Abstract
The critical role of Janus kinase-2 (JAK2) in regulation of myelopoiesis was established 2 decades ago, but identification of mutations in the pseudokinase domain of JAK2 in myeloproliferative neoplasms (MPNs) and in other hematologic malignancies highlighted the role of JAK2 in human disease. These findings have revolutionized the diagnostics of MPNs and led to development of novel JAK2 therapeutics. However, the molecular mechanisms by which mutations in the pseudokinase domain lead to hyperactivation of JAK2 and clinical disease have been unclear. Here, we describe recent advances in the molecular characterization of the JAK2 pseudokinase domain and how pathogenic mutations lead to constitutive activation of JAK2.
Introduction
Myeloproliferative neoplasms (MPNs) are clonal malignancies characterized by overproduction of one or more differentiated myeloid lineages. In 1951, Dameshek recognized the relationship between chronic myelogenous leukemia, polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), and thus characterized the MPN disease entity.1 The molecular basis for the most common MPNs (ie, PV, ET, and PMF) remained unknown until 2005, when 4 groups using different strategies all identified a single somatic mutation, V617F, in the pseudokinase domain of Janus kinase-2 (JAK2), a cytoplasmic tyrosine kinase.2-5 JAK2 V617F was found to be constitutively active and lead to MPNs in several mouse models.6
Identification of the JAK2 mutation V617F (exon 14) stimulated research efforts on JAKs in hematologic diseases and has led to a rather complete picture of genetic alterations in MPN. In PV, JAK2 V617F is found in >95% of patients and JAK2 exon 12 mutations in ∼4%. In ET and PMF, ∼60% of patients harbor JAK2 V617F, 20% to 25% calreticulin mutations, ∼5% thrombopoietin receptor (MPL) mutations, and 5% to 10% of patients lack mutations in any of these genes.7-9 These mutations in JAK2, MPL, and calreticulin are driver mutations, and they all activate the JAK2 pathway, but additional recurrent somatic mutations in several genes (TET2, ASXL1, DNMT3A, CBL, LNK, IDH1/2, IKF1, EZH2, TP53, SRSF2), encoding transcriptional and epigenetic regulators and signaling proteins, occur in MPNs.10 These additional mutations modulate disease progression and can also occur as a primary mutation, but it is now convincingly demonstrated that MPNs can be initiated from a single JAK2 V617F hematopoietic stem cell.11
JAK mutations have also emerged in other hematologic diseases, and the majority of the pathogenic mutations in JAK2 (also in JAK1 and JAK3) localize in or near the pseudokinase domain.12 However, the molecular mechanisms by which different mutations in the pseudokinase domain lead to constitutive tyrosine kinase activity and human disease have remained elusive.
Regulation of JAK2
The JAK tyrosine kinases (JAK1-3, TYK2) mediate signaling of approximately 60 cytokines and hormones.13 The biological functions of JAKs are determined by their interaction with specific cytokine receptors. JAK2 has a critical role in signaling by hematopoietic cytokines involved in myelopoiesis, and it associates via N-terminal band 4.1, ezrin, radixin, moesin (FERM), and Src homology-2 (SH2)-like domains to the cytoplasmic region of erythropoietin (Epo), thrombopoietin, granulocyte colony stimulating factor, IL-3, and granulocyte macrophage colony stimulating factor receptors.14-16 Ligand-induced receptor rearrangement allows trans-phosphorylation of the tyrosine kinase domain, resulting in stimulation of kinase activity and progression of signal transduction though signal transducers and activators of transcriptions (STATs), Ras–mitogen-activated protein kinase (MAPK), and phosphoinositide 3-kinase pathways, which regulate proliferation, apoptosis, and differentiation in myeloid cells.
