Key Points
ASXL2 was mutated in 22.7% (25/110) of adult and pediatric t(8;21)/RUNX1-RUNX1T1 acute myeloid leukemia patients.
ASXL2 mutations are mutually exclusive with ASXL1 mutations and occur in t(8;21) but not inv(16)/t(16;16) or RUNX1-mutant AML.
Abstract
Acute myeloid leukemia (AML) with t(8;21) (q22;q22) is considered to have favorable risk; however, nearly half of t(8;21) patients are not cured, and recent studies have highlighted remarkable genetic heterogeneity in this subset of AML. Here we identify somatic mutations in additional sex combs-like 2 (ASXL2) in 22.7% (25/110) of patients with t(8;21), but not in patients with inv(16)/t(16;16) (0/60) or RUNX1-mutated AML (0/26). ASXL2 mutations were similarly frequent in adults and children t(8;21) and were mutually exclusive with ASXL1 mutations. Although overall survival was similar between ASXL1 and ASXL2 mutant t(8;21) AML patients and their wild-type counterparts, patients with ASXL1 or ASXL2 mutations had a cumulative incidence of relapse of 54.6% and 36.0%, respectively, compared with 25% in ASXL1/2 wild-type counterparts (P = .226). These results identify a high-frequency mutation in t(8;21) AML and identify the need for future studies to investigate the clinical and biological relevance of ASXL2 mutations in this unique subset of AML.
Introduction
Acute myeloid leukemia (AML) with t(8;21) (q22;q22) is recognized by the World Health Organization1 as a unique subtype of AML within the category of “AML with recurrent genetic abnormalities.” Compared with other cytogenetic subsets of AML, patients with t(8;21) are considered a favorable risk group according to their high remission and survival rates2 . At the same time, nearly half of t(8;21) AML patients are not cured,3,4 and there is a need for markers to identify patients unlikely to respond to current therapies and develop novel therapeutic approaches based on better understanding of the pathophysiology of this subset of AML.
The t(8;21) results in fusion of RUNX1 with RUNX1T1, and considerable experimental evidence reveals that full-length RUNX1-RUNX1T1 is not sufficient to induce leukemic transformation on its own.5 It is therefore posited that additional genetic alterations cooperate with RUNX1-RUNX1T1 translocations to induce overt leukemia. Identification of these additional genetic abnormalities in patients with core-binding factor (CBF) translocations has been helpful in predicting outcome and hopefully serving as novel therapeutic targets for CBF AML. Indeed, more than 50% of t(8;21) AML patients have been shown to have a mutation in KIT, FLT3, N-RAS, or K-RAS,6-10 all of which have been pursued as potential therapeutic targets. Moreover, work by Krauth et al identified that mutations in the polycomb-associated gene ASXL1 occur in 11.5% of t(8;21) adult AML patients and are associated with adverse event-free survival.11 Intriguingly, Huether et al recently identified mutations in additional sex combs-like 2 (ASXL2) in several pediatric leukemia patients.12 Here we identify a high frequency of ASXL2 mutations among CBF AML patients specifically bearing RUNX1-RUNX1T1 translocations and use data from uniformly treated CBF AML adult and pediatric patient populations to identify clinical correlates and mutational cooccurrence of ASXL2 mutations in this unique subset of AML.
Methods
Patients and treatments
One hundred ten t(8;21) and 60 inv(16)/t(16;16) AML patients were included, including 75 t(8;21) and 32 inv(16)/t(16;16) adults from the CBF-2006 trial13 (A Phase 3 Trial of Systematic Versus Response-adapted Timed-Sequential Induction in Patients with Core Binding Factor [CBF] Acute Myeloid Leukemia [AML]; EudraCT 2006 005163-26; ClinicalTrials.gov NCT00428558), as well as 35 t(8;21) and 28 inv(16)/t(16;16) children from the ELAM02 trial (Treating Patients with Childhood Acute Myeloid Leukemia with Interleukin-2; ClinicalTrials.gov NCT00149162). Twenty-six de novo adult AML patients with RUNX1 mutations were also included (from the ALFA-0701 trial14 [A Randomized Study of Gemtuzumab Ozogamicin (GO) with Daunorubicine and Cytarabine in Untreated AML Aged of 50-70 Years Old]). Studies were approved by the Ethics Committee of Nimes University Hospital and by the Institutional Review Board of the French Regulatory Agency and were conducted in accordance with the Declaration of Helsinki protocol. Outcome data were updated as of December 2013, for a median follow-up of 44.8 months.
Mutational and minimal residual disease analysis
Whole-exome sequencing of DNA was performed from pretreatment as well as paired remission bone marrow mononuclear cells from 3 t(8;21) AML patients (supplemental Methods, available on the Blood Web site). All coding exons of ASXL2 were sequenced by next-generation sequencing for all patients at a median depth of 1153× (supplemental Table 1; supplemental Figure 1; supplemental Methods). All additional molecular and cytogenetic analyses are described in the supplemental Methods.
