Key Points
Both overexpression and knockout of miR-126 result in enhanced leukemogenesis.
Overexpression and knockout of miR-126 activate distinct gene signaling and are associated with different biological consequences.
Abstract
It is generally assumed that gain- and loss-of-function manipulations of a functionally important gene should lead to the opposite phenotypes. We show in this study that both overexpression and knockout of microRNA (miR)-126 surprisingly result in enhanced leukemogenesis in cooperation with the t(8;21) fusion genes AML1-ETO/RUNX1-RUNX1T1 and AML1-ETO9a (a potent oncogenic isoform of AML1-ETO). In accordance with our observation that increased expression of miR-126 is associated with unfavorable survival in patients with t(8;21) acute myeloid leukemia (AML), we show that miR-126 overexpression exhibits a stronger effect on long-term survival and progression of AML1-ETO9a-mediated leukemia stem cells/leukemia initiating cells (LSCs/LICs) in mice than does miR-126 knockout. Furthermore, miR-126 knockout substantially enhances responsiveness of leukemia cells to standard chemotherapy. Mechanistically, miR-126 overexpression activates genes that are highly expressed in LSCs/LICs and/or primitive hematopoietic stem/progenitor cells, likely through targeting ERRFI1 and SPRED1, whereas miR-126 knockout activates genes that are highly expressed in committed, more differentiated hematopoietic progenitor cells, presumably through inducing FZD7 expression. Our data demonstrate that miR-126 plays a critical but 2-faceted role in leukemia and thereby uncover a new layer of miRNA regulation in cancer. Moreover, because miR-126 depletion can sensitize AML cells to standard chemotherapy, our data also suggest that miR-126 represents a promising therapeutic target.
Introduction
MicroRNAs (miRNAs) have been implicated in the pathogenesis of various types of cancers.1-8 Some miRNAs play distinct roles in different types of cancers. For example, miRNA (miR)-126, originally identified as an endothelial-specific miRNA playing an essential role in angiogenesis and vascular integrity,9-11 has been shown to function as a critical tumor suppressor in various types of solid tumors.5,12-19 In contrast, we have shown that miR-126 is aberrantly overexpressed and likely plays an oncogenic role in core binding factor (CBF) leukemia.20 CBF leukemia is characterized by the presence of a t(8;21)(q22;q22) or an inv(16)(p13.1q22) chromosomal rearrangement, which accounts for ∼20% to 30% of primary acute myeloid leukemia (AML) cases.21-23 The potential oncogenic role of miR-126 in AML was further confirmed by other groups.24,25 However, it was reported that attenuation of miR-126 expression in normal hematopoietic stem/progenitor cells (HSPCs) resulted in expansion of long-term repopulating hematopoietic stem cells.26 Thus, the definitive role of miR-126 in the hematopoietic system warrants further investigation.
To precisely define the function of a given gene, it is often suggested that both gain- and loss-of-function studies be conducted.27,28 Gain- and loss-of-function studies are considered logical counterparts, and it is commonly believed that their phenotypes should be opposite.29-31 To further define the pathological role of miR-126 in leukemia, we first conducted both gain- and loss-of-function in vivo studies of miR-126 in mouse models of t(8;21) AML, the AML subtype that expresses miR-126 at the highest level among all AML subtypes. Surprisingly, both forced expression and knockout of miR-126 substantially promoted development of t(8;21) AML in mice but were associated with different consequences with regard to the long-term self-renewal and progression of leukemia stem cells/leukemia initiating cells (LSCs/LICs) and to the responsiveness of leukemia cells to standard chemotherapy. Second, we investigated the underlying molecular mechanisms.
Methods
Serial bone marrow transplantation (in vivo reconstitution) assays
For primary bone marrow transplantation (BMT) assays, mouse bone marrow (BM) progenitor (lineage negative) cells (ie, HSPCs) were isolated from 4- to 6-week-old wild-type (C57BL/6J CD45.2 [B6]) or miR-126 knockout (miR-126−/−, miR-126ΚΟ)9 mice 5 days after 5-fluorouracil (5-FU) treatment. The progenitor cells were retrovirally transduced with MSCV-PIG3 –based constructs through 2 rounds of “spinoculation” as described previously.6,20,32-34 After 5 days of selection with 2 μg/mL of puromycin, retrovirally transduced donor cells were injected by tail vein into lethally irradiated (960 rad) 8- to 10-week-old B6.SJL (CD45.1) recipient mice with 0.5 × 106 donor cells plus a radioprotective dose of whole BM cells (1 × 106; freshly harvested from a B6.SJL mouse) per recipient mouse. For secondary BMT assays, primary leukemic mouse BM cells (CD45.2+) from the groups of MSCV-AML1-ETO9a (AE9a), MSCV-PIG-AE9a-miR-126 (AE9a+miR-126), and miR-126KO+AE9a were collected and sorted by flow cytometry when the mice developed full-blown AML, and were then injected through tail vein into lethally irradiated secondary recipient mice with 1 × 106 donor cells per mouse. Primary empty vector control mouse BM cells were transplanted into secondary recipient mice as normal controls. In the tertiary and quaternary BMT assay, sorted leukemic mouse BM cells (CD45.2+) from the secondary and tertiary BMT recipients were collected and injected into lethally irradiated tertiary recipient mice, respectively, with 0.5 × 106 (for tertiary BMT) or 0.2 × 106 (for quaternary BMT) donor cells plus 1 × 106 of radioprotective wild-type whole BM cells per mouse.
Limiting dilution assays
BM or spleen leukemic cells (CD45.2+; sorted by flow cytometry) collected from secondary BMT recipients were transplanted into lethally irradiated recipients with 4 different doses of donor cells for each group. The numbers of recipient mice that developed full-blown leukemia within 15 weeks posttransplant were counted. Extreme limiting dilution assay software35 was used to estimate the frequency of LSCs/LICs.
Chemotherapy treatment
Cytarabine (Ara-C; Bedford Laboratories) and doxorubicin (Bedford Laboratories) were reconstituted with phosphate-buffered saline, filtered, and stored in aliquots at −20°C. Drugs were delivered by tail vein and intraperitoneal injection. The schedule consisted of 50 mg/kg Ara-C and 1.5 mg/kg doxorubicin daily during the first 3 days of treatment, followed by 50 mg/kg Ara-C daily for 2 additional days (ie, “5+3” regimen36 ). Treatment began at day 15 posttransplant in the tertiary BMT mice.
Affymetrix gene arrays of mouse samples
A total of 39 mouse BM samples were analyzed with GeneChip Mouse Gene 2.0 ST Array (Affymetrix). For each sample, the CD45.2+ cells (ie, transplanted donor cells) were sorted with flow cytometry from whole BM cells collected from BMT recipients at the end point. The microarray data have been deposited into the Gene Expression Omnibus database (accession number GSE65939).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA)37 was used to analyze gene sets enriched in genes dysregulated in mouse AE9a+miR-126 or miR-126KO+AE9a samples. The Kyoto Encyclopedia of Genes and Genomes Pathway Database gene set38 and the chemical and genetic perturbation gene set obtained from the Molecular Signatures Database37 were used as the gene set input for GSEA.