Because JAKs are the triggering kinases for cytokine receptors, their activities are tightly regulated on several levels. Cytokine stimulation induces phosphorylation of JAK2 at multiple residues, which regulate its activity either positively or negatively through still largely undefined mechanisms. Prior to stimulation, JAK2 is constitutively phosphorylated only on Ser523, which negatively regulates basal (no cytokine) JAK2 activity.17,18 Upon cytokine stimulation, Tyr1007 and Tyr1008 in the activation loop of the tyrosine kinase domain are rapidly autophosphorylated in trans, which upregulates catalytic activity and facilitates the phosphorylation of additional sites, leading to full catalytic activation.19 Finally, the extent of JAK2 activity is limited by phosphorylation of inhibitory sites, including Tyr570,20 concomitant with dephosphorylation of the stimulatory sites by protein tyrosine phosphatases (SHP1, SHP2, PTP1B, TCPTP, CD45). In addition, SOCS1 and SOCS3 proteins and the SH2B family of proteins (including LNK, which is mutated in a subset of MPNs) provide important regulation of JAK2 activity (reviewed in Vainchaker et al21 and Babon et al22 ).
In cytoplasmic Src and Abl tyrosine kinases, intramolecular interactions mediated by the SH2 and SH3 domains serve important regulatory functions. JAKs lack an SH3 domain, and their SH2-like domain does not bind phosphotyrosine. However, the kinase-like or pseudokinase domain (JAK homology-2 [JH2]), just upstream of the tyrosine kinase domain (JH1), is a critical regulatory domain (Figure 1A). The first evidence that JH2 plays a negative regulatory role in JAK2 signaling came from the gain-of-function mutation tumorous lethal (tuml) in the Drosophila JAK, hopscotch.23 A positive regulatory role for JH2 was indicated by mutations in JAK3 JH2 that are loss-of-function and cause severe combined immunodeficiency.24,25 Similarly, mutations in TYK2 JH2 were found to impair the activity of the kinase domain.26 Analysis of JAK2 showed that deletion of JH2 increases basal activity, but abrogates cytokine-induced signaling.27-29 Taken together, these results indicate that JH2 has a dual regulatory function: maintain a low basal activity of JAK in the absence of cytokines and facilitate JAK activation upon cytokine binding to the receptor.
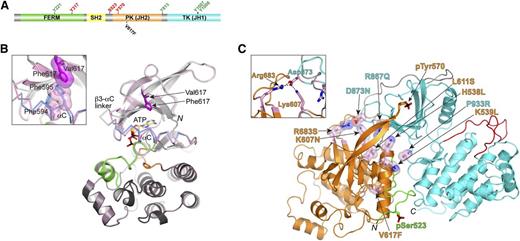
JAK2 domain organization and structural data for JH2 pseudokinase domain. (A) Domain organization of JAK2 shown to linear scale (human, 1132 residues). Select residues mapped as positive- and negative-regulatory phosphorylation sites are labeled in green and red, respectively. The position of the most prevalent MPN mutation, V617F, is also indicated. (B) Crystal structure of JAK2 JH2, wild-type and V617F.33 For wild-type JH2, the N lobe is colored light gray and the C lobe is dark gray. The C helix (αC) is colored blue, the activation loop is colored green, and the catalytic loop is colored orange. The structure of JH2 V617F is superimposed on the wild-type structure and is colored pink throughout. For αC and the preceding linker (β3-αC linker), where the differences between the 2 structures are found, Cα traces are shown. The positions of Val617 (dark gray) and Phe617 (magenta) are indicated. N, N-terminus (residue 536). The C-terminus is not in view. The inset shows a magnified area of the region near Phe617, in particular, the π-stacking interactions between Phe617, Phe594 (αC), and Phe595 (αC) in V617F. The side chains of Phe594 and Phe595 in wild-type JH2 are colored blue. (C) The autoinhibitory pose of JAK2 JH2-JH1 that was derived from MD simulations.39 JH2 is colored orange, JH1 is colored cyan, the SH2-JH2 linker is colored green, and the JH2-JH1 linker is colored gray. Select MPN or leukemia mutations are shown in stick-and-sphere representation (side chains) and colored pink (carbon atoms). Phosphorylated Ser523 and Tyr570 are shown in stick representation. Red, oxygen atoms; blue, nitrogen atoms; yellow, sulfur atoms; black, phosphorus atoms. N, N-terminus (residue 520); C, C-terminus (residue 1132). The inset shows a magnified area of the region near Arg683 (JH2) and Asp873 (JH1). Salt bridges are indicated by dashed black lines. FERM, N-terminal band 4.1, ezrin, radixin, moesin; PK, pseudokinase; TK, tyrosine kinase.
