzes, binds to and propagates the trimethylation of H3K27. This histone modification generally functions as a gene silencer.68,69
Somatic EZH2 gene mutations are frequently found (∼10%) in patients with MDS and related myeloproliferative neoplasms, as well as secondary AML patients, and are an independent unfavorable prognostic factor.51,70,71 In contrast, EZH2 mutations are rarely found in de novo AML samples.70,72 Mutations identified in patients with myeloid diseases are dispersed throughout the EZH2 gene and result in loss of methyltransferase activity.73 EZH2 mutations are found in either monoallelic or biallelic states.51,70,71 The latter are associated with copy-neutral loss of heterozygosity or uniparental disomy that is frequently present in the distal portion of 7q in myeloid malignancy.38 These findings raised the possibility that −7/del(7q) could play a causative role through haploinsufficiency of EZH2. However, conditional deletion of EZH2 in HSPCs using EZH2flox/flox mice increased the frequency of spontaneous γδ T-cell leukemia but did not induce myeloid tumors.74
More than half the MDS patients who harbor an EZH2 mutation also have a tet methylcytosine dioxygenase 2 (TET2; 4q24) mutation.75,76 Accordingly, the potential to develop MDS in individuals with an EZH2 deletion with or without alterations of other genes was tested using mice reconstituted with EZH2flox/flox hematopoietic cells in which EZH2 was deleted after transplantation. These mice developed MDS-like disease at low frequency but when TET2 was knocked down, the incidence of myeloid disease increased and the phenotype became more aggressive.76 Similar results were obtained when a Runx1 point mutant (S291fs) was induced.77 These data suggest that loss of EZH2 expression by −7/del(7q) contributes to MDS development in cooperation with TET2 or RUNX1 mutations. Transcriptomic analyses of HSPCs in these mice revealed that upon deletion of EZH2, key developmental regulator genes were kept repressed, suggesting compensation by EZH1,78 whereas a range of oncogenic direct and indirect polycomb targets became derepressed.76 In addition, RUNX1(S291fs)/EZH2-null MDS cells secreted inflammatory cytokines such as interleukin-6 and tumor necrosis factor α that depleted surrounding normal HSPCs, allowing propagation of MDS clones.77
MLL3
MLL3 belongs to the MLL protein family possessing histone methyltransferase activity for lysine 4 of histone 3 (H3K4), a histone mark associated with active transcription.79,80 As with the EZH2 gene, loss-of-function mutations of MLL3 were first identified in MDS and AML.81,82 Of note, these cases frequently exhibited Ras pathway mutations (NF1 deletions or N-Ras, K-Ras, and Ptpn11 activation) and TP53 inactivation,52 suggesting that MLL3 loss may cooperate with these aberrations to cause myeloid malignancies.
The effects of MLL3 suppression on leukemogenesis was tested using short hairpin (sh) RNA for MLL3 and p53−/− HSPCs.52 Mice transplanted with p53−/− HSPCs develop hematopoietic diseases, mainly thymic lymphoma. Although the introduction of shMLL3 into p53−/− HSPCs did not affect disease latency, introduction of both shMLL3 and shNF1 caused the development of aggressive myeloid malignancies. In addition, shNF1-transduced p53−/− HSPCs with one MLL3 allele disrupted by CRISPR/Cas9-mediated genome editing displayed similar phenotypes to those transduced with both shNF1 and shMLL3. These results indicate that inactivation of MLL3 and NF1 cooperatively promotes myeloid leukemogenesis in a p53-null background and MLL3 acts as a haploinsufficient tumor suppressor in this context. Of note, although transduction of the shMLL3 alone into p53−/− recipients did not affect survival, transplanted mice exhibited a temporary reduction in peripheral blood counts due to maturation arrest and dysplasia in multiple hematopoietic lineages. This suggests that a defect of NF1 plays a role in sustaining the MDS-like phenotype.