RNA extraction, quantitative real-time polymerase chain reaction, flow cytometry, western blot analysis, and luciferase reporter and mutagenesis assays
Results
miR-126 is expressed at a significantly higher level in t(8;21) AML than in non-CBF AML and inv(16) AML, and its increased expression is associated with poor prognosis in patients with t(8;21) AML
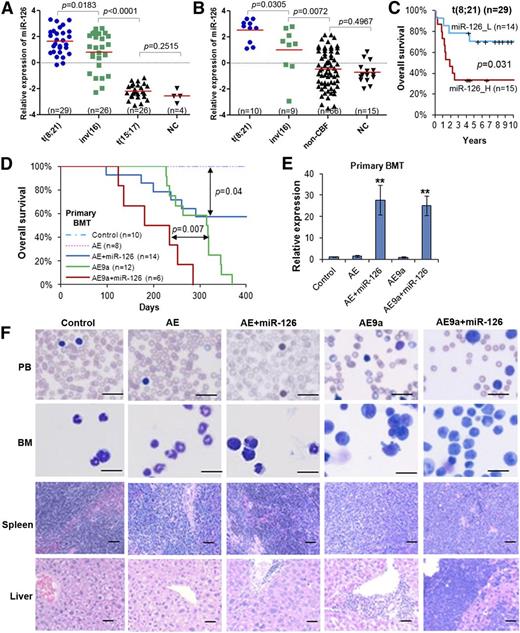
Consistent with our previous report from a bead-based miRNA expression profiling analysis,20 in our 2 independent new miRNA profiling assays of 85 samples (In-house 85S data set; 81 AML and 4 normal control samples) and 100 samples (In-house 100S data set; 85 AML and 15 normal control samples)6,32,34 (see supplemental Table 1, available on the Blood Web site) using Exiqon miRNA microarray platforms, we confirmed that both t(8;21) and inv(16) AML samples exhibit significantly higher miR-126 expression levels (P < .01) than non-CBF AML and normal hematopoietic cells (Figure 1A-B). Notably, miR-126 was also expressed at significantly higher levels (P < .05) in t(8;21) AML than in inv(16) AML; all t(8;21) AML cases, but not all inv(16) cases, exhibited higher levels of miR-126 expression than all normal control cases (Figure 1A-B).
miR-126 expression patterns in human AML subtypes, its prognosis impact in t(8;21) AML, and its pathological effect in leukemogenesis induced by AE and AE9a fusion genes. Expression patterns of miR-126 in the In-house 85S data set (A) or the In-house 100S data set (B). The P values were calculated by 2-tailed Student t test. (C) Comparison of OS between t(8;21) AML patients with higher or lower levels of miR-126 expression (median OS, 1.9 years vs 7.0 years, respectively; P = .031) in the In-house 85S data set. Kaplan-Meier survival curves are shown. The P values were calculated by log-rank test. (D) Effect of miR-126 on AE- and AE9a-induced primary leukemogenesis. Kaplan-Meier curves are shown for 5 cohorts of transplanted mice: MSCV-PIG (control), MSCV-PIG-AML1-ETO (AE), MSCV-PIG-AML1-ETO-miR-126 (AE+miR-126), MSCV-PIG-AML1-ETO9a (AE9a), and MSCV-PIG-AML1-ETO9a-miR-126 (AE9a+miR-126). P values were calculated by log-rank test. (E) Expression of miR-126 level was detected by qRT-PCR assay in the primary BMT mouse BM cells, which were isolated and sorted by flow cytometry for CD45.2+ cells at the end point. **P < .01. (F) Wright-Giemsa–stained PB and BM, and H&E-stained spleen and liver of the primary BMT recipient mice at the end point. Bars represent 25 μm for PB and BM; 100 μm for spleen and liver. H&E, hematoxylin and eosin; _H, high; _L, low; n, number of mice studied; NC, normal hematopoietic cells; OS, overall survival; PB, peripheral blood; qRT-PCR, quantitative real-time polymerase chain reaction.
miR-126 expression patterns in human AML subtypes, its prognosis impact in t(8;21) AML, and its pathological effect in leukemogenesis induced by AE and AE9a fusion genes. Expression patterns of miR-126 in the In-house 85S data set (A) or the In-house 100S data set (B). The P values were calculated by 2-tailed Student t test. (C) Comparison of OS between t(8;21) AML patients with higher or lower levels of miR-126 expression (median OS, 1.9 years vs 7.0 years, respectively; P = .031) in the In-house 85S data set. Kaplan-Meier survival curves are shown. The P values were calculated by log-rank test. (D) Effect of miR-126 on AE- and AE9a-induced primary leukemogenesis. Kaplan-Meier curves are shown for 5 cohorts of transplanted mice: MSCV-PIG (control), MSCV-PIG-AML1-ETO (AE), MSCV-PIG-AML1-ETO-miR-126 (AE+miR-126), MSCV-PIG-AML1-ETO9a (AE9a), and MSCV-PIG-AML1-ETO9a-miR-126 (AE9a+miR-126). P values were calculated by log-rank test. (E) Expression of miR-126 level was detected by qRT-PCR assay in the primary BMT mouse BM cells, which were isolated and sorted by flow cytometry for CD45.2+ cells at the end point. **P < .01. (F) Wright-Giemsa–stained PB and BM, and H&E-stained spleen and liver of the primary BMT recipient mice at the end point. Bars represent 25 μm for PB and BM; 100 μm for spleen and liver. H&E, hematoxylin and eosin; _H, high; _L, low; n, number of mice studied; NC, normal hematopoietic cells; OS, overall survival; PB, peripheral blood; qRT-PCR, quantitative real-time polymerase chain reaction.
We then analyzed the prognostic impact of miR-126 expression on survival of AML patients. In analysis of the 81 de novo AML cases of the In-house 85S data set (supplemental Table 1; Figure 1A), we found that AML patients with higher levels of miR-126 had a significantly shorter OS than those with lower levels (supplemental Figure 1A). However, by analysis of subcohorts of patients with CBF, t(8;21), inv(16), or t(15;17) (non-CBF) AML, we found that miR-126 expression had a significant prognostic impact only in t(8;21) AML (Figure 1C; supplemental Figure 1A). Our univariate analysis showed that increased expression levels of miR-126 is a predictor of shorter OS in patients with t(8;21) AML (supplemental Table 2). There was a strong positive correlation between increased expression levels of miR-126 and increased blast cell percentages in PB and BM (supplemental Figure 1B-C). Our multivariate analysis indicated that the prognostic impact of miR-126 expression on OS was independent of age, sex, white blood cell counts, hemoglobin level, and platelet counts of the t(8;21) patients (supplemental Table 3).
Among the 85 AML samples of the In-house 100S data set, 70 were collected from patients with de novo AML at diagnosis before treatment (supplemental Table 1). The miR-126 expression level exhibited no significantly prognostic impact on OS of the whole set or individual subsets of AML, except for the t(8;21) AML subset (supplemental Figure 1D).
Forced expression of miR-126 promotes leukemogenesis induced by t(8;21) fusion genes
To investigate the biological function of miR-126 in the pathogenesis of t(8;21) AML, we performed in vivo gain-of-function studies through mouse BMT assays. Briefly, lethally irradiated recipient mice were reconstituted with wild-type HSPCs retrovirally transduced with MSCV-PIG3,32 (Control), MSCV-PIG-AML1-ETO (AE); AE9a, a potent oncogenic isoform of AE39 ), MSCV-PIG-AE-miR-126 (AE+miR-126), or AE9a+miR-126. Consistent with previous reports that AE9a alone, but not AE alone, can cause AML in mice,39-41 we observed that all recipient mice of the AE9a group developed AML within 400 days, whereas the AE group did not develop AML (Figure 1D). Forced expression of miR-126 significantly accelerated AE9a-mediated leukemogenesis (median survival of AE9a+miR-126 vs AE9a alone was 234 days vs 317 days, respectively; P < .01). Forced expression of miR-126 also cooperated with AE and induced leukemia in 43% (6/14) of AE+miR-126 mice (Figure 1D-E). Forced expression of miR-126 in the presence of AE or AE9a caused a higher percentage of leukemic blast cells in the PB, and, particularly in the BM, associated with a more severe leukemic infiltration in spleen and liver of the recipient mice compared with their counterparts without miR-126 overexpression (Figure 1F). Thus, our data suggest that miR-126 can promote leukemogenesis through cooperation with AE and AE9a.