Statistical methods
Details of statistical analysis are located in supplemental Methods.
Results and discussion
Discovery and recurrence of ASXL2 mutations
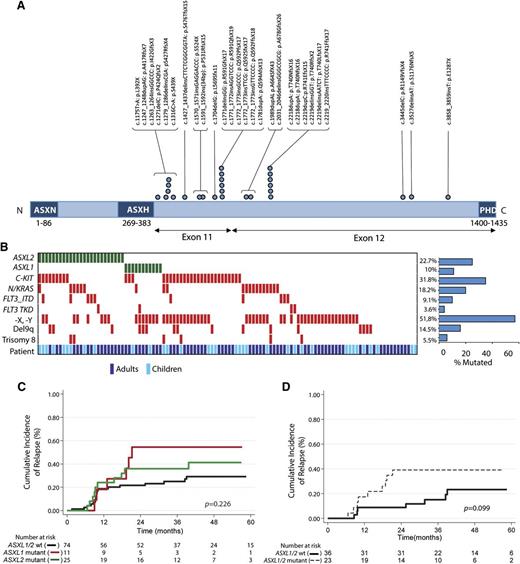
To identify novel mutations cooperating with RUNX1-RUNX1T1 fusions, we performed whole-exome sequencing on 3 diagnostic t(8;21) AML samples paired with a remission sample for each patient. This analysis revealed a somatic ASXL2 p.R741EfsX15 mutation restricted to the diagnosis sample of 1 patient with a variant allele frequency of 39.5% (supplemental Figure 2A) and a somatic ASXL1 p.L823X mutation in a second patient (variant allele frequency, 39%; supplemental Table 2). After validating the mutation in ASXL2 by Sanger sequencing (supplemental Figure 2B), we next performed targeted sequencing of ASXL2 across 110 pediatric and adult t(8;21) patients. Overall, 22.7% of t(8;21) patients (25/110) harbored an ASXL2 mutation (Figure 1A). The majority of ASXL2 mutations (85%) were out-of-frame frameshift mutations (supplemental Table 3). The somatic nature of these mutations in ASXL2 was verified by sequencing DNA from time of presentation and at remission in 6 patients total. Similar to previously described mutations in ASXL1,15 ASXL2 mutations were exclusively heterozygous, with the exception of a single patient with 2 different truncating ASXL2 mutations (supplemental Table 3), and were enriched in the 3′ region of the gene, occurring exclusively in exons 11 and 12. Interestingly, ASXL1 mutations (supplemental Table 3) were mutually exclusive with ASXL2 mutations (Figure 1B). Analysis of mutational cooccurrences revealed a significant cooccurrence of ASXL2 and FLT3-ITD mutations (P = .046), a feature not shared by ASXL1 mutant t(8;21) patients.
ASXL2 mutations are frequent in AML patients bearing the t(8;21) translocation and are associated with a trend for increased risk for relapse. (A) Gene diagram depicting ASXL2 mutations in adult and pediatric patients with t(8;21) AML. (B) Pattern of molecular and cytogenetic lesions in patients with t(8;21) AML. Each column represents 1 of the 110 t(8;21) AML samples sequenced here. Patient demographics (adult vs pediatric sample) are also included. Cumulative incidence of relapse according to enrollment ASXL2 and ASXL1 mutation status in (C) the entire t(8;21) AML cohort and (D) adult patients who achieved more than 3 log-fold reduction in RUNX1-RUNX1T1 transcripts before initiation of second consolidation course.
ASXL2 mutations are frequent in AML patients bearing the t(8;21) translocation and are associated with a trend for increased risk for relapse. (A) Gene diagram depicting ASXL2 mutations in adult and pediatric patients with t(8;21) AML. (B) Pattern of molecular and cytogenetic lesions in patients with t(8;21) AML. Each column represents 1 of the 110 t(8;21) AML samples sequenced here. Patient demographics (adult vs pediatric sample) are also included. Cumulative incidence of relapse according to enrollment ASXL2 and ASXL1 mutation status in (C) the entire t(8;21) AML cohort and (D) adult patients who achieved more than 3 log-fold reduction in RUNX1-RUNX1T1 transcripts before initiation of second consolidation course.
No ASXL1 or ASXL2 mutations were found in 60 inv(16)/t(16;16) AML patients (supplemental Figure 3). Likewise, although ASXL1 mutations were seen in 30.8% of RUNX1-mutated de novo AML patients, 0/26 RUNX1-mutant AML samples had an ASXL2 mutation (supplemental Figure 3).
Implications of ASXL2 mutations
Unlike mutations in ASXL1, which have been associated with advanced age in AML,16 ASXL2 mutations were similarly frequent among pediatric and adult t(8;21) AML patients (9/35 vs 16/75, respectively; P = .6; supplemental Figure 2C). Patients with ASXL2 or ASXL1 mutations had a significantly higher median white blood cell count at presentation than their wild-type counterparts (20.3 vs 15.8 vs 10.7 × 109 cells/μL, respectively; P = .029) (Table 1). Data for adult and pediatric t(8;21) AML patients are reported separately in supplemental Table 4.
