Key Points
NR4A1/3 nuclear receptors suppress hyperproliferation and DNA damage of HSCs.
NR4A1/3 act as transcriptional activators of C/EBPα while repressing a proliferative inflammatory response in HSCs.
Abstract
Members of the NR4A subfamily of nuclear receptors have complex, overlapping roles during hematopoietic cell development and also function as tumor suppressors of hematologic malignancies. We previously identified NR4A1 and NR4A3 (NR4A1/3) as functionally redundant suppressors of acute myeloid leukemia (AML) development. However, their role in hematopoietic stem cell (HSC) homeostasis remains to be disclosed. Using a conditional Nr4a1/Nr4a3 knockout mouse (CDKO), we show that codepletion of NR4A1/3 promotes acute changes in HSC homeostasis including loss of HSC quiescence, accumulation of oxidative stress, and DNA damage while maintaining stem cell regenerative and differentiation capacity. Molecular profiling of CDKO HSCs revealed widespread upregulation of genetic programs governing cell cycle and inflammation and an aberrant activation of the interferon and NF-κB signaling pathways in the absence of stimuli. Mechanistically, we demonstrate that NR4A1/3 restrict HSC proliferation in part through activation of a C/EBPα-driven antiproliferative network by directly binding to a hematopoietic-specific Cebpa enhancer and activating Cebpa transcription. In addition, NR4A1/3 occupy the regulatory regions of NF-κB-regulated inflammatory cytokines, antagonizing the activation of NF-κB signaling. Taken together, our results reveal a novel coordinate control of HSC quiescence by NR4A1/3 through direct activation of C/EBPα and suppression of activation of NF-κB-driven proliferative inflammatory responses.
Introduction
NR4As (NR4A1, NR4A2, NR4A3) represent a highly conserved subgroup of nuclear receptor (NR) transcription factors that control expression of overlapping sets of transcriptional targets in a ligand-independent manner.1 They function as stimulus-responsive immediate early gene products whose expression is induced in response to environmental stimuli including stress and metabolic, mitogenic, inflammatory, hormonal, and apoptotic signals.2-4 During hematopoiesis, NR4As are essential and diverse regulators of differentiation and function of distinct subsets of lymphoid and myeloid cells.5-11 NR4As also function as tumor suppressors of both myeloid and lymphoid malignancies. We previously reported that NR4A1 and NR4A3 (NR4A1/3) are essential functionally redundant tumor suppressors of acute myeloid leukemia (AML) and pre-AML malignancies including myelodysplastic/myeloproliferative disorders.12,13 Germ line and conditional depletion of NR4A1/3 in mice leads to AML development, whereas reduction in NR4A1/NR4A3 gene dosage in hypoallelic mice leads to myelodysplastic/myeloproliferative disease.12-14 Expression of NR4A1/3 is silenced in patients with AML across diverse cytogenetic backgrounds and is reduced in myelodysplastic disease.12,15,16 Suppression of NR4A1/3 is also observed in B-cell lymphomas, and reexpression of NR4A1 suppresses tumor growth in mouse xenograft models of human B-cell lymphoma.17 In normal murine hematopoietic stem and progenitor cells (HSPCs), NR4A1 and NR4A2 are preferentially expressed in HSCs, and forced expression of NR4A2 drives early hematopoietic progenitors into quiescence.18,19 Together, these findings underscore a key role for NR4As as essential suppressors of hematopoietic neoplasms. However, the functional role of NR4A1/3 in HSCs and the molecular mechanisms by which they control normal HSPC homeostasis remain unclear.
Here we address the functions of NR4A1/3 in HSPC development, using a conditional Nr4a1/Nr4a3 double-knockout (CDKO) mouse model to temporally restrict combined deletion of Nr4a1/3 in adult mice. We show that NR4A1/3 act upstream of key HSC transcriptional networks to promote HSC quiescence, and their depletion leads to chronic unscheduled proliferation of HSCs, marked by increased inflammation and DNA damage, culminating in the malignant transformation of the HSC compartment.
Materials and methods
Mice
The CDKO mouse (Nr4a1fl/fl; Nr4a3−/− ; Rosa26-CreERT2) was generated by crossing a Nr4a1fl/fl mouse with a Nr4a3−/− mouse.14 The Nr4a1fl/fl; Nr4a3−/− mouse was subsequently crossed with a Rosa26-CreERT2 mouse to target tamoxifen-inducible Cre recombinase expression in all tissues. Cre-negative Nr4a1fl/fl; Nr4a3−/− mice were used as control. To achieve Nr4a1 excision, CDKO mice (8-12 weeks) were treated with 4 or 5 daily intraperitoneal doses of tamoxifen (3 mg) in corn oil, as indicated. Mouse experiments were performed under approval of the Baylor College of Medicine Institutional Animal Care and Use Committee.
Flow cytometry
Flow cytometry analyses and cell sorting were performed in LSRII machine and BD Biosciences Aria sorter. For live cells analysis, immunophenotyping was performed using direct conjugated monoclonal antibodies against cell surface markers. The nuclear dye propidium iodide (Sigma-Aldrich) was used to identify dead cells. For intracellular staining, cell surface staining in live cells was followed by fixation and permeabilization using the BD Cytofix/Cytoperm kit (BD Bioscience). Antibodies for flow cytometry are listed in supplemental Experimental Procedures, available on the Blood Web site.
Transcriptome profiling
To measure global transcriptional changes after Nr4a1/3 codepletion, we treated CDKO or control mice with 4 daily tamoxifen injections and performed a RNA-Seq analysis in 10 000 sorted HSCs (Lin−Sca1+Kit+CD150−CD48+). Details of the library preparation and bioinformatics analysis are described in supplemental Experimental Procedures. Raw and processed data were deposited in the Gene Expression Omnibus database (accession number: GSE100990).