JAK2 domain organization and structural data for JH2 pseudokinase domain. (A) Domain organization of JAK2 shown to linear scale (human, 1132 residues). Select residues mapped as positive- and negative-regulatory phosphorylation sites are labeled in green and red, respectively. The position of the most prevalent MPN mutation, V617F, is also indicated. (B) Crystal structure of JAK2 JH2, wild-type and V617F.33 For wild-type JH2, the N lobe is colored light gray and the C lobe is dark gray. The C helix (αC) is colored blue, the activation loop is colored green, and the catalytic loop is colored orange. The structure of JH2 V617F is superimposed on the wild-type structure and is colored pink throughout. For αC and the preceding linker (β3-αC linker), where the differences between the 2 structures are found, Cα traces are shown. The positions of Val617 (dark gray) and Phe617 (magenta) are indicated. N, N-terminus (residue 536). The C-terminus is not in view. The inset shows a magnified area of the region near Phe617, in particular, the π-stacking interactions between Phe617, Phe594 (αC), and Phe595 (αC) in V617F. The side chains of Phe594 and Phe595 in wild-type JH2 are colored blue. (C) The autoinhibitory pose of JAK2 JH2-JH1 that was derived from MD simulations.39 JH2 is colored orange, JH1 is colored cyan, the SH2-JH2 linker is colored green, and the JH2-JH1 linker is colored gray. Select MPN or leukemia mutations are shown in stick-and-sphere representation (side chains) and colored pink (carbon atoms). Phosphorylated Ser523 and Tyr570 are shown in stick representation. Red, oxygen atoms; blue, nitrogen atoms; yellow, sulfur atoms; black, phosphorus atoms. N, N-terminus (residue 520); C, C-terminus (residue 1132). The inset shows a magnified area of the region near Arg683 (JH2) and Asp873 (JH1). Salt bridges are indicated by dashed black lines. FERM, N-terminal band 4.1, ezrin, radixin, moesin; PK, pseudokinase; TK, tyrosine kinase.
JAK2 mutations in hematological diseases
JAK2 is found in oncogenic fusion proteins in rare cases of lymphoid and myeloid leukemias, where JH1 alone or JH1 and JH2 are fused with a dimerizing domain derived from TEL, PCM1, BCR, or PAX5 genes.21 However, human diseases are more frequently associated with JAK2 point mutations, and JH2 has been shown to be a mutational hotspot. V617F is the predominant (∼80%) somatic mutation in MPNs, and a homologous constitutively active mutation in JAK1, V658F, is found in T-acute lymphoblastic leukemia.30,31 How a single V617F mutation can lead to 3 different MPNs is an intriguing question and appears to be controlled by several factors. The hyperactive phenotype of JAK2-V617F is dependent on the expression of type I cytokine receptors (EpoR, MPL, or G-CSFR), and the disease phenotype is also influenced by the intensity of JAK2-V617F signaling (PMF greater than ET greater than PV) and by the predominant STAT activation signature (reviewed in Vainchaker et al21 ).
JAK2-V617F results from a single nucleotide change, which likely contributes to its high incidence, but over 30 different point mutations, small deletions, or insertions in JH2 of JAK2 have been shown to cause or be linked to hematologic diseases.12 The mutations in JH2 are concentrated in 3 regions encoded by exon 14 (V617F, majority of MPNs), exon 16 (B-cell leukemias), and in the linker between SH2 and JH2 domain encoded by exon 12 (found exclusively in 4% of PV cases). Disease-associated mutations have also been identified in the tyrosine kinase domain (JH1), as reviewed in Haan et al12 and Vainchaker et al,21 but these are far less common. These findings demonstrate the critical regulatory function of JH2, but at the same time, raise the question of how different mutations can lead to similar hyperactive phenotypes.