The in vivo effect of miR-126 overexpression on hematopoiesis was also investigated. Notably, 5 of 10 (50%) recipient mice transplanted with MSCV-PIG-miR-126–transduced HSPCs, but none of the 6 control mice, developed disease within 400 days (supplemental Figure 2A-B). The miR-126–overexpressing diseased mice were associated with syndromes of BM failure (supplemental Figure 2C-D). Thus, our data suggest that forced expression of miR-126 alone can cause some hematopoietic abnormalities.
Depletion of miR-126 expression also promotes leukemogenesis induced by t(8;21) fusion genes
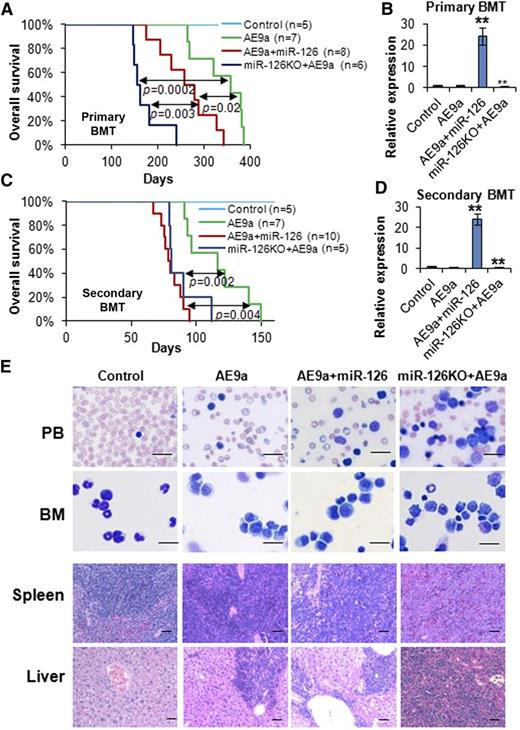
We also performed loss-of-function studies in vivo with an miR-126−/− mouse model.9 Surprisingly, AE-transduced miR-126−/− (miR-126KO+AE) HSPCs caused AML in all recipient mice within 300 days (median survival, 122 days), in contrast to no leukemia caused by AE-transduced wild-type HSPCs (supplemental Figure 3A-B). Similarly, AE9a-transduced miR-126−/− (miR-126KO+AE9a) HSPCs caused leukemia significantly faster than AE9a-transduced wild-type HSPCs in recipients (median survival, 130 days vs 317 days, respectively; P < .0001) (supplemental Figure 3A-B). miR-126KO+AE mice and miR-126KO+AE9a mice showed a high proportion of immature blast cells in the PB and BM, as well as severe leukemia cell infiltration in spleen and liver (supplemental Figure 3C). We found that miR-126−/− and wild-type mice responded similarly to 5-FU (supplemental Figure 4), suggesting that the acceleration of leukemogenesis by miR-126 knockout was not due to an unusual effect of 5-FU on miR-126−/− BM cells. Notably, miR-126KO+AE mice developed leukemia significantly faster than AE+miR-126 mice (supplemental Figure 5A). Similarly, miR-126KO+AE9a mice developed leukemia significantly faster than AE9a+miR-126 mice, and both developed leukemia faster than AE9a-alone mice (supplemental Figure 5B). To validate our observations, we repeated primary BMT assays with the AE9a model. Consistent with our first assays (supplemental Figure 5B), the miR-126KO+AE9a group developed AML significantly faster than the AE9a+miR-126 group (median survival, 158 days vs 267 days, respectively; P = .0033), and both developed AML significantly faster (P < .05) than the AE9a-alone group (median survival, 368 days) (Figure 2A-B). Thus, our data demonstrated that complete depletion of miR-126 expression had an even greater effect than miR-126 overexpression on promoting the primary leukemogenesis.
Knockout and overexpression of miR-126 promote AE9a-induced leukemogenesis in primary and secondary BMT. (A) The effect of the overexpression and knockout of miR-126 on AE9a-induced primary leukemogenesis in primary BMT recipient mice. Kaplan-Meier curves and P values (log-rank test) are shown. (B) Expression of miR-126 was detected by qRT-PCR assay in leukemic or control BM cells (CD45.2+) isolated from primary BMT recipient mice at the end point. **P < .01. Survival curves (C) and relative miR-126 expression levels (D) in secondary BMT recipient mice are shown. **P < .01. (E) Wright-Giemsa–stained PB and BM, and H&E-stained spleen and liver of the secondary BMT recipient mice at the end point. Bars represent 25 μm for PB and BM; 100 μm for spleen and liver.
Knockout and overexpression of miR-126 promote AE9a-induced leukemogenesis in primary and secondary BMT. (A) The effect of the overexpression and knockout of miR-126 on AE9a-induced primary leukemogenesis in primary BMT recipient mice. Kaplan-Meier curves and P values (log-rank test) are shown. (B) Expression of miR-126 was detected by qRT-PCR assay in leukemic or control BM cells (CD45.2+) isolated from primary BMT recipient mice at the end point. **P < .01. Survival curves (C) and relative miR-126 expression levels (D) in secondary BMT recipient mice are shown. **P < .01. (E) Wright-Giemsa–stained PB and BM, and H&E-stained spleen and liver of the secondary BMT recipient mice at the end point. Bars represent 25 μm for PB and BM; 100 μm for spleen and liver.
Notably, forced expression of AE or AE9a did not upregulate endogenous expression of miR-126 in the BM cells (Figures 1E and 2B; supplemental Figure 3B), or in colony-forming cells (supplemental Figure 5C). Similarly, internal tandem duplications of FLT3 and KIT mutations, 2 major secondary mutations in CBF leukemia,41-45 did not upregulate expression of miR-126, either alone or together with AE (supplemental Figure 5D). Thus, the especially high expression of miR-126 in human t(8;21) AML (Figure 1A-B) is unlikely induced by the fusion genes and/or secondary mutations directly. The demethylation of the miR-126 promoter region20 is still likely a major cause of its overexpression.
The depletion and, in particular, overexpression of miR-126 enhanced long-term self-renewal of the leukemia cells
To assess the impact of miR-126 overexpression and depletion on long-term self-renewal of t(8;21) LSCs/LICs, we performed serial mouse BMT assays. In the secondary mouse BMT assay, leukemic or control BM cells collected from primary BMT recipient mice were transplanted to lethally irradiated wild-type secondary recipient mice. All the groups, except for the negative control group, developed leukemia much faster than the corresponding primary BMT groups (Figure 2C vs 2A). Notably, the AE9a+miR-126 group showed similar leukemia development to the miR-126KO+AE9a group in secondary BMT (median survival, 79 days vs 81 days, respectively; P = .22), and both groups had significantly shorter survival (P < .01) than the AE9a group (median survival, 116 days) (Figure 2C-D). Both AE9a+miR-126 and miR-126KO+AE9a mice exhibited a more aggressive leukemic phenotype than AE9a mice (Figure 2E).