Although overall survival was similar between ASXL1 or ASXL2 mutant t(8;21) AML patients and wild-type counterparts (supplemental Figure 4A-C), patients with ASXL1 or ASXL2 mutations had a 3-year cumulative incidence of relapse of 54.6% and 36.0%, respectively, compared with 25% in ASXL1/2 wild-type counterparts (P = .226; Figure 1C and supplemental Figure 4D). Previous analysis of the CBF-2006 trial identified achievement of more than a 3 log-fold reduction in the RUNX1-RUNX1T1 transcript before initiation of second consolidation (minimal residual disease 2 [MRD2]) as the sole factor significantly influencing outcome in multivariate analysis of CBF patients.13 Among adults achieving more than 3 log-fold reduction at MRD2, the 3-year cumulative incidence of relapse was 44.4%; it was 35.7% for ASXL1 and ASXL2 mutated patients, respectively, compared with 15.2% in ASXL1/2 wild-type patients (P = .128; Figure 1D and supplemental Figure 4E).
Huether et al first noted ASXL2 mutations in 7.2% of pediatric CBF AML patients.12 Here we extend those findings and identify that ASXL2 mutations are specifically recurrent in adult as well as pediatric CBF AML patients bearing t(8;21), occurring in nearly 25% of such patients compared with 0% of inv(16)/t(16;16) patients (P = .00005). The fact that ASXL2 mutations were not identified in prior whole-exome/genome sequencing studies in AML17-21 suggests that genomic studies focused on specific clinical and genetic subsets of AML may identify additional novel mutations enriched within defined subsets of AML patients. Moreover, the discovery that t(8;21) AML patients who achieve a more than 3 log-fold reduction at MRD2 but harbor ASXL1 or ASXL2 mutations have a potentially higher risk for relapse suggests the need for further studies addressing the prognostic relevance of ASXL2 mutations in t(8;21).13 The fact that ASXL1 and ASXL2 mutations were mutually exclusive with one another, combined with the multiple unique domains in common between ASXL1 and ASXL2,22 suggests a shared mechanism of transformation for mutations in these 2 genes. Indeed, 32.7% of t(8;21) patients harbor an ASXL1 or ASLX2 mutation, making ASXL gene family alterations among the most common genetic alterations in t(8;21) AML patients. Understanding the functional basis for this frequent cooccurrence of RUNX1-RUNX1T1 translocation and mutations in ASXL1 or ASXL2 may be a critical step in furthering our knowledge of CBF AML pathogenesis and therapy.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Karine Celli-Lebras, Corinne Duguet, and Virginie Bacca for their help with data management; Julie Lejeune for data handling and statistical help; and Lamya Haddaoui (Tumour Bank for the Groupe Ouest Est d'Etude des Leucémies aiguës et Autres Maladies du Sang, Hôpital Cochin, Paris) and Christophe Roumier (Tumour Bank for the Acute Leukemia French Association (ALFA) group, Centre Hospitalier Universitaire de Lille) for handling and storing patient samples. The work of all clinical research assistants of the Groupe Ouest Est d'Etude des Leucémies aiguës et Autres Maladies du Sang and ALFA groups is also acknowledged here. J.-B.M. thanks Stéphane de Botton and Eric Solary (Gustave Roussy Cancer Campus Grand Paris) for their support.
The work was supported by the French National Cancer Institute, the French Ministry of Health (programme hospitalier de recherche clinique, 2006-0213), and a grant from the Association of Laurette Fugain. J.-B.M. is supported by grants from the Fondation de France and Philippe Foundation. O.A.-W. is supported by a National Institutes of Health K08 Clinical Investigator Award (1K08CA160647-01), a US Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (W81XWH-12-1-0041), the Josie Roberston Clinical Investigator Program, and a Damon Runyon Clinical Investigator Award with Support from the Evans Foundation.
Authorship
Contribution: E.J. was the principal investigator of the CBF-2006 study; G.L. is the principal investigator of the Elam02 study; H.D. and C.P created the patient database; N.B. performed statistical analysis; E.J., N.B., J.-B.M, N.I., H.D., G.L., and A.P. enrolled patients in the studies; N.D., O.N., S.G., C.P., C.L., H.L, P.E., A.R., and S.C. performed genetic analysis and analyzed mutational data; M.F. interpreted next-generation sequencing results; and J.-B.M., N.D., N.B., and O.A.-W. wrote the manuscript, which was approved by all the authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Omar Abdel-Wahab, Memorial Sloan-Kettering Cancer Center, Zuckerman 802, 408 East 69th Street, New York, NY 10065; e-mail: abdelwao@mskcc.org; and Eric Jourdan, Hôpital Universitaire Carémeau, Place du Pr. Robert Debré, 30029 Nîmes Cedex 9, France; e-mail: eric.jourdan@chu-nimes.fr.
References
Author notes
J.-B.M., N.D., N.B., O.A.-W., and E.J. contributed equally to this study.