Key Points
Integrating cytogenetic and genomic data in pediatric ALL reveals 2 subgroups with different outcomes independent of other risk factors.
A total of 75% of children on UKALL2003 had a good-risk genetic profile, which predicted an EFS and OS of 94% and 97% at 5 years.
Abstract
Recent genomic studies have provided a refined genetic map of acute lymphoblastic leukemia (ALL) and increased the number of potential prognostic markers. Therefore, we integrated copy-number alteration data from the 8 most commonly deleted genes, subordinately, with established chromosomal abnormalities to derive a 2-tier genetic classification. The classification was developed using 809 ALL97/99 patients and validated using 742 United Kingdom (UK)ALL2003 patients. Good-risk (GR) genetic features included ETV6-RUNX1, high hyperdiploidy, normal copy-number status for all 8 genes, isolated deletions affecting ETV6/PAX5/BTG1, and ETV6 deletions with a single additional deletion of BTG1/PAX5/CDKN2A/B. All other genetic features were classified as poor risk (PR). Three-quarters of UKALL2003 patients had a GR genetic profile and a significantly improved event-free survival (EFS) (94%) compared with patients with a PR genetic profile (79%). This difference was driven by a lower relapse rate (4% vs 17%), was seen across all patient subgroups, and was independent of other risk factors. Even genetic GR patients with minimal residual disease (>0.01%) at day 29 had an EFS in excess of 90%. In conclusion, the integration of genomic and cytogenetic data defines 2 subgroups with distinct responses to treatment and identifies a large subset of children suitable for treatment deintensification.
Introduction
Somatic genetic alterations that initiate and drive carcinogenesis are the hallmarks of cancer. Genomic profiling has revolutionized our understanding of cancer and refined the classification of patients into clinically relevant subgroups.1 This is exemplified in pediatric B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) by significant chromosomal abnormalities that provide important diagnostic and prognostic information that is routinely used in risk stratification for treatment. Patients harboring ETV6-RUNX1 or high hyperdiploidy (51-65 chromosomes) typically have a favorable outcome, while those with t(9;22)(q34;q11.2)/BCR-ABL1, MLL rearrangements, near haploidy (<30 chromosomes), low hypodiploidy (30-39 chromosomes), intrachromosomal amplification of chromosome 21 (iAMP21), or t(17;19)(q23;p13) have an inferior outcome when treated with standard therapy.2 Modern treatment protocols stratify patients on factors such as genetics, age, white cell count (WCC), and treatment response. High-risk (HR) patients receive more intensive chemotherapy, stem cell transplantation, or targeted therapy, whereas low-risk (LR) patients are treated less intensively.
Genomic technologies have identified a plethora of novel copy-number alterations (CNAs) and sequence mutations that typically affect genes involved in lymphoid differentiation, proliferation, cell cycle, and transcription.3 In contrast to chromosomal abnormalities, which are commonly initiating events, these CNAs are usually cooperating aberrations that correlate with specific cytogenetic subtypes.4 Several studies have reported a poor outcome for patients with these aberrations, especially IKZF1 deletions5-8 and P2RY8-CRLF2 fusion,9-12 but a consensus view of how best to incorporate the results into current stratification algorithms has failed to emerge. The major complicating factor is the frequency with which these so-called poor-risk (PR) secondary abnormalities coexist with each other and with other HR genetic subtypes, such as iAMP21, BCR-ABL1, and, particularly, the BCR-ABL1–like (or Philadelphia chromosome–like) subtype.4,13 The BCR-ABL1–like subtype is defined by a gene expression signature that is similar to that observed for BCR-ABL1 cases but occurs in patients lacking this gene fusion. BCR-ABL1–like is mutually exclusive of established chromosomal abnormalities but is enriched for IKZF1 deletions, CDKN2A/B deletions, JAK2 mutations, and CRLF2 rearrangements.4,13 However, to date, only 2 groups have published expression data on this subtype; the Dutch Childhood Oncology Group14,15 described 2 unselected cohorts of patients, whereas the US Children’s Oncology Group reported on an HR cohort of patients.13 Although both groups reported inferior outcomes for their BCR-ABL1–like patients, the key drivers of relapse have yet to be elucidated. Moreover, as it is unclear if the 2 profiles identify the same subset of patients, other groups cannot reliably adopt this approach for risk stratification. Therefore, there remains an urgent clinical need to optimally integrate genomic data into an informative classification system.
In this study, we have adopted a novel approach to overcome this problem by integrating CNA data from 8 key genes/loci with established cytogenetic risk groups to generate a novel overarching genetic risk classification of pediatric BCP-ALL. We have validated this new classification using an independent patient cohort treated on a modern protocol and demonstrated how it interacts with other risk factors.
Patients and methods
Patients were diagnosed with BCP-ALL by standard flow-cytometric criteria and were treated on Medical Research Council (MRC) ALL97/99 (1997–2002) or United Kingdom (UK)ALL2003 (2003–2011) as previously described.16,17 Local ethical committee approval was obtained for ALL97 by individual treatment centers, whereas approval for UKALL2003 was obtained from the Scottish Multi-Centre Research Ethics Committee. Informed consent was given by parents and patients in accordance with the Declaration of Helsinki. In UKALL2003, minimal residual disease (MRD) was evaluated by real-time quantitative polymerase chain reaction (PCR) analysis of immunoglobulin and T-cell receptor gene rearrangements as defined by the European MRD Study Group.18 Patients were classified into 1 of 3 groups based on their MRD results: (1) LR (patients with undetectable MRD at the end of induction [day 29] or patients with low-level MRD [<0.01%] at day 29 and undetectable MRD at the start of interim maintenance); (2) HR (patients with MRD >0.01% at day 29); or (3) indeterminate group (patients with no MRD results due to no or poor sample and patients with persistent low-level disease [<0.01% MRD] before the start of interim maintenance).16 Full details of the treatment regimens have been published.16,17 Briefly, in ALL99 and UKALL2003, patients were assigned to regimen A or B based on whether they were National Cancer Institute (NCI) standard (<10 years old and WCC <50 × 109/L) or HR (≥10 years old or WCC >50 × 109/L), respectively. Regimen A comprised a 3-drug induction (vincristine, steroids, and asparaginase) followed by consolidation (daily mercaptopurine and weekly intrathecal methotrexate), central nervous system–directed therapy, interim maintenance (daily mercaptopurine, weekly methotrexate, monthly vincristine, and steroid pulses), delayed intensification