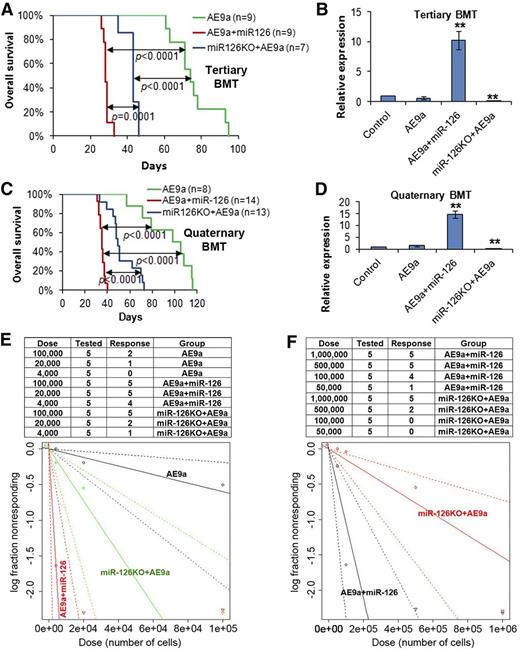
We then performed tertiary mouse BMT assays with secondary leukemia BM cells as donor cells. All groups of mice developed AML much faster than the secondary BMT groups (Figure 3A vs Figure 2C). Strikingly, the AE9a+miR-126 group developed leukemia significantly faster than the miR-126KO+AE9a group in tertiary BMT (median survival, 28 days vs 43 days, respectively; P = .0001), and both groups developed leukemia significantly faster (P < .0001) than the AE9a group (median survival, 74 days) (Figure 3A-B; supplemental Figure 5E). Quaternary mouse BMT assays were further conducted with tertiary leukemia BM cells as donors. Again, the AE9a+miR-126 group developed AML significantly faster than the miR-126KO+AE9a group (median survival, 35 days vs 49 days, respectively; P < .0001), whereas both groups developed leukemia significantly faster (P < .0001) than the AE9a group (median survival, 98 days) (Figure 3C-D; supplemental Figure 6A).
Overexpression of miR-126 exhibited a more potent effect than knockout of miR-126 on long-term LSC/LIC self-renewal and frequency. (A) The tertiary BMT recipients were transplanted with leukemic BM cells isolated from the secondary BMT recipient mice of the AE9a, AE9a+miR-126, and miR-126KO+AE9a groups. Kaplan-Meier curves and P values (log-rank test) are shown. (B) Expression of miR-126 was detected by qRT-PCR assay in leukemic BM cells (CD45.2+) of tertiary BMT recipients. **P < .01. Survival curves (C) and relative miR-126 expression levels (D) of the 3 groups in quaternary BMT are shown. (E) Mouse BM leukemic cells from secondary BMT recipient mice were used as donor cells for BMT in the limiting dilution assays. The estimated LSC/LIC frequencies of the AE9a, AE9a+miR-126, and miR-126KO+AE9a groups are 1/166 619 (95% CI, 1/529 517-1/52 429), 1/2476 (95% CI, 1/7237-1/847), and 1/27 399 (95% CI, 1/66 212-1/11 338), respectively. Significance of the frequency difference: AE9a vs AE9a+miR-126, P = 5.35 × 10−9 (χ2-test); AE9a vs miR-126KO+AE9a, P = .0092; AE9a+miR-126 vs miR-126KO+AE9a, P =.00072. (F) Mouse spleen leukemic cells from secondary BMT recipient mice were used as donor cells. The estimated LSC/LIC frequency of the AE9a+miR-126 group is 1/95 040 (95% CI, 1/215 902-1/41 837), significantly greater (P = .00088) than that of the miR-126KO+AE9a group at 1/653 224 (95% CI, 1/1 380 618-1/309 066). CI, confidence interval; Dose, number of donor cells; Tested, total number of mice used as BMT recipients in the limiting dilution assay; Response, mice that developed leukemia within 15 weeks post BMT.
Overexpression of miR-126 exhibited a more potent effect than knockout of miR-126 on long-term LSC/LIC self-renewal and frequency. (A) The tertiary BMT recipients were transplanted with leukemic BM cells isolated from the secondary BMT recipient mice of the AE9a, AE9a+miR-126, and miR-126KO+AE9a groups. Kaplan-Meier curves and P values (log-rank test) are shown. (B) Expression of miR-126 was detected by qRT-PCR assay in leukemic BM cells (CD45.2+) of tertiary BMT recipients. **P < .01. Survival curves (C) and relative miR-126 expression levels (D) of the 3 groups in quaternary BMT are shown. (E) Mouse BM leukemic cells from secondary BMT recipient mice were used as donor cells for BMT in the limiting dilution assays. The estimated LSC/LIC frequencies of the AE9a, AE9a+miR-126, and miR-126KO+AE9a groups are 1/166 619 (95% CI, 1/529 517-1/52 429), 1/2476 (95% CI, 1/7237-1/847), and 1/27 399 (95% CI, 1/66 212-1/11 338), respectively. Significance of the frequency difference: AE9a vs AE9a+miR-126, P = 5.35 × 10−9 (χ2-test); AE9a vs miR-126KO+AE9a, P = .0092; AE9a+miR-126 vs miR-126KO+AE9a, P =.00072. (F) Mouse spleen leukemic cells from secondary BMT recipient mice were used as donor cells. The estimated LSC/LIC frequency of the AE9a+miR-126 group is 1/95 040 (95% CI, 1/215 902-1/41 837), significantly greater (P = .00088) than that of the miR-126KO+AE9a group at 1/653 224 (95% CI, 1/1 380 618-1/309 066). CI, confidence interval; Dose, number of donor cells; Tested, total number of mice used as BMT recipients in the limiting dilution assay; Response, mice that developed leukemia within 15 weeks post BMT.
To validate the above observations, we repeated the serial BMT assays with leukemia BM cells collected from primary recipient mice of a different BMT experiment. Similar patterns were observed (supplemental Figure 6B). Collectively, depletion and especially forced expression of miR-126 substantially enhanced long-term leukemia progression. Consistent with the significantly positive correlation between miR-126 expression and PB or BM blast percentages of t(8;21) AML patients (supplemental Figure 1B-C), the AE9a+miR-126 mice exhibited much higher engraftment degrees (median, ∼80% to >95%) of donor leukemic cells in the BM than AE9a or miR-126KO+AE9a mice (median, ∼30% to ∼60% for both groups) in all 4 BMT passages (supplemental Figure 7).
To directly compare the effects of miR-126 overexpression and depletion on the frequency of LSCs/LICs, we conducted limiting dilution assays with mouse BM leukemic cells collected from secondary BMT recipients (Figure 2C) as donor cells. As expected, the estimated LSC/LIC frequency of the AE9a+miR-126 group was significantly greater (P < .001) than that of the miR-126KO+AE9a group (1/2476 vs 1/27 399, respectively; >10-fold), and both were significantly higher (P < .01) than that of the AE9a group (1/166 619) (Figure 3E). A similar pattern was observed when spleen leukemic cells were used as donor cells in the limiting dilution assays; the estimated LSC/LIC frequency of the AE9a+miR-126 group was more than sevenfold greater than that of the miR-126KO+AE9a group (1/95 040 vs 1/653 224; P < .001) (Figure 3F).
Depletion of miR-126 expression sensitizes AE9a-mediated leukemia to standard chemotherapy
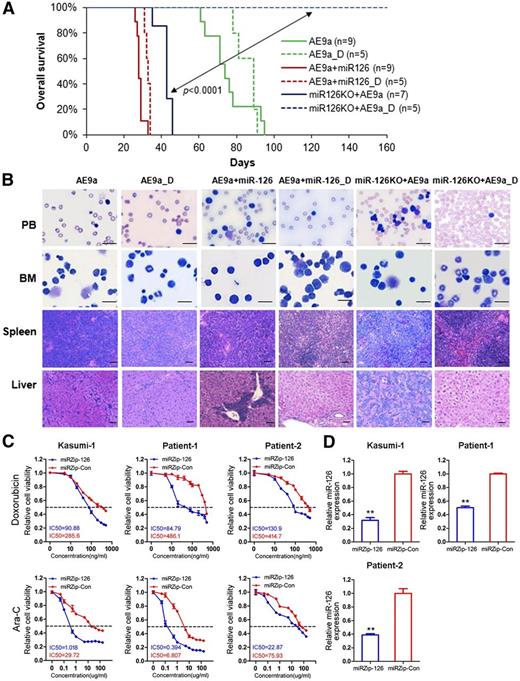
Because increased expression of miR-126 is an independent predictor of poor survival of AML patients with t(8;21) (Figure 1C), we hypothesized that the expression level of miR126 might be associated with response to standard chemotherapy. To test this, we treated mice transplanted with AE9a, AE9a+miR-126, or miR-126KO+AE9a leukemic BM cells with phosphate-buffered saline or Ara-C plus doxorubicin (5+3 regimen)36 after the onset of AML in mice. It was reported previously that t(8;21) patients with a higher abundance of AE9a transcripts are resistant to standard chemotherapy and associated with poor survival.46 Consistently, we found that both AE9a AML and AE9a+miR-126 AML were largely resistant to the standard chemotherapy, and all the treated and untreated mice died of AML within 100 days and 40 days, respectively (Figure 4A). Remarkably, however, miR-126KO+AE9a AML was very responsive to the treatment, and all the treated mice survived without leukemia development for more than 160 days, whereas all the untreated mice died of AML within 50 days (Figure 4A). White blood cells in the PB and BM of miR-126KO+AE9a mice with chemotherapy were well differentiated, and the organ architectures of spleen and liver were well preserved (Figure 4B), suggesting that these AML mice were largely cured by the chemotherapy.