Characteristics of JH2 and JH2 V617F
The pseudokinase designation for JAK2 JH2 was based primarily on a sequence comparison with canonical protein kinases, which showed that several conserved catalytic residues were substituted, particularly aspartic acid by asparagine (Asn673) in the catalytic loop.14 Other substitutions include alanine (Ala597) for glutamic acid in α-helix C (αC) in the N lobe, proline (Pro700) instead of phenylalanine in the DFG motif at the start of the activation loop, and a threonine (Thr557) instead of glycine in the nucleotide binding loop. Recent biochemical and structural studies have provided new insights into JH2 function. A major development was the demonstration that JAK2 JH2 binds ATP and possesses a low level of catalytic activity, phosphorylating Ser523 in the SH2-JH2 linker and Tyr570 in JH2 itself,32 both of which function as negative regulatory sites for JAK2 activity.17,18,20
Despite the sequence variations described above, the crystal structure determination of JAK2 JH2 showed that JH2 adopts a prototypical protein kinase fold33 (Figure 1B). JH2 binds adenosine (ATP) with micromolar affinity (tighter than canonical protein kinases), but with a noncanonical binding mode. The major differences are the presence of one (not 2) Mg2+ ion and the role of the aspartic acid (Asp699) of the DPG sequence, which forms a salt bridge with the β3 lysine (Lys581). In canonical protein kinases, this aspartate coordinates 2 Mg2+ ions, and the β3 lysine is salt-bridged with a glutamic acid from αC.
The structures of JH2 of JAK134 and TYK2 (PDB code 3ZON and Lupardus et al35 ) have also been determined and are highly similar to that of JAK2. These 3 JAK JH2s bind ATP (Murphy et al36 and unpublished data, O.S.), and whether JAK3 JH2 binds ATP is not known, but only JAK2 JH2 has been shown to have catalytic activity. Because JAK2 is the only JAK to associate with predimerized type I cytokine receptors (eg, EpoR), one plausible explanation is that an additional negative regulatory layer (phosphorylation of Ser523 and Tyr570; see below) is required to keep basal activity low.
The crystal structure of the pathogenic JAK2 JH2 mutant V617F (situated in the β4-β5 loop) is highly similar to that of wild-type JH2, but structural perturbations are apparent in αC, an important regulatory element in protein kinases, and in the nearby (spatially) β3-αC and β4-β5 loops.33 In the wild-type JH2 structure, αC contains a kink in the middle, whereas in the V617F structure, π-stacking interactions involving Phe617 (V617F), Phe594, and Phe595 in αC result in straightening of the helix. Molecular dynamics (MD) simulations also indicated that αC is stabilized in V617F,33 and the functional significance of the π-stacking interactions is supported by mutagenesis data, in which substitution of either Phe594 or Phe595 with alanine suppressed the hyperactivity of V617F while retaining cytokine-inducibility.33,37,38 However, the structural differences between wild-type JAK2 JH2 and V617F do not provide a ready explanation for the mechanism of pathogenic activation by V617F.
JH2-JH1 autoinhibitory interaction and hyperactivity of JAK2 mutations
In collaboration with Yibing Shan at D.E. Shaw Research, we used unbiased, long time-scale MD simulations to derive a molecular model for the interaction between JH2 and JH1 of JAK2.39 About the same time, the crystal structure of TYK2 JH2-JH1 was reported.35 The model for JAK2 and the crystal structure for TYK2 JH2-JH1 are in striking agreement and reveal an extensive interface between the 2 domains. The N lobe of JH2 contacts the N lobe and kinase hinge region of JH1, and the SH2-JH2 linker bridges the 2 domains (Figure 1C). Importantly, nearly all of the pathogenic mutations map to the JH2-JH1 interface, including the exon 12 mutations (eg, K539L), exon 16 mutations (eg, R683G/S), and mutations in the β2-β3 loop of JH1 (eg, D873N)12 (Figure 1C).