Results
Codepletion of NR4A1/3 leads to unscheduled HSC proliferation, oxidative stress, and increased DNA damage
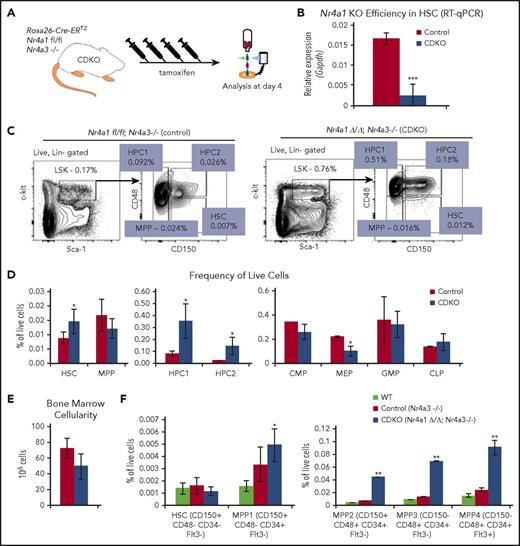
To study the acute effects of NR4A1/3 codepletion on the functional behavior of HSPCs, we used mice carrying a floxed Nr4a1 gene in an Nr4a3-null background intercrossed with mice carrying a Rosa 26 locus-targeted Cre recombinase–estrogen receptor T2 fusion gene (CDKO mice, Rosa26Cre-ERT2/Nr4a1fl/fl/Nr4a3−/−) to regulate tamoxifen-dependent codepletion of NR4A1/3 in adult mice. To address the consequences of tamoxifen treatment on HSPC homeostasis, we first compared the frequencies and proliferative status of HSPCs in vehicle-treated and tamoxifen-treated (4 daily injections) wild-type mice (supplemental Figure 1A). Tamoxifen treatment had no significant effects on HSPC homeostasis, including proliferation of HSCs (supplemental Figure 1B-C). Therefore, for subsequent experiments, we used this treatment regimen to compare the consequences of codepletion of NR4A1/3 on HSPC homeostasis in tamoxifen-treated Cre-positive Nr4a1fl/fl;Nr4a3−/− (CDKO) mice vs Cre-negative Nr4a1fl/fl; Nr4a3−/− control mice (Figure 1A). At this time, NR4A1 expression is reduced more than 85% in CDKO HSCs (Lin−Sca1+Kit+CD48−CD150+) (Figure 1B) compared with control animals. The frequencies of HSCs and the early progenitor populations, HPC1 (Lin−Sca1+Kit+CD48+CD150−) and HPC2 (Lin−Sca1+Kit+CD48+CD150+), were significantly increased in CDKO mice, whereas lineage-committed progenitors showed no significant changes, with the exception of a reduction in the frequency of megakaryocyte/erythrocyte progenitors (Figure 1C-D). Total bone marrow cellularity showed a slight reduction in CDKO relative to control mice, although it was not statistically significant (Figure 1E).
Acute codepletion of NR4A1/3 leads to loss of quiescence and increased proliferation of HCSs. (A) Experimental design for tamoxifen-dependent depletion of Nr4a1 in CDKO mice, using 4 daily treatments with tamoxifen (Nr4a1fl/fl; Nr4a3−/−; Rosa26-ERT2). (B) NR4A1 depletion efficiency measured by quantitative reverse transcription polymerase chain reaction (RT-qPCR) in HSCs after 4 daily tamoxifen injections (n = 3). (C) Representative flow cytometry plots illustrating subfractionation strategy of the HSPC (LSK) compartment to identify HSCs and early progenitors (MPP, HPC1, HPC2), based on surface expression of CD150 and CD48.65 (D) Quantitation of HSPC (left and center, n = 5 CDKO and 3 control) and lineage-restricted progenitor cell frequencies (right, n = 3) at 4 days after acute NR4A1/3 codepletion. (E) Bone marrow cellularity after acute NR4A1/3 codepletion (n = 3). (F) Frequency of HSPCs using CD34, Flt3, CD48, and CD150 markers (n = 4). For F, statistical significance was calculated between wild-type and CDKO samples. Results expressed as mean ± SD. *P ≤ .05; **P ≤ .01; ***P ≤ .001. CLP, common lymphoid progenitors (Lin−CD11c−CD27+IL7r+Flt3+Ly6d−); CMP, common myeloid progenitors (Lin−Sca1−Kit+CD16/32−CD34+); GMP, granulocyte-macrophage progenitors (Lin−Sca1−Kit+CD16/32+CD34+); MEP, megakaryocyte-erythrocyte progenitors (Lin−Sca1−Kit+CD16/32−CD34−).
Acute codepletion of NR4A1/3 leads to loss of quiescence and increased proliferation of HCSs. (A) Experimental design for tamoxifen-dependent depletion of Nr4a1 in CDKO mice, using 4 daily treatments with tamoxifen (Nr4a1fl/fl; Nr4a3−/−; Rosa26-ERT2). (B) NR4A1 depletion efficiency measured by quantitative reverse transcription polymerase chain reaction (RT-qPCR) in HSCs after 4 daily tamoxifen injections (n = 3). (C) Representative flow cytometry plots illustrating subfractionation strategy of the HSPC (LSK) compartment to identify HSCs and early progenitors (MPP, HPC1, HPC2), based on surface expression of CD150 and CD48.65 (D) Quantitation of HSPC (left and center, n = 5 CDKO and 3 control) and lineage-restricted progenitor cell frequencies (right, n = 3) at 4 days after acute NR4A1/3 codepletion. (E) Bone marrow cellularity after acute NR4A1/3 codepletion (n = 3). (F) Frequency of HSPCs using CD34, Flt3, CD48, and CD150 markers (n = 4). For F, statistical significance was calculated between wild-type and CDKO samples. Results expressed as mean ± SD. *P ≤ .05; **P ≤ .01; ***P ≤ .001. CLP, common lymphoid progenitors (Lin−CD11c−CD27+IL7r+Flt3+Ly6d−); CMP, common myeloid progenitors (Lin−Sca1−Kit+CD16/32−CD34+); GMP, granulocyte-macrophage progenitors (Lin−Sca1−Kit+CD16/32+CD34+); MEP, megakaryocyte-erythrocyte progenitors (Lin−Sca1−Kit+CD16/32−CD34−).
To better separate long-term HSCs from short-term progenitors, we also used a more rigorous gating scheme, which fractionates the LSK compartment using the CD34, Flt3, CD150, and CD48 markers.20 We observed no change in the frequency of long-term HSCs, defined as Lin−Sca1+Kit+CD48−CD150+CD34−Flt3− on Nr4a1/3 CDKO, but the frequencies of all other multipotent progenitors within the LSK compartment were increased (Figure 1F). Notably, the frequencies of the myeloid-biased MPP3 (Lin−Sca1+Kit+CD48+CD150−CD34+Flt3−) and the lymphoid-biased MPP4 (Lin−Sca1+Kit+CD48+CD150−CD34+Flt3+) populations were increased at similar levels, indicating a lack of lineage-skewing on Nr4a1/3 CDKO. In contrast, we observed no significant differences in HSPC frequencies between wild-type and control (Nr4a3-null).