Depletion of miR-126 increases sensitivity to chemotherapy treatment in mice carrying AE9a-induced AML. (A) AE9a, AE9a+miR-126, and miR-126KO+AE9a tertiary leukemic recipient mice were treated with phosphate-buffered saline (control; solid line), or a daily dose of 100 mg/kg Ara-C for 5 days along with a daily dose of 3 mg/kg doxorubicin during the first 3 days of Ara-C treatment (indicated by _D and dashed line; 5+3 regimen36 ). Kaplan-Meier curves are shown. (B) Wright-Giemsa–stained PB and BM, and H&E-stained spleen and liver of the treated tertiary BMT recipient mice at the end point. Bars represents 25 μm for PB and BM; 100 μm for spleen and liver. (C) The relative viability of Kasumi-1 and primary leukemia cells of t(8;21) AML patients treated with serial dilutions of doxorubicin and Ara-C for 48 hours. (D) Lentivirus-mediated inhibition of miR-126 was detected by qRT-PCR. The data shown are the means of 3 biological replicates. **P < .01. Error bars represent standard deviation. IC50, half-maximum inhibitory concentration miRZip-Con, miRZip™ control lentivirus (System Biosciences); miRZip-126, miRZip™ anti-miR-126 lentivirus (System Biosciences, Mountain View, CA).
Depletion of miR-126 increases sensitivity to chemotherapy treatment in mice carrying AE9a-induced AML. (A) AE9a, AE9a+miR-126, and miR-126KO+AE9a tertiary leukemic recipient mice were treated with phosphate-buffered saline (control; solid line), or a daily dose of 100 mg/kg Ara-C for 5 days along with a daily dose of 3 mg/kg doxorubicin during the first 3 days of Ara-C treatment (indicated by _D and dashed line; 5+3 regimen36 ). Kaplan-Meier curves are shown. (B) Wright-Giemsa–stained PB and BM, and H&E-stained spleen and liver of the treated tertiary BMT recipient mice at the end point. Bars represents 25 μm for PB and BM; 100 μm for spleen and liver. (C) The relative viability of Kasumi-1 and primary leukemia cells of t(8;21) AML patients treated with serial dilutions of doxorubicin and Ara-C for 48 hours. (D) Lentivirus-mediated inhibition of miR-126 was detected by qRT-PCR. The data shown are the means of 3 biological replicates. **P < .01. Error bars represent standard deviation. IC50, half-maximum inhibitory concentration miRZip-Con, miRZip™ control lentivirus (System Biosciences); miRZip-126, miRZip™ anti-miR-126 lentivirus (System Biosciences, Mountain View, CA).
Next, we investigated whether suppression of miR-126 could sensitize human t(8;21) AML cells to standard chemotherapy agents. As shown in Figure 4C, lentivirus-mediated inhibition of miR-126 (Figure 4D) dramatically sensitized Kasumi-1 (a t[8;21] AML cell line) and primary t(8;21) AML cells to doxorubicin and Ara-C. Thus, our data suggest that the depletion of miR-126 expression substantially increases sensitivity of t(8;21) AML cells to standard chemotherapy.
Overexpression and knockout of miR-126 trigger distinct gene signaling
To elucidate the molecular mechanisms underlying the biological functions of miR-126 in AML, we conducted messenger RNA microarray analysis for 36 mouse AML samples with AE9a, AE9a+miR-126, and miR-126KO+AE9a collected from the first, second, third, and fourth passages of BMT recipient mice (3 mice per group per passage), along with 3 normal controls from the first passage of BMT (supplemental Figure 8). Strikingly, through GSEA,37 we found that the upregulated genes in AE9a+miR-126 samples were significantly enriched with genes highly expressed in LSCs or normal HSPCs (supplemental Figure 9A). In contrast, the upregulated genes in miR-126KO+AE9a samples were enriched with genes highly expressed in mature hematopoietic cells or later progenitors, as well as with genes related to apoptosis, cell cycle, and/or DNA replication (supplemental Figure 9B).
Through significance analysis of microarrays,47 we identified 625 and 1443 genes that were expressed at significantly (P < .05; false discovery rate <0.01) higher and lower levels, respectively, in AE9a+miR-126 than in both AE9a and miR-126KO+AE9a AML cells; of them, 57 (AE9a+miR-126_High) and 313 (AE9a+miR-126_Low) genes were significantly positively and negatively correlated, respectively, with miR-126 in expression across the 29 t(8;21) patient samples in which we have both miRNA and messenger RNA expression data (supplemental Table 4; supplemental Figure 10). Similarly, we identified 201 and 94 genes that were expressed at significantly (P < .05; false discovery rate <0.01) higher and lower levels, respectively, in miR-126KO+AE9a than in both AE9a and AE9a+miR-126 AML cells; of them, 88 (miR-126KO+AE9a_High) and 24 (miR-126KO+AE9a_Low) genes significantly negatively and positively correlated, respectively, with miR-126 in expression across human t(8;21) AML samples (supplemental Table 5; supplemental Figure 11).
Consistent with our GSEA analyses (supplemental Figure 9), in analysis of the GSE24006 data set,48 we found that most of the 57 AE9a+miR-126_High genes were expressed at significantly higher levels in primitive HSPCs (including hematopoietic stem cells and multipotent progenitors) than in committed progenitors (including common myeloid progenitors, granulocyte/monocyte progenitors, and megakaryocyte/erythrocyte progenitors) (Figure 5A). The opposite pattern was observed for the 313 AE9a+miR-126_Low genes (supplemental Figure 12). Conversely, most of the 88 miR-126KO+AE9a_High genes were expressed at significantly higher levels in committed progenitors than in primitive HSPCs (Figure 5B), whereas roughly half of the 24 miR-126KO+AE9a_Low genes were highly expressed in primitive HSPCs (supplemental Figure 13). These data suggest that overexpression and knockout of miR-126 trigger distinct gene signaling that is highly expressed in different hematopoietic cell populations (ie, in primitive HSPCs and in committed/differentiated progenitors, respectively) (Figure 5C).