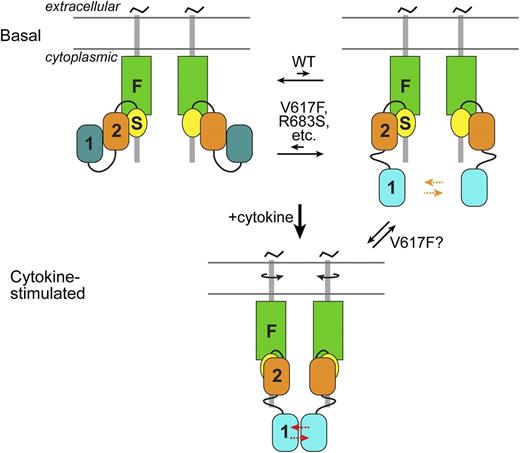
The interaction between JH2 and JH1 is thought to be inhibitory on JH1 through 2 mechanisms: sequestration of JH1 and stabilization of an inactive state of JH1 (Figure 2). For activation-loop trans-phosphorylation of JH1 to occur (on Tyr1007-Tyr1008), which is the key activating event, the 2 JH1s in the receptor JAK2 dimer must access one another. The interaction with JH2 would limit the “comingling” of JH1s in the basal state on predimerized type I receptors. Furthermore, the MD simulations suggest that the interaction with JH2 hinders formation of the critical (for catalytic activity) salt bridge in JH1 between the β3 lysine (Lys882) and the αC glutamic acid (Glu898).39
Schematic of JAK2 activation. (Top) In the basal state, 2 JAK2 molecules associate via their N-terminal band 4.1, ezrin, radixin, moesin (FERM or F) and SH2 (S) domains with predimerized type I cytokine receptors. The autoinhibitory interaction between JH2 (2) and JH1 (1) sequesters JH1 from each other and stabilizes an inactive state of JH1 (dark cyan) (left). This is in equilibrium with a state (right) in which JH1 is disengaged from JH2 (JH1 cyan, higher activity), which increases the probability of trans-phosphorylation (orange arrows) on the activation loop of JH1. By destabilizing the autoinhibitory interaction, pathogenic mutations, such as V617F and R683S, shift the equilibrium to the partially active state. (Bottom) Cytokine binding to the extracellular region of the cytokine receptors induces a structural rearrangement in the cytoplasmic region (possibly through rotation of the transmembrane helices), which greatly facilitates (red arrows) JH1 trans-phosphorylation. JH2 is also necessary for cytokine-induced JAK2 activation, but the molecular interaction(s) responsible for this is not known (and not shown). As indicated, in addition to destabilizing the autoinhibitory interaction, V617F might promote the JH2-mediated positive interaction normally induced by cytokine (or possibly promote an interaction distinct from the cytokine-mediated one).
Schematic of JAK2 activation. (Top) In the basal state, 2 JAK2 molecules associate via their N-terminal band 4.1, ezrin, radixin, moesin (FERM or F) and SH2 (S) domains with predimerized type I cytokine receptors. The autoinhibitory interaction between JH2 (2) and JH1 (1) sequesters JH1 from each other and stabilizes an inactive state of JH1 (dark cyan) (left). This is in equilibrium with a state (right) in which JH1 is disengaged from JH2 (JH1 cyan, higher activity), which increases the probability of trans-phosphorylation (orange arrows) on the activation loop of JH1. By destabilizing the autoinhibitory interaction, pathogenic mutations, such as V617F and R683S, shift the equilibrium to the partially active state. (Bottom) Cytokine binding to the extracellular region of the cytokine receptors induces a structural rearrangement in the cytoplasmic region (possibly through rotation of the transmembrane helices), which greatly facilitates (red arrows) JH1 trans-phosphorylation. JH2 is also necessary for cytokine-induced JAK2 activation, but the molecular interaction(s) responsible for this is not known (and not shown). As indicated, in addition to destabilizing the autoinhibitory interaction, V617F might promote the JH2-mediated positive interaction normally induced by cytokine (or possibly promote an interaction distinct from the cytokine-mediated one).