Comparison of transcriptional profiles between p53−/− alone and shMLL3, p53−/− HSPCs revealed that gene expression in shMLL3, p53−/− HSPCs was enriched for those identified as a leukemia stem cell signature83 and that genes suppressed by shMLL3 were significantly correlated with genes downregulated in MDS HSCs.84 Because MLL3 functions as a chromatin modulator, it is suggested that loss of function of MLL3 induces altered gene expression patterns in HSPCs, which eventually perturbs normal hematopoietic development/differentiation and results in development of MDS-like disease.
Other candidate genes proposed to be responsible for myeloid malignancies
A number of genes have been proposed as candidates responsible for myeloid malignancies bearing −7/del(7q). One well-studied example is CUX1 (7q22.1),85 the mammalian ortholog of the Drosophila melanogaster cut (ct) gene,86 which encodes a homeobox transcription factor. Loss of one allele of this gene is frequently detected not only in myeloid tumors but also in uterine leiomyomas and breast cancers (see references in Ramdzan and Nepveu87 ). Inactivating point mutations in one allele are also frequently found in cancers of the endometrium, large intestine, and lung.88 These data suggest that CUX1 is a haploinsufficient tumor suppressor. This is further supported by pseudotumor formation in D melanogaster induced by RNA interference–mediated knockdown of ct.88 Although CUX1 heterozygous mice are indistinguishable from wild-type mice, CUX1−/− homozygous mice exhibit myeloid hyperplasia during limited observation periods, because only few mice survive to weaning.89-91 Other examples include DOCK4 (7q31.1), which encodes a guanine exchange factor and is disrupted in murine osteosarcoma cells,92 and low expression has been linked to erythroid dysplasia.93 LUC7L2 (7q34), a putative RNA-splicing gene, was proposed to encode an MDS-related splicesomal protein.38,39,42
Concluding remarks and future prospects
It is probable that −7/del(7q) contributes to myeloid tumorigenesis through two distinct mechanisms. First, HSPCs/−7 emerge at an early stage; the subsequent loss of SAMD9/SAMD9L confers a relative advantage on HSPCs over surrounding cells to maintain the −7/del(7q) clone, which then has time to accumulate further genetic/epigenetic alterations. If the bone marrow is already greatly perturbed by aplastic anemia, SCN (possibly requiring high levels of cytokines), or diseases caused by g/f SAMD9/SAMD9L mutations, then HSPCs/−7 probably have the potential to develop into MDS even without additional genetic/epigenetic alterations. Regarding the second mechanism, −7/del(7q) occurs in the late phase of disease as an additional chromosomal aberration, and loss of EZH2 and/or MLL3 disturbs the epigenetic control of already-abnormal or even already-leukemic HSPCs in cooperation with preexisting gene alterations to promote disease.
The broad deletion, a unique feature of del(7q), suggests that the lack of >2 responsible genes is necessary to develop/promote myeloid tumors. For instance, haploinsufficiency of the Samd9L gene (7q21.3) causes the development of AML with EVI-1 overexpression in mice,53 whereas a pinpoint Ezh2 loss (7q36.1) has been detected in the human AML genome.50,51 Nevertheless, preponderance of −7 over del(7q) in AML with EVI-1 overexpression (refs28-30 ) might indicate that lack of both Samd9/Samd9L and Ezh2 (and possibly other tumor suppressors located at 7q) is necessary to effectively promote human AML. In this regard, analysis of the biological effects of large segment loss (see references in Kotini et al45 and Wong et al,46,47 ) is challenging but will be useful not only for identifying novel responsible genes but also for dissecting the roles of already-known tumor suppressors in combination. This will provide a precise indicator for prognosis and a strategy to develop tailor-made therapies for patients carrying −7 and improve our understanding of myeloid tumors, particularly MDS, which currently remains an enigma in hematology.
Acknowledgments
The authors thank Akiko Nagamachi, Atsushi Iwama, Seishi Ogawa, and Hideki Makishima for helpful discussions.
Authorship
Contribution: T.I., H.M., and H.H. wrote the paper.
Conflict-of-interest disclosure: The authors declare competing financial interests.
Correspondence: Toshiya Inaba, Department of Molecular Oncology and Leukemia Program Project, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi, Minami-ku, Hiroshima 734-8553, Japan; e-mail: tinaba@hiroshima-u.ac.jp.