Comparison of expression levels between Pri HSPCs and CPs for the genes dysregulated in AE9a+miR-126 and miR-126KO+AE9a leukemic cells. Expression patterns between Pri HSPCs and CPs from the GSE24006 data set48 for the 57 AE9a+miR-126_High genes (A) and the 88 miR-126KO+AE9a_High genes (B) are shown. Expression data were mean centered, and the relative value for each sample is represented by a color, with red and green representing a high and low expression, respectively (scale is shown). (C) Within each gene set (ie, AE9a+miR-126_High, AE9a+miR-126_Low, miR-126KO+AE9a_High, or miR-126KO+AE9a_Low), the proportion of genes that are expressed at a significantly higher level in Pri HSPCs than in CPs (Pri HSPCs_High; blue), a significantly higher level in CPs than in Pri HSPCs (CPs_High; red), or a comparable level between Pri HSPCs and CPs (nonsignificant [NS]; green) is shown. Gene expression levels were compared between Pri HSPCs (including BM_HSC and BM_MPP samples) and CPs (including BM_CMP, BM_GMP, and BM_MEP samples) in the GSE24006 data set48 through significance analysis of microarrays,47 with a q value <0.05 and a false discovery rate <0.001 as criteria for statistical significance. CB_HSC and CB_MPP samples were not included in the significance analysis of microarrays due to their tissue differences from other BM samples. CB, core blood; CMP, common myeloid progenitor; CPs, committed progenitors; GMP, granulocyte/monocyte progenitor; HSC, hematopoietic stem cell; MEP, megakaryocyte/erythrocyte progenitor; MPP, multipotent progenitor; Pri HSPCs, primitive HSPCs.
Comparison of expression levels between Pri HSPCs and CPs for the genes dysregulated in AE9a+miR-126 and miR-126KO+AE9a leukemic cells. Expression patterns between Pri HSPCs and CPs from the GSE24006 data set48 for the 57 AE9a+miR-126_High genes (A) and the 88 miR-126KO+AE9a_High genes (B) are shown. Expression data were mean centered, and the relative value for each sample is represented by a color, with red and green representing a high and low expression, respectively (scale is shown). (C) Within each gene set (ie, AE9a+miR-126_High, AE9a+miR-126_Low, miR-126KO+AE9a_High, or miR-126KO+AE9a_Low), the proportion of genes that are expressed at a significantly higher level in Pri HSPCs than in CPs (Pri HSPCs_High; blue), a significantly higher level in CPs than in Pri HSPCs (CPs_High; red), or a comparable level between Pri HSPCs and CPs (nonsignificant [NS]; green) is shown. Gene expression levels were compared between Pri HSPCs (including BM_HSC and BM_MPP samples) and CPs (including BM_CMP, BM_GMP, and BM_MEP samples) in the GSE24006 data set48 through significance analysis of microarrays,47 with a q value <0.05 and a false discovery rate <0.001 as criteria for statistical significance. CB_HSC and CB_MPP samples were not included in the significance analysis of microarrays due to their tissue differences from other BM samples. CB, core blood; CMP, common myeloid progenitor; CPs, committed progenitors; GMP, granulocyte/monocyte progenitor; HSC, hematopoietic stem cell; MEP, megakaryocyte/erythrocyte progenitor; MPP, multipotent progenitor; Pri HSPCs, primitive HSPCs.
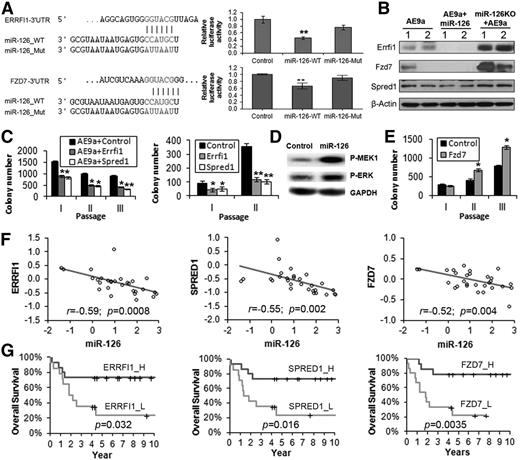
ERRFI1, SPRED1, and FZD7 are critical direct targets of miR-126
Among the 313 AE9a+miR-126_Low genes (supplemental Table 4), ERRFI1 (MIG6) and SPRED1 are predicted targets of miR-126 in both human and mouse genomes (SPRED1 has been experimentally validated9,10 ) and function as critical tumor suppressors in various cancers by inhibiting the mitogen-activated protein kinase (MAPK) pathway (ie, the RAS-RAF-MEK-ERK cascade).49-55 Among the 88 miR-126KO+AE9a_High genes (supplemental Table 5), Fzd7, a Wnt receptor involved in the activation of the Wnt signaling cascade,56-58 is also a predicted target of miR-126. Our luciferase reporter and western blot assays demonstrate that both ERRFI1 and FZD7 are direct targets of miR-126 (Figure 6A-B). Forced expression of Errfi1 or Spred1 can significantly inhibit the replating capacity of AE9a-transduced fresh mouse HSPCs or AE9a+miR-126 mouse leukemic BM cells (Figure 6C), suggesting that they are functionally important targets of miR-126. As expected, forced expression of miR-126 led to a substantial increase of phosphorylated MEK and ERK (Figure 6D). In contrast, ectopic expression of Fzd7 dramatically enhanced the colony-forming/replating capacity of AE9a AML cells (Figure 6E). As expected, the 3 target genes significantly negatively correlated with miR-126 in expression in t(8;21) AML patient samples (Figure 6F), and their increased expression was associated with longer OS in t(8;21) AML patients (Figure 6G), opposite to that of miR-126 (Figure 1C). Their prognostic impacts were significant in univariable (supplemental Table 2) and mutivariable (supplemental Table 3) testing models.
Direct target genes of miR-126. (A) ERRFI1 and FZD7 3′UTR and the wild-type (_WT) and mutant (_Mut) sequences of mature miR-126 (left), and luciferase reporter and mutagenesis assay results (right). Whereas wild-type miR-126 significantly inhibits luciferase activity of the reporter plasmid carrying the 3′UTR of ERRFI1and FZD7, mutation in the miR-126 seed sequence rescues the inhibitory effect. (B) Western blot analysis of Errfi1, Fzd7, and Spred1 levels in AE9a, AE9a+miR-126, and miR-126KO+AE9a AML cells collected from quaternary BMT recipients (2 samples per group). β-Actin was used as an endogenous control. (C) Colony-forming/replating assays with cotransduction of MSCVneo-AE9a together with MSCV-PIG (Control), MSCV-PIG-Errfi1, or MSCV-PIG-Spred1 into normal mouse BM progenitor cells (left), or with transduction of MSCV-PIG (Control), MSCV-PIG-Errfi1, or MSCV-PIG-Spred1 into AE9a+miR-126 BM leukemia cells collected from tertiary BMT recipients (right). (D) Western blot analysis of phosphorylated (P)-MEK1 or P-ERK in human 293T cells transfected with MSCV-PIG (Control) or MSCV-PIG-miR-126 (miR-126) 48 hours posttransfection. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. (E) Colony-forming/replating assays with transduction of MSCV-PIG (Control) and MSCV-PIG-Fzd7 (Fzd7) into AE9a BM leukemia cells collected from tertiary BMT recipients. (F) Correlation between the expression levels of miR-126 and ERRFI1, SPRED1, or FZD7 in the set of 29 AML patients carrying t(8;21). All expression data were log2 transformed and mean centered. The correlation coefficient (r) and P values were detected by Pearson correlation, and the correlation regression lines were drawn with the linear regression algorithm. (G) Comparison of the OS in t(8;21) AML patients with higher or lower levels of ERRFI1, SPRED1, or FZD7 expression. Kaplan-Meier survival curves and P values (log-rank test) are shown.