As outlined above, nearly all of the known gain-of-function, disease-causing mutations in JAK2 JH2 and JH1 are localized in or near the JH2-JH1 interface. One illustrative example is the interacting pair of residues Arg683 in JH2 (β7) and Asp873 in JH1 (β2-β3 loop), which form a salt bridge, both in the JAK2 model and in the TYK2 crystal structure (Arg744 and Asp921) (Figure 1C). Mutations in both of these residues (R683G/S and D873N) have been linked to B-ALL.12,21 Loss of this salt bridge is predicted to destabilize the JH2-JH1 autoinhibitory interaction, leading to facile trans-phosphorylation of JH1 (Figure 2). The MD-derived model also predicts that pSer523 and pTyr570, the former phosphorylated by JAK2 JH2 and the latter by JH2 or JH1 (postactivation), interact with specific basic residues in JH1 and JH2 to fortify the autoinhibitory interaction39 (Figure 1C).
In the model of JAK2 JH2-JH1, the predominant MPN mutation, V617F (exon 14), does not interact directly with residues in JH1, but instead makes contacts with the SH2-JH2 linker. MD simulations of JH2-JH1 V617F indicate that V617F destabilizes the position of the SH2-JH2 linker, which in turn would destabilize the JH2-JH1 interaction.39 However, the high constitutive activity caused by V617F suggests an additional mechanism: hyperstabilization of the positive regulatory interaction mediated by JH2, but the molecular nature of this interaction is currently unknown (Figure 2).
Conclusions and future perspectives
Recent structural studies36,39 have provided major insights into the mechanisms by which pathogenic mutations lead to constitutive JAK2 activation. In the unstimulated basal state, the function of JH2 is to prevent JH1 trans-phosphorylation, which is achieved through the JH2-JH1 autoinhibitory interaction described above. Most of the pathogenic JAK2 mutations are localized in the interdomain interface and are predicted to disrupt the autoinhibitory interaction, resulting in JAK2 hyperactivation. However, there are still important questions to be answered. For example, the classical view of cytokine receptor activation is that cytokine binding brings the JAK2 molecules into proximity to facilitate JH1 trans-phosphorylation. This view was recently challenged by a study that indicates that the cytoplasmic regions of growth hormone receptors are juxtaposed in the basal state and separate upon ligand binding.40 In this model, the JH2-JH1 autoinhibitory interaction would occur in trans, between 2 JAK2 molecules. The described JH2-JH1 interaction models are compatible with either a cis or trans interaction. Another important issue to resolve is the molecular characterization of the JH2-mediated positive regulatory interaction, as this probably also plays an important role in V617F hyperactivation. A detailed understanding of the pathogenic activation of JAK2 may afford an opportunity to develop novel small-molecule therapeutics for MPNs, which would act to inhibit the basal hyperactivity of V617F, but allow normal cytokine stimulation of wild-type JAK2. Although a difficult challenge, several regions of JAK2 JH2, including the ATP-binding pocket41 and αC,33,37-39 are potential targets for such compounds.
Acknowledgments
This work was supported by grants from the Medical Research Council of Academy of Finland, Sigrid Juselius Foundation, Medical Research Fund of Tampere University Hospital, Novo Nordisk Foundation, Finnish Cancer Foundation, Tampere Tuberculosis Foundation, and the National Institutes of Health National Institute of Allergy and Infectious Diseases (R21 AI095808).
Authorship
Contribution: O.S. and S.R.H. wrote the paper and approved the final version.
Conflict-of-interest disclosure: O.S. and S.R.H. have a pending patent application on the use of the JAK2 JH2 crystal structure for therapeutic targeting.
Correspondence: Olli Silvennoinen, School of Medicine, University of Tampere, Biokatu 8, FI-33014 Tampere, Finland; e-mail: olli.silvennoinen@uta.fi; and Stevan Hubbard, Kimmel Center for Biology and Medicine at the Skirball Institute and Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, 540 First Ave, New York, NY 10016; e-mail: stevan.hubbard@med.nyu.edu.