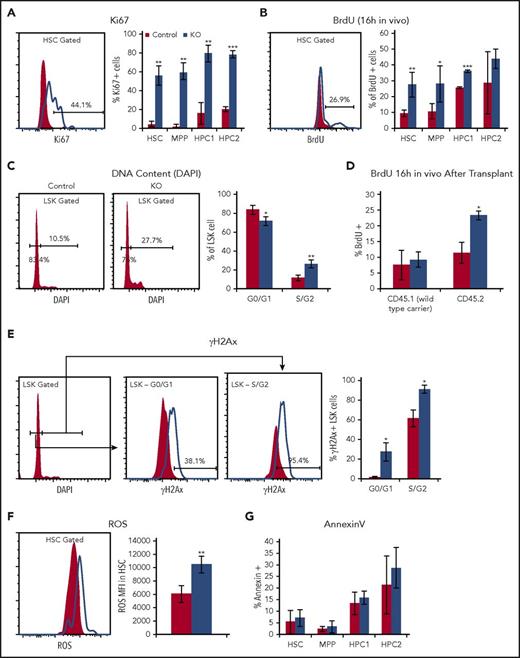
To address the causes of HSPC accumulation in CDKO bone marrow, we analyzed their proliferative and cell cycle status. Cell cycle analysis using the Ki67 marker, BrdU incorporation, and DNA content analysis showed that all HSPC populations within the LSK compartment (Lin−Sca1+Kit+) underwent an abrupt increase of cell cycle entry on acute NR4A1/3 codepletion (Figure 2A-C; supplemental Figure 2). The observation that HSPC proliferation is generally increased, but HSPC accumulation is more pronounced in the downstream HPC1/2 progenitors compared with HSCs and MPPs (Figure 1D), suggests that proliferative CDKO HSCs and MPPs may have a greater tendency for differentiation into HPC1/2 progenitors.
Nr4a1/3-null HSCs have increased cell cycle with increased oxidative stress and DNA damage. (A) Ki67 and (B) BrdU incorporation (16 hours in vivo), (C) DNA content analysis after acute NR4A1/3 codepletion. (D) BrdU incorporation analysis in HSCs from wild-type CD45.1 recipient mice transplanted with 2 million CD45.2 surface marked control or CDKO bone marrow together with CD45.1 competitor at a 1:1 ratio. Daily tamoxifen treatment was initiated 6 weeks after transplant, and BrdU incorporation was analyzed 4 days after tamoxifen treatment. (E) γH2Ax in levels in LSK cells separated by cell cycle stage using 4′,6-diamidino-2-phenylindole (DAPI), (F) reactive oxidative species levels in HSCs measured by CellROX fluorescent dye, and (G) frequency of AnnexinV-positive cells in HSPCs after acute codepletion of NR4A1/3. Results expressed as mean ± SD, n = 3 to 5 mice per group. *P ≤ .05; **P ≤ .01; ***P ≤ .001.
Nr4a1/3-null HSCs have increased cell cycle with increased oxidative stress and DNA damage. (A) Ki67 and (B) BrdU incorporation (16 hours in vivo), (C) DNA content analysis after acute NR4A1/3 codepletion. (D) BrdU incorporation analysis in HSCs from wild-type CD45.1 recipient mice transplanted with 2 million CD45.2 surface marked control or CDKO bone marrow together with CD45.1 competitor at a 1:1 ratio. Daily tamoxifen treatment was initiated 6 weeks after transplant, and BrdU incorporation was analyzed 4 days after tamoxifen treatment. (E) γH2Ax in levels in LSK cells separated by cell cycle stage using 4′,6-diamidino-2-phenylindole (DAPI), (F) reactive oxidative species levels in HSCs measured by CellROX fluorescent dye, and (G) frequency of AnnexinV-positive cells in HSPCs after acute codepletion of NR4A1/3. Results expressed as mean ± SD, n = 3 to 5 mice per group. *P ≤ .05; **P ≤ .01; ***P ≤ .001.
To determine whether the hyperproliferative phenotype in CDKO mice is cell-autonomous, we transplanted unfractionated bone marrow from CDKO or control mice carrying a CD45.2 surface marker together with CD45.1 wild-type bone marrow cells at a 1:1 ratio into lethally irradiated CD45.1 recipients. After 6 weeks of engraftment, we treated all mice with tamoxifen for 4 days and analyzed HSC cell cycle using a 16-hour BrdU pulse in vivo. We found that HSC hyperproliferation was restricted to CD45.2-marked CDKO-derived HSCs in transplanted mice, demonstrating that this phenotype was intrinsic to Nr4a1/3-null HSCs (Figure 2D).
Loss of quiescence in HSCs is associated with increased replication stress, and HSCs carrying proliferation-inducing leukemic lesions have higher levels of genomic instability, which may contribute to acquisition of secondary mutations.21-25 To assess the effects of acute NR4A1/3 codepletion on HSPC genomic stability, we examined the extent of double-stranded DNA breakage by measuring the frequency of γH2Ax-immunopositive LSK cells at 4 days after tamoxifen treatment. Given the higher intrinsic levels of γH2Ax in cycling cells, we analyzed G0/G1 and S/G2 cells separately, using DAPI.26 We observed a marked increase in the frequency of γH2Ax-positive CDKO LSK cells regardless of the cell cycle stage (Figure 2E). In addition, we observed a significant elevation in reactive oxidative species in CDKO vs control HSCs (Figure 2F). Despite increased DNA damage, however, the frequency of apoptotic cells was unchanged (Figure 2G). Thus, codepletion of NR4A1/3 in HSCs led to a hyperproliferative state marked by increased levels of oxidative stress and DNA damage.
Nr4a1/3-null HSCs retain stem cell capacity
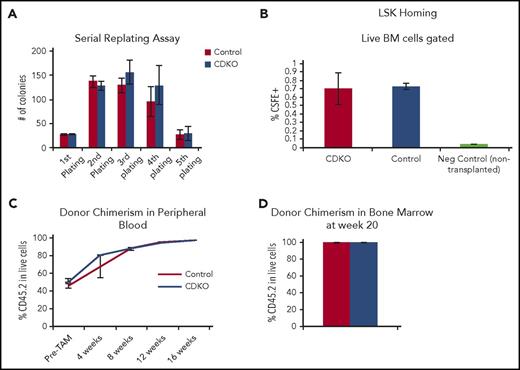
To test in vitro self-renewal capacity of CDKO HSCs, we performed a serial replating assay by initially plating 100 control or Nr4a1/3-null HSCs in methylcellulose, followed by 4 rounds of weekly replating. Nr4a1/3-null and control HSCs displayed similar colony-forming capacity, which was reduced after 5 rounds of replating (Figure 3A). Next, we examined the homing ability of CDKO HSCs by transplanting 50 000 LSK cells labeled with carboxyfluorescein succinimidyl ester (CFSE) into lethally irradiated mice. Donor control or CDKO cells were transplanted 4 days after tamoxifen treatment. Homing of LSK CSFE+ cells to the bone marrow was analyzed 16 hours after transplant, and no statistically significant difference was observed between control and CDKO cells (Figure 3B). Finally, we analyzed the long-term engraftment capacity of Nr4a1/3-null HSCs by transplanting 200 HSCs from untreated CD45.2 CDKO or control mice into lethally irradiated CD45.1 recipients together with 200 000 CD45.1 bone marrow cells as carriers. Six weeks after transplant, peripheral blood analysis confirmed a similar initial engraftment of both CKDO and control donor-derived CD45.2 cells (Figure 3C). Recipient mice were treated with tamoxifen (5 days), and CD45.2 cell frequencies in peripheral blood were analyzed every 4 weeks. No significant difference was observed in engraftment of CDKO or control HSCs up to 16 weeks after treatment. This similar engraftment was also confirmed in bone marrow at week 20 (Figure 3D).
Nr4a1/3-null HSCs have normal self-renewal, homing and engraftment capacity. (A) Serial replating of 100 sorted HSCs after acute Nr4a1/3 codepletion. From the second to the fifth round of replating, 10 000 cells were seeded. (B) LSK homing assay. CDKO and control mice (n = 3) were treated with 4 doses of tamoxifen, and LSK cells were sorted and subsequently labeled with CSFE in vitro. LSK CSFE+ cells were then transplanted into wild-type mice (n = 3 recipients per genotype) that were lethally irradiated the day before. Sixteen hours after transplant, recipient mice were killed and bone marrow was analyzed by flow cytometry for CSFE detection. Bone marrow from nontransplanted mice was used as negative control. (C) Flow cytometry analysis of CD45.2 donor cell chimerism in mice transplanted with 200 CDKO or control HSCs. Recipient mice were analyzed 6 weeks after transplant to confirm initial engraftment (Pre-TAM) and immediately treated with tamoxifen and analyzed every 4 weeks after tamoxifen treatment. (D) CD45.2 donor cell chimerism in the bone marrow at week 20. Results expressed as mean ± SD, n = 3 (A-B) or n = 3 control and 5 CDKO (C-D).
Nr4a1/3-null HSCs have normal self-renewal, homing and engraftment capacity. (A) Serial replating of 100 sorted HSCs after acute Nr4a1/3 codepletion. From the second to the fifth round of replating, 10 000 cells were seeded. (B) LSK homing assay. CDKO and control mice (n = 3) were treated with 4 doses of tamoxifen, and LSK cells were sorted and subsequently labeled with CSFE in vitro. LSK CSFE+ cells were then transplanted into wild-type mice (n = 3 recipients per genotype) that were lethally irradiated the day before. Sixteen hours after transplant, recipient mice were killed and bone marrow was analyzed by flow cytometry for CSFE detection. Bone marrow from nontransplanted mice was used as negative control. (C) Flow cytometry analysis of CD45.2 donor cell chimerism in mice transplanted with 200 CDKO or control HSCs. Recipient mice were analyzed 6 weeks after transplant to confirm initial engraftment (Pre-TAM) and immediately treated with tamoxifen and analyzed every 4 weeks after tamoxifen treatment. (D) CD45.2 donor cell chimerism in the bone marrow at week 20. Results expressed as mean ± SD, n = 3 (A-B) or n = 3 control and 5 CDKO (C-D).
Long-term codepletion of NR4A1/3 leads to chronic activation of the HSC cell cycle and reduced bone marrow cellularity
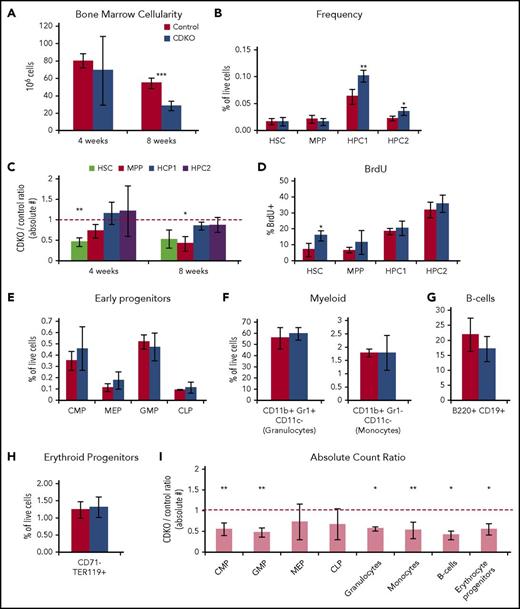
To analyze the long-term effects of NR4A1/3 codepletion on HSC differentiation potential in vivo, we analyzed the changes in HSPCs at 4 and 8 weeks after NR4A1/3 codepletion at points preceding myelocytosis and appearance of AML blasts.14 A marked reduction in bone marrow cellularity (∼50%) was observed in CDKO mice at 8 weeks after Nr4a1/3 ablation (Figure 4A). Flow cytometry analysis showed a significant increase in relative frequencies of HPC1/2 progenitors (Figure 4B). Taking into account the decrease in bone marrow cellularity, we quantified the absolute numbers of each HSPC population and observed a ∼50% reduction of CDKO HSCs at 4 weeks after NR4A1/3 codepletion, whereas HPC1/2 population abundance remained at levels similar to control mice at both 4- and 8-week points (Figure 4C). Furthermore, analysis of BrdU incorporation at 8 weeks showed that changes in HSPC frequencies are accompanied by a chronic increase of cell cycle in CDKO HSCs (Figure 4D). Despite the accumulation of HPC1/2 in CDKO mice, lineage-committed bone marrow progenitors had similar frequencies to controls, suggesting that long-term NR4A1/3 codepletion leads to an early progenitor accumulation that does not extend beyond the HPC1/2 stage (Figure 4E-H), with the exception of Ly6− monocytes, which showed a reduced frequency and are known to be dependent on NR4A1 expression (supplemental Figure 3A-B).9 Consistent with the decrease in bone marrow cellularity, estimation of absolute numbers of lineage-committed bone marrow progenitors indicated a widespread reduction in absolute counts regardless of lineage downstream of the HPC1/2 stage (Figure 4I).
Long-term codepletion of NR4A1/3 leads to a chronic activation of HSC cell cycle and reduced bone marrow cellularity. (A) Bone marrow cellularity at 4 and 8 weeks after NR4A1/3 codepletion. (B) Flow cytometry analysis of HSPC populations at 8 weeks after tamoxifen treatment. (C) Ratio of absolute counts of HSPC populations between CDKO and control mice at 4 and 8 weeks after NR4A1/3 codepletion. The red line represents 1:1 ratio. (D) BrdU analysis after a 16-hour BrdU uptake in HSPCs after an 8-week NR4A1/3 codepletion. (E-H) Flow cytometry analysis at 8 weeks after tamoxifen treatment of (E) early myeloid and lymphoid progenitors, (F) myeloid, (G) B cells, and (H) erythroid bone marrow cells. (I) Ratio of absolute counts of lineage-committed bone marrow cells between CDKO and control mice at 8 weeks after NR4A1/3 codepletion. The red line represents 1:1 ratio. Results expressed as mean ± SD, n = 3 to 5 mice per group. *P ≤ .05; **P ≤ .01; *** P ≤ .001.
Long-term codepletion of NR4A1/3 leads to a chronic activation of HSC cell cycle and reduced bone marrow cellularity. (A) Bone marrow cellularity at 4 and 8 weeks after NR4A1/3 codepletion. (B) Flow cytometry analysis of HSPC populations at 8 weeks after tamoxifen treatment. (C) Ratio of absolute counts of HSPC populations between CDKO and control mice at 4 and 8 weeks after NR4A1/3 codepletion. The red line represents 1:1 ratio. (D) BrdU analysis after a 16-hour BrdU uptake in HSPCs after an 8-week NR4A1/3 codepletion. (E-H) Flow cytometry analysis at 8 weeks after tamoxifen treatment of (E) early myeloid and lymphoid progenitors, (F) myeloid, (G) B cells, and (H) erythroid bone marrow cells. (I) Ratio of absolute counts of lineage-committed bone marrow cells between CDKO and control mice at 8 weeks after NR4A1/3 codepletion. The red line represents 1:1 ratio. Results expressed as mean ± SD, n = 3 to 5 mice per group. *P ≤ .05; **P ≤ .01; *** P ≤ .001.
Together, these results indicate that long-term codepletion of NR4A1/3 leads to HSC overproliferation and an increase in the frequency of early HSPCs, resulting in a reduction of overall bone marrow cellularity. With the exception of Ly6− monocytes, the Nr4a1/3 CDKO phenotype was not lineage-restricted, suggesting the disruption of myelopoiesis previously observed in Nr4a1/3-null mice occurs at later stages during the AML progression.14
Gene expression profiling of Nr4a1/3 CDKO HSCs reveals dysregulated proliferative and inflammatory gene signatures
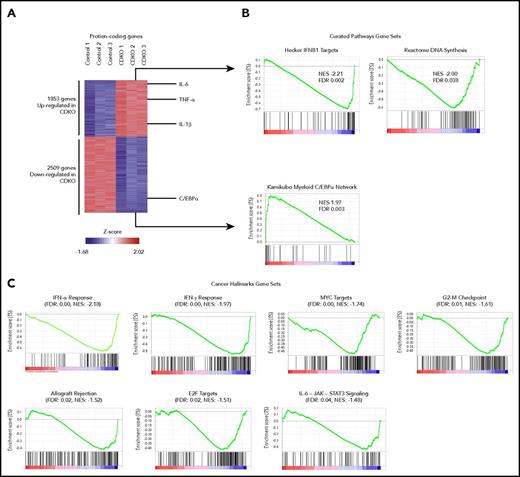
To analyze transcriptional changes in HSCs after acute codepletion of NR4A1/3, we isolated 10 000 HSCs (LSK CD150+CD48-) from CDKO and control mice after 4 daily tamoxifen injections and performed a global transcriptome analysis using RNA-Seq. A total of 4362 protein-coding genes were differentially expressed (Figures 5A; fold change >1.5; adjusted P value <.05). Gene set enrichment analysis identified a transcriptional signature associated with the AML tumor suppressor C/EBPα to be the most enriched gene set among downregulated genes on Nr4a1/3 CDKO, including the Cebpa gene itself. Among upregulated genes, we identified enrichment of multiple gene sets associated with cell proliferation such as MYC targets and DNA replication, as well as gene sets associated with inflammation including type I/II interferon (IFN) response and interleukin (IL)-6-JAK2-STAT signaling (Figure 5B-C). The inflammatory cytokines IL-1β, IL-6, IL-10, and TNF-α were themselves upregulated on Nr4a1/3 CDKO. Importantly, the activation of different inflammatory pathways such as type I and II IFN, IL-1β, and IL-6 can promote HSC and early progenitor proliferation during stress-induced hematopoiesis to increase production of mature blood cells.27-32 In summary, these findings indicate that NR4A1/3 exert coordinate control of HSC quiescence by acting as upstream activators of a C/EBPα-centered transcriptional network while repressing inflammatory proliferative responses.
Global transcriptional changes in CDKO relative to control HSCs after acute Nr4a1/3 codepletion. (A) Heat map of differentially expressed (P < .05 and fold change greater than 1.5 or less than −1.5) genes in CDKO relative to control HSCs (LSK CD48−CD150+) at 4 days after tamoxifen treatment. HSCs were sorted from 3 pools of bone marrow from control and CDKO mice (5 mice per pool) and subjected to RNA-seq. (B) Gene set enrichment analysis using the “Curated Pathways” gene sets as reference database. (C) Gene set enrichment analysis using the “Hallmarks of Cancer” gene sets. FDR, false discovery rate; NES, normalized enrichment score.
Global transcriptional changes in CDKO relative to control HSCs after acute Nr4a1/3 codepletion. (A) Heat map of differentially expressed (P < .05 and fold change greater than 1.5 or less than −1.5) genes in CDKO relative to control HSCs (LSK CD48−CD150+) at 4 days after tamoxifen treatment. HSCs were sorted from 3 pools of bone marrow from control and CDKO mice (5 mice per pool) and subjected to RNA-seq. (B) Gene set enrichment analysis using the “Curated Pathways” gene sets as reference database. (C) Gene set enrichment analysis using the “Hallmarks of Cancer” gene sets. FDR, false discovery rate; NES, normalized enrichment score.
C/EBPα is a direct transcriptional target of NR4As and key mediator of NR4A-dependent control of HSC proliferation
To confirm NR4A-dependent regulation of C/EBPα, we measured the consequences of NR4A1/3 codepletion on C/EBPα mRNA expression in sorted HSCs after 4 daily tamoxifen injections. We detected an 8-fold reduction in C/EBPα mRNA in CDKO HSCs relative to controls (Figure 6A). Because C/EBPα is also required for granulocytic differentiation, we also examined its expression in CDKO GMPs.24 We found that downregulation of C/EBPα after NR4A1/3 CDKO was not detected in the GMP compartment, in which the NR4As are expressed at low levels (supplemental Figure 4A-B), reinforcing the previous observation that the NR4A1/3 codepletion phenotype is restricted to HSCs and early hematopoietic progenitors.
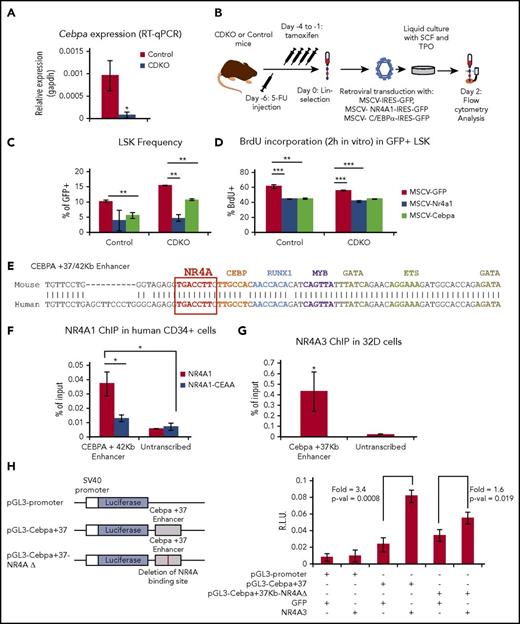
NR4A act upstream of a C/EBPα transcriptional network in HSCs. (A) RT-qPCR confirmation of C/EBPα downregulation in CDKO relative to control HSCs after acute NR4A1/3 codepletion. (B) Experimental design for overexpression of NR4A1 and C/EBPα in bone marrow Lin− cells. (C) Frequency of LSKs and (D) 2 hours in vitro BrdU incorporation after 48 hours in culture after retroviral transduction in Lin− cells. (E) Pairwise alignment of mouse and human +37/42 Kb CEBPA enhancer containing the NR4A binding element. Modified from Cooper et al.35 (F) ChIP-qPCR analysis of NR4A1 and NR4A1CEAA (DNA-binding mutant) recruitment to the CEBPA +42 Kb enhancer in expanded CD34+ bone marrow cells from healthy donors. (G) ChIP-qPCR analysis of NR4A3 binding to the Cebpa +37 Kb enhancer in murine 32Dcl3 cell line. (H) Luciferase reporter assay of the activation of the Cebpa +37 Kb enhancer by NR4A1/3 in 32Dcl3 cells. Vector schemes are shown on the left, and luciferase measurements on the right. RLU, relative light units. Results expressed as mean ± SD, n = 3. *P ≤ .05; **P ≤ .01; ***P ≤ .001. ChIP, chromatin immunoprecipitation.
NR4A act upstream of a C/EBPα transcriptional network in HSCs. (A) RT-qPCR confirmation of C/EBPα downregulation in CDKO relative to control HSCs after acute NR4A1/3 codepletion. (B) Experimental design for overexpression of NR4A1 and C/EBPα in bone marrow Lin− cells. (C) Frequency of LSKs and (D) 2 hours in vitro BrdU incorporation after 48 hours in culture after retroviral transduction in Lin− cells. (E) Pairwise alignment of mouse and human +37/42 Kb CEBPA enhancer containing the NR4A binding element. Modified from Cooper et al.35 (F) ChIP-qPCR analysis of NR4A1 and NR4A1CEAA (DNA-binding mutant) recruitment to the CEBPA +42 Kb enhancer in expanded CD34+ bone marrow cells from healthy donors. (G) ChIP-qPCR analysis of NR4A3 binding to the Cebpa +37 Kb enhancer in murine 32Dcl3 cell line. (H) Luciferase reporter assay of the activation of the Cebpa +37 Kb enhancer by NR4A1/3 in 32Dcl3 cells. Vector schemes are shown on the left, and luciferase measurements on the right. RLU, relative light units. Results expressed as mean ± SD, n = 3. *P ≤ .05; **P ≤ .01; ***P ≤ .001. ChIP, chromatin immunoprecipitation.
Next, we asked whether overexpression of C/EBPα can suppress the expansion and overproliferation of CDKO HSPCs. Using MSCV-IRES-GFP retroviral vectors, we overexpressed NR4A1 and C/EBPα in Lin− cells from CDKO and control mice after a 4-day tamoxifen treatment and analyzed the changes in LSK frequency and cell cycle by measuring in vitro BrdU incorporation after 2 days in culture (Figure 6B). The results showed that overexpression of C/EBPα in CDKO HSPCs significantly suppresses LSK frequency, albeit less efficiently than NR4A1 (Figure 6C). Analysis of BrdU incorporation indicated that overexpression of either NR4A1 or C/EBPα led to a reduction of actively cycling cells within the LSK compartment in both control and CDKO mice (Figure 6D). Notably, the 5-FU treatment required to promote HSPC cell cycle and allow retroviral integration is known to cause a downregulation of NR4As (supplemental Figure 4C).18,33 Thus, although C/EBPα is capable of suppressing the cell cycle of both CDKO and control HSCs, the proliferation requirement for retroviral integration precludes the observation of a loss-of-quiescence phenotype among CDKO LSK cells. Finally, overexpression of NR4A1 in Lin− cells from wild-type mice led to an upregulation of endogenous Cebpa transcription (supplemental Figure 4D).
Similar to mouse HSPCs, culturing of human CD34+ bone marrow cells led to silencing of all 3 NR4As and a decrease in C/EBPα expression relative to uncultured HSPCs (supplemental Figure 5A). Reexpression of NR4A1 caused a ∼50% reduction in cells in S/G2 cell cycle phase (supplemental Figure 5B) and a statistically significant increase C/EBPα expression (supplemental Figure 5C).
Expression of C/EBPα in HSCs is driven by a conserved enhancer element at +37/42 Kb (mouse/human) that is controlled by key hematopoietic transcription factors including RUNX1, PU.1, GATA2, MYB, and SCL.34,35 This enhancer contains a canonical NR4A response element (NBRE: AAGGTCA), suggesting CEBPA may be a direct transcriptional target of NR4As (Figure 6E).36 Query of our publicly available ChIP-Seq dataset in human AML cells revealed that NR4A1 is recruited to this enhancer (supplemental Figure 6A-B).37 To determine whether NR4As also bind the +37/42 Kb enhancer in normal HSPCs, we used human bone marrow CD34+ HSPCs and the murine bone marrow-derived 32Dcl3 cell line, which are more amenable to electroporation than mouse HSPCs. We electroporated CD34+ HSPCs with in vitro-transcribed NR4A1 RNA and observed a significant recruitment of NR4A1 to the CEBPA +42 KB enhancer (Figure 6F; supplemental Figure 6C-D). NR4A1 recruitment was abrogated in CD34+ cells electroporated with an NR4A1 mutant unable to bind the AAGGTCA motif (NR4A1CEAA).7,14 In addition, we detected a weaker recruitment of NR4A1 to a CEBPA +9 Kb enhancer, which also contains the NBRE and is active in CD34+ cells (supplemental Figure 6C).38 For analyzing NR4A binding in mouse 32Dcl3 cells, we electroporated NR4A3 IVT-RNA rather than NR4A1 because of the higher stability of NR4A3 protein in 32Dcl3 cells (supplemental Figure 5E), and similar to human CD34+ cells, we detected a significant recruitment of NR4A3 to the mouse Cebpa +37 Kb enhancer (Figure 6G).
To measure the effects of NR4A expression on the activity of the murine Cebpa +37 Kb enhancer, we performed a luciferase reporter assay in 32Dcl3 cells using a vector containing a 450-bp region of the Cebpa +37 Kb enhancer linked to luciferase (Figure 6H). Cotransfection of NR4A3 led to a significant increase in luciferase expression relative to GFP. In addition, deletion of the NBRE significantly decreased activation of the Cebpa +37 Kb enhancer by NR4A3. In summary, our results indicate that C/EBPα is a direct transcriptional target of NR4A1/3 in normal HSPCs and contributes to the antiproliferative activity of NR4As in HSCs.
Acute codepletion of NR4A1/3 leads to a proliferative inflammatory phenotype in HSCs
Analysis of the transcriptome after Nr4a1/3 deletion in HSCs revealed that NR4As suppress multiple gene programs related to inflammation and type I and II IFN signatures, and inflammatory cytokines were among the most upregulated gene sets on NR4A1/3 codepletion. Acute exposure to IFN-α or IFN-γ leads to a rapid proliferative response in HSCs that is dependent on STAT1 and causes subsequent activation of the PKB/AKT pathway.27,28 Measurement of IFN-γ protein levels in HSCs after a 4-day tamoxifen treatment showed a significant increase in Nr4a1/3-null HSCs (Figure 7A). We also detected a marked increase in the levels of Sca1 in the Lin−Nr4a1/3 CDKO cells (Figure 7B), which is a well-described consequence of IFN-1 activation in bone marrow cells.27 Next, we measured the phosphorylation level of STAT1 and AKT by flow cytometry and found both significantly elevated in Nr4a1/3-null HSCs (Figure 7C). Phosphorylation of ERK1/2, which acts downstream of IFN-1 in T cells,39 was also increased on Nr4a1/3 CDKO (Figure 7D). IFN-1 is also known to increase MYC protein levels in HSCs via posttranscriptional mechanisms.40 Accordingly, we found MYC protein levels were increased in Nr4a1/3 CDKO HSCs without a correspondent increase in Myc transcripts (Figure 7D).
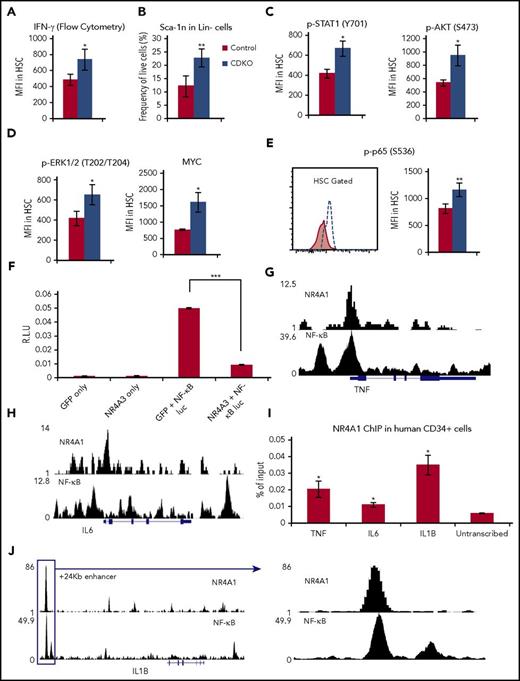
NR4A1/3 suppress an inflammatory proliferative response in HSCs. (A) Flow cytometry quantification of IFNγ levels in HSCs. (B) Sca-1 frequency in Lin− cells. (C-D) Flow cytometry quantification of protein levels in HSCs of (C) p-STAT1, p-AKT, and (D) p-ERK1/2 and MYC. (E) Flow cytometry quantification of phospho-p65 levels in HSCs. (F) Luciferase reporter assay after cotransfection of NR4A3 of GFP expression vectors with a luciferase reporter vector driven by a 5× NF-κB response elements in 32Dcl3 cells. (G-H) ChIP-Seq overlap between NR4A1 in Kasumi-1 cells and NF-κB in TNF-α-treated GM10847 cells in the (G) TNF locus and (H) IL6 locus. (I) ChIP-qPCR analysis of NR4A1 binding to NF-κB-regulated inflammatory cytokines in expanded human CD34+ bone marrow cells. (J) ChIP-Seq overlap between NR4A1 in Kasumi-1 cells and NF-κB in TNF-α-treated GM10847 in the IL1B +24 Kb enhancer. NR4A1 ChIP-Seq profile was extracted from Duren et al.37 NF-κB ChIP-Seq profile was extracted from the publically available ENCODE dataset.66 All flow cytometry analysis in A-E were performed at 4 days after first tamoxifen treatment. Results expressed as mean ± SD, n = 4 (A,G-I) or 3 (B-C). *P ≤ .05; **P ≤ .01; ***P ≤ .001.
NR4A1/3 suppress an inflammatory proliferative response in HSCs. (A) Flow cytometry quantification of IFNγ levels in HSCs. (B) Sca-1 frequency in Lin− cells. (C-D) Flow cytometry quantification of protein levels in HSCs of (C) p-STAT1, p-AKT, and (D) p-ERK1/2 and MYC. (E) Flow cytometry quantification of phospho-p65 levels in HSCs. (F) Luciferase reporter assay after cotransfection of NR4A3 of GFP expression vectors with a luciferase reporter vector driven by a 5× NF-κB response elements in 32Dcl3 cells. (G-H) ChIP-Seq overlap between NR4A1 in Kasumi-1 cells and NF-κB in TNF-α-treated GM10847 cells in the (G) TNF locus and (H) IL6 locus. (I) ChIP-qPCR analysis of NR4A1 binding to NF-κB-regulated inflammatory cytokines in expanded human CD34+ bone marrow cells. (J) ChIP-Seq overlap between NR4A1 in Kasumi-1 cells and NF-κB in TNF-α-treated GM10847 in the IL1B +24 Kb enhancer. NR4A1 ChIP-Seq profile was extracted from Duren et al.37 NF-κB ChIP-Seq profile was extracted from the publically available ENCODE dataset.66 All flow cytometry analysis in A-E were performed at 4 days after first tamoxifen treatment. Results expressed as mean ± SD, n = 4 (A,G-I) or 3 (B-C). *P ≤ .05; **P ≤ .01; ***P ≤ .001.