Direct target genes of miR-126. (A) ERRFI1 and FZD7 3′UTR and the wild-type (_WT) and mutant (_Mut) sequences of mature miR-126 (left), and luciferase reporter and mutagenesis assay results (right). Whereas wild-type miR-126 significantly inhibits luciferase activity of the reporter plasmid carrying the 3′UTR of ERRFI1and FZD7, mutation in the miR-126 seed sequence rescues the inhibitory effect. (B) Western blot analysis of Errfi1, Fzd7, and Spred1 levels in AE9a, AE9a+miR-126, and miR-126KO+AE9a AML cells collected from quaternary BMT recipients (2 samples per group). β-Actin was used as an endogenous control. (C) Colony-forming/replating assays with cotransduction of MSCVneo-AE9a together with MSCV-PIG (Control), MSCV-PIG-Errfi1, or MSCV-PIG-Spred1 into normal mouse BM progenitor cells (left), or with transduction of MSCV-PIG (Control), MSCV-PIG-Errfi1, or MSCV-PIG-Spred1 into AE9a+miR-126 BM leukemia cells collected from tertiary BMT recipients (right). (D) Western blot analysis of phosphorylated (P)-MEK1 or P-ERK in human 293T cells transfected with MSCV-PIG (Control) or MSCV-PIG-miR-126 (miR-126) 48 hours posttransfection. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. (E) Colony-forming/replating assays with transduction of MSCV-PIG (Control) and MSCV-PIG-Fzd7 (Fzd7) into AE9a BM leukemia cells collected from tertiary BMT recipients. (F) Correlation between the expression levels of miR-126 and ERRFI1, SPRED1, or FZD7 in the set of 29 AML patients carrying t(8;21). All expression data were log2 transformed and mean centered. The correlation coefficient (r) and P values were detected by Pearson correlation, and the correlation regression lines were drawn with the linear regression algorithm. (G) Comparison of the OS in t(8;21) AML patients with higher or lower levels of ERRFI1, SPRED1, or FZD7 expression. Kaplan-Meier survival curves and P values (log-rank test) are shown.
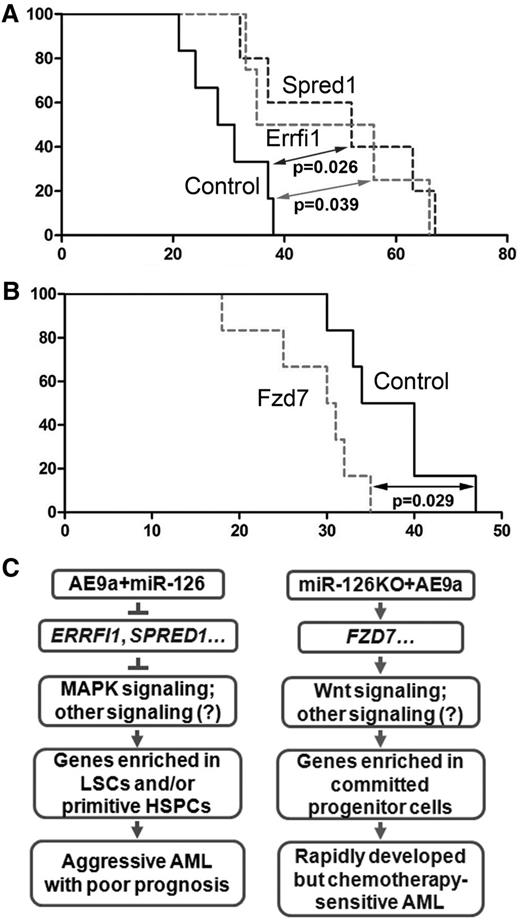
The function of the miR-126 targets Errfi1, Spred1, and Fzd7 in leukemogenesis was further investigated in vivo. Our BMT assays showed that forced expression of Errfi1 and Spred1 significantly delayed the progression of AE9a+miR-126 AML (Figure 7A) and was associated with a decrease in c-Kit+ cell population in the BM (supplemental Figure 14A). In contrast, forced expression of Fzd7 significantly accelerated the progression of AE9a AML (Figure 7B) and was associated with an increase in c-Kit+ cell population in the BM (supplemental Figure 14B). The increased expression of each of these miR-126 targets was confirmed by quantitative real-time polymerase chain reaction (supplemental Figure 14C).
The in vivo function of ERRFI1, SPRED1, and FZD7 in AE9a-mediated leukemogenesis. (A) Kaplan-Meier curves are shown for 3 groups of transplanted mice: MSCV-PIG (Control, n = 5), MSCV-PIG-Errfi1 (Errfi1, n = 4), and MSCV-PIG-Spred1 (Spred1, n = 5). The leukemic BM cells collected from tertiary miR-126+AE9a BMT recipients were used as the donor cells. The P values were calculated by log-rank test. (B) Kaplan-Meier curves are shown for 2 groups of transplanted mice: MSCV-PIG (Control, n = 5) and MSCV-PIG-Fzd7 (Fzd7, n = 6). The leukemia BM cells collected from tertiary AE9a BMT recipients were used as the donor cells. The P value was calculated by log-rank test. (C) Schematic model of target genes and signaling pathways activated in AE9a+miR-126 cells (left) and miR-126KO+AE9a cells (right).
The in vivo function of ERRFI1, SPRED1, and FZD7 in AE9a-mediated leukemogenesis. (A) Kaplan-Meier curves are shown for 3 groups of transplanted mice: MSCV-PIG (Control, n = 5), MSCV-PIG-Errfi1 (Errfi1, n = 4), and MSCV-PIG-Spred1 (Spred1, n = 5). The leukemic BM cells collected from tertiary miR-126+AE9a BMT recipients were used as the donor cells. The P values were calculated by log-rank test. (B) Kaplan-Meier curves are shown for 2 groups of transplanted mice: MSCV-PIG (Control, n = 5) and MSCV-PIG-Fzd7 (Fzd7, n = 6). The leukemia BM cells collected from tertiary AE9a BMT recipients were used as the donor cells. The P value was calculated by log-rank test. (C) Schematic model of target genes and signaling pathways activated in AE9a+miR-126 cells (left) and miR-126KO+AE9a cells (right).
Collectively, highly expressed miR-126 likely substantially represses expression of a set of tumor-suppressor targets, including ERRFI1 and SPRED1, which in turn activates MAPK signaling and probably other pathways, leading to the upregulation of genes that are usually highly expressed in primitive HSPCs and/or LSCs and eventually resulting in the development of a very aggressive form of t(8;21) AML (Figure 7C, left panel). In contrast, knockout of miR-126 induces expression of FZD7 (and likely other targets), which in turn activates the Wnt signaling and probably other pathways, leading to a rapidly developed but chemotherapy-sensitive AML (Figure 7C, right panel).
Discussion
Although miR-126 is overexpressed in both t(8;21) and inv(16) AML,20 here we show that it is expressed at a particularly higher level in t(8;21) AML, where its increased expression is associated with a poor prognosis. Although KIT mutations and internal tandem duplications of FLT3 are predictors of unfavorable outcome of patients with CBF leukemia, especially t(8;21) AML,44,59-62 the unfavorable impact of miR-126 on prognosis is likely not related to these mutations because they do not regulate miR-126 expression.
Because forced expression of miR-126 enhances t(8;21) fusion–mediated cell transformation in vitro,20 we originally expected that overexpression and knockout of miR-126 should accelerate and inhibit, respectively, t(8;21) fusion–mediated leukemogenesis in vivo. Surprisingly, however, we found that both gain- and loss-of-function of miR-126 substantially promoted leukemogenesis. Notably, in primary BMT, miR-126 knockout exhibited an even more potent effect than miR-126 overexpression on promoting t(8;21) fusion–mediated leukemogenesis. Nevertheless, miR-126 overexpression exhibited a more significant enhancement effect than miR-126 knockout on long-term self-renewal and progression of LSCs/LICs, as evidenced by a faster progression of leukemia in tertiary and quaternary BMT, as well as a much higher frequency of LSCs/LICs in leukemic BM cells from secondary BMT recipients. Notably, miR-126 knockout substantially enhanced responsiveness of leukemia cells to standard chemotherapy.
Our mechanistic studies show that miR-126 overexpression activates genes that are highly expressed in LSCs/LICs and/or primitive HSPCs, whereas miR-126 knockout activates genes that are highly expressed in committed, more differentiated hematopoietic cells. Thus, it is possible that miR-126 overexpression mainly targets primitive HSPCs and that miR-126 knockout mainly targets committed, more differentiated progenitor cells. As a result, the original population of miR-126KO+AE9a–targeted cells is much larger than the population of AE9a+miR-126–targeted cells within the BM donor cells. Therefore, miR-126KO+AE9a causes leukemia significantly faster than AE9a+miR-126 in primary BMT. Nonetheless, when AE9a+miR-126 LSCs/LICs gradually become enriched, they surpass miR-126KO+AE9a cells (mostly transformed committed progenitors) in leukemia progression in later passages of serial BMT. In addition, consistent with the observations from us and others that depleted expression of miR-126 promotes apoptosis, whereas miR-126 overexpression inhibits apoptosis,20,24 the apoptosis gene set in the Kyoto Encyclopedia of Genes and Genomes Pathway Database is significantly enriched in genes that are highly expressed in miR-126KO+AE9a AML cells and genes that are downregulated in AE9a+miR-126 AML cells. Interestingly, both cell cycle and DNA replication gene sets exhibit similar enrichment patterns to the apoptosis gene set in miR-126KO+AE9a and AE9a+miR-126 AML cells, suggesting that miR-126KO+AE9a cells may be more proliferative than AE9a+miR-126 cells. The property of miR-126KO+AE9a AML cells of a higher degree of differentiation and proliferation compared to AE9a+miR-126 AML cells likely contributes to their higher sensitivity to standard chemotherapy, because standard chemotherapy is more effective in more mature and proliferative AML cells compared to primitive AML cells, which are characterized by a lack of cell cycle activity.63
Furthermore, our data suggest that ERRFI1 and SPRED1 are 2 critical tumor-suppressor targets of miR-126, and their repression contributes to the accelerated progression of t(8;21) AML when miR-126 is overexpressed, likely through activating the MAPK pathway. On the other hand, FZD7 appears to be a major oncogenic target of miR-126 that contributes to the enhanced leukemogenesis mediated by t(8;21) fusion proteins when miR-126 expression is depleted, likely through activating the Wnt signaling.
In summary, we show in this study that both gain- and loss-of-function of a single miRNA (eg, miR-126) can promote tumorigenesis and enhance long-term self-renewal/progression of cancer stem cells through targeting distinct gene signaling and thereby associating with different biological consequences. Our data suggest that although miRNAs serve as “fine tuners” of expression of their target genes, their own expression in normal cells must be finely tuned to prevent tumorigenesis. Collectively, our work provides novel insight into the complexity and biological importance of miRNA regulation in tumorigenesis and drug resistance.
Our discoveries also suggest that miR-126 represents a unique and promising therapeutic target. Indeed, although t(8;21) AML is associated with a relatively favorable prognosis, only ∼60% of the patients are “cured” using contemporary treatment.42,64 Moreover, current treatment frequently involves intensive postremission treatment with multiple cycles of high-dose Ara-C that impairs the quality of life of the patients.65 Thus, a combination of miR-126 knockdown with standard chemotherapy could provide an improved therapeutic strategy with a higher efficacy and minimal side effects to cure t(8;21) AML, especially for the present therapy-resistant subgroup, and to improve the quality of life of t(8;21) patients.
The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE65939).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their late mentor and colleague, Dr Janet D. Rowley, for her long-term support; Ms Jennifer Strong for manuscript editing; and Drs Dong-Er Zhang, Gregory Hannon, and Lin He for providing retroviral constructs. The authors also appreciate the constructive suggestions from the anonymous reviewers.
This work was supported in part by an American Cancer Society Research Scholar grant (J.C.); a Leukemia & Lymphoma Society Special Fellowship (Z.L.); National Institutes of Health National Cancer Institute grants R01 CA178454, R01 CA182528, and R01 CA127277 (J.C.); a Leukemia & Lymphoma Society Translational Research Grant (J.C.); an American Cancer Society Illinois Division Research Scholar grant (Z.L.); Gabrielle’s Angel Foundation for Cancer Research (J.C., Z.L., X.J., and H.H.); and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (A.G.E. and P.P.L.).
Authorship
Contribution: Z.L. and J.C. conceived the project and designed the research; Z.L., P.C., R.S., Y.L., C.H., Y.W., S.A., M.H., S.G., Z.Z., A.G.E., S.L., H.W., H.H., M.B.N., X.J., and J.C. performed the experiments and/or data analyses; Z.L., A.G.E., H.H., S.W., E.N.O., R.A.L., M.M.L.B., J.Z., X.J., M.W., J.J., P.P.L., and J.C. contributed reagents/analytic tools and/or grant support; and Z.L. and J.C. wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Z.L. is Department of Human Genetics, University of Chicago, Chicago, IL.
Correspondence: Jianjun Chen, University of Cincinnati College of Medicine, 3125 Eden Ave, Room 3316, Cincinnati, OH 45219; e-mail: chen3jj@uc.edu; and Zejuan Li, University of Chicago, 5841 S. Maryland Ave, G705C, Chicago, IL 60637; e-mail: zjli@uchicago.edu.
References
Author notes
Z.L., P.C., and R.S. contributed equally to this study.

![Figure 5. Comparison of expression levels between Pri HSPCs and CPs for the genes dysregulated in AE9a+miR-126 and miR-126KO+AE9a leukemic cells. Expression patterns between Pri HSPCs and CPs from the GSE24006 data set48 for the 57 AE9a+miR-126_High genes (A) and the 88 miR-126KO+AE9a_High genes (B) are shown. Expression data were mean centered, and the relative value for each sample is represented by a color, with red and green representing a high and low expression, respectively (scale is shown). (C) Within each gene set (ie, AE9a+miR-126_High, AE9a+miR-126_Low, miR-126KO+AE9a_High, or miR-126KO+AE9a_Low), the proportion of genes that are expressed at a significantly higher level in Pri HSPCs than in CPs (Pri HSPCs_High; blue), a significantly higher level in CPs than in Pri HSPCs (CPs_High; red), or a comparable level between Pri HSPCs and CPs (nonsignificant [NS]; green) is shown. Gene expression levels were compared between Pri HSPCs (including BM_HSC and BM_MPP samples) and CPs (including BM_CMP, BM_GMP, and BM_MEP samples) in the GSE24006 data set48 through significance analysis of microarrays,47 with a q value <0.05 and a false discovery rate <0.001 as criteria for statistical significance. CB_HSC and CB_MPP samples were not included in the significance analysis of microarrays due to their tissue differences from other BM samples. CB, core blood; CMP, common myeloid progenitor; CPs, committed progenitors; GMP, granulocyte/monocyte progenitor; HSC, hematopoietic stem cell; MEP, megakaryocyte/erythrocyte progenitor; MPP, multipotent progenitor; Pri HSPCs, primitive HSPCs.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/17/10.1182_blood-2015-04-639062/4/m_2005f5.jpeg?Expires=1769133423&Signature=Upwto0gFGV9jQKdgsRQhyd9Mu1mU0xFKUKNCrR08yODUMxEmzDNRfjMMpD8Oorhgkp2zcySVM~DSGyqzq6~UshNcT6d-FKAn1jF2KhX9j6JxPj16BxD0eVNpj30wQQh0aDiU3HSBCbMSWqFIX4MueHNU6L5KsoDi7DpDlD1dMc1jiSpWNKd3e7NRoBYh30JjYEiDmLOvAxKtE1ibkuPILdznUHU0G9TqQTIoYnYHgpqum6wdQ3-V72pjS1rW~AymbYANLJpRujqONYm-eXFJdLG3COnWokWd3rMz5OoUDJ1i-JpIkFrj0l8uY2GAhrYuYk~ug5e-GNd0F9-DxAKh9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)