NR4As are known transcriptional regulators of inflammation in macrophages, where they form a negative feedback loop with NF-κB transcription factors.7,41,42 Exposure to inflammatory signals drives the rapid expression of the NR4As in a NF-κB-dependent manner. NR4As subsequently bind to p65 NF-κB and recruit CoREST co-repressor, antagonizing expression of inflammatory NF-κB targets including TNF-α, iNOS, and IL-1β7. Although NF-κB signaling appears to be dispensable for HSC function in the absence of stress, NF-κB is directly activated on acute exposure to inflammatory signals during stress-induced hematopoiesis, promoting the secretion of inflammatory cytokines, which leads to the expansion of HSCs and early hematopoietic progenitors.29,43 Given the known role of NR4A in antagonizing NF-κB activity in inflammatory cells and the increased levels of NF-κB-regulated inflammatory cytokines such as IL-6, TNF-α, and IL-1β on Nr4a1/3 CDKO, we hypothesized that acute depletion of NR4A1/3 may lead to an aberrant constitutive activation of NF-κB in HSCs. To test this hypothesis, we measured the levels of p65 activation (phospho-p65) in HSCs and found a significant increase of p65 activation in Nr4a1/3-null HSCs (Figure 7E). To determine whether NR4As can repress NF-κB in mouse hematopoietic progenitors, we cotransfected a luciferase reporter vector driven by 5 copies of NF-κB response element together with NR4A3 or GFP (control) into 32Dcl3 cells. We observed an 81% reduction of NF-κB activation in the presence of NR4A3 (Figure 7F).
Next, we analyzed binding of NR4A1 to TNF, IL1B, and IL6 and observed significant recruitment to the NF-κB regulatory region of TNF and, to a lesser extent, IL6 in human CD34+ cells (Figure 7G-I). In the case of IL1B, we focused on the recently described +24 Kb enhancer, which is activated by LPS, binds NF-κB, and contains an NBRE that is occupied by NR4A1 in Kasumi-1 cells44,45 (Figure 7J). We observed a significant recruitment of NR4A1 to this region (Figure 7I).
Discussion
We have demonstrated that codepletion of NR4A1/3 in adult mice causes chronic HSC hyperproliferation, with accumulation of oxidative stress and DNA damage leading to a preleukemic state that predisposes HSCs to AML transformation. Despite their cell cycle defects, Nr4a1/3-null HSCs retain multilineage differentiation and self-renewal capabilities. The antiproliferative mechanisms of NR4A action are partially mediated by intrinsic transcriptional regulation C/EBPα through direct binding of NR4A to a conserved CEBPA enhancer. Furthermore, we demonstrated that NR4A1/3 suppress proliferative inflammatory responses driven by IFN and NF-κB signaling. Although the proliferative phenotype is cell-intrinsic to HSCs, it is also conceivable, given the complex regulatory roles of NR4As in late-stage lymphoid and myeloid development and function, that extrinsic hematopoietic and/or niche-derived factors may also contribute to the overall phenotype observed in CDKO mice.
The hyperproliferative phenotype in Nr4a1/3-null HSCs recapitulates key features of the preleukemic state in acute myeloid leukemias, in which the unscheduled proliferation of HSCs leads to a mutagenic state that drives the acquisition of secondary mutations and subsequent leukemia progression.46 Mutations, translocations, and/or dysregulation of several key hematopoietic transcription factors including RUNX1, C/EBPα, and GATA2 are found as driver mutations in preleukemic HSCs.47-51 Further, gene-targeted knockin of AML-associated C/EBPα mutants into murine HSCs causes hyperproliferation and increased DNA double-strand breaks followed by the onset of AML, resembling the phenotype of Nr4a1/3-null preleukemic HSCs.52 AML-initiating chromosomal translocations have also been shown to increase DNA double-strand breaks in the absence of apoptosis, allowing mutant HSC clones to persist in bone marrow and drive AML transformation.53
Analysis of the transcriptional changes after acute codepletion of NR4A1/3 in HSCs revealed that the NR4As function as upstream activators of a C/EBPα-centered transcriptional network via direct transcriptional activation of C/EBPα. C/EBPα is a known driver of HSC quiescence and myeloid differentiation, and mice lacking Cebpa have increased HSC proliferation and excessive DNA double-strand breaks, but display myeloid differentiation blockade at the CMP progenitor stage and do not develop AML.24,54,55 Hence, virtually all CEBPA mutations in human patients with AML are hypomorphic and result in excessive HSPC proliferation without impairing myeloid differentiation, and ultimately develop AML.52,56 In this regard, the incomplete downregulation of Cebpa in CDKO mice may recapitulate a Cebpa hypomorphic mutation, rather than a complete loss of function.
Transcriptome analysis of Nr4a1/3-null HSCs also revealed significant activation of inflammatory signatures, including those activated by type I and type II IFNs and NF-κB-driven inflammatory cytokines that may also contribute to the proliferative phenotype of CDKO HSCs. Previous studies have demonstrated that acute treatment with type I (IFN-α/β) and type II (IFN-γ) IFNs results in a rapid cell cycle entry of HSCs.27,28 Conversely, suppression of type I and II IFN is essential for HSC homeostasis. Germ line deletion of the type I IFN transcriptional repressor Irf2 leads to HSC overproliferation and exhaustion.57 Similarly, deletion of IFN-γ repressors Adar1 and Irgm1 leads to HSC overproliferation and reduced repopulation potential.58-60 Our observations that codepletion of NR4A1/3 is sufficient to activate IFN signaling responses in HSCs indicates that direct or indirect repression of an IFN transcriptional response by NR4A1/3 may represent an alternative mechanism to prevent unscheduled HSC proliferation during steady-state hematopoiesis.
Although the mechanisms underlying NR4A-dependent repression of IFN signaling remain unclear, our findings reveal that several key NF-κB-regulated cytokines are direct transcriptional targets of NR4As in HSPCs. Attenuation of NF-κB signaling is essential for HSC homeostasis during steady-state hematopoiesis, and chronic stress activation of NF-κB through deletion of the negative regulator miR-146a results in low-grade inflammation that can give rise to both myeloid and lymphoid malignancies.61 Constitutive activation of NF-κB-dependent transcription and increased cytokine secretion are also key features of AML leukemia-initiating cells that contribute to their maintenance, and NF-κB–associated cytokines are elevated in the plasma of patients with AML.62-64 Our findings indicate that aberrant NF-κB activation is an early preleukemic event in CDKO HSCs that may predispose to leukemic transformation. However, the functional contribution of NF-κB activation to the preleukemic phenotype in CDKO HSCs remains to be established.
In summary, our study reveals a previously unsuspected role of the NR4A nuclear receptors in coordinating HSC quiescence and suppressing HSC inflammatory responses, in part through direct transcriptional activation of a C/EBPα-driven antiproliferative network while suppressing NF-κB-driven inflammatory signaling.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Loc Nguyen for mouse maintenance.
This work was supported by grant RO1 CA160747 from the National Institutes of Health, National Cancer Institute (O.M.C.), and by the Genomic and RNA Profiling, and the Cytometry and Cell Sorting Shared Resources of the Dan L. Duncan Cancer Center (National Cancer Institute grant P30CA125123).
Authorship
Contribution: P.R.F. and O.M.C conceived the experiments and wrote the manuscript; and P.R.F. performed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Orla M. Conneely, Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030; e-mail: orlac@bcm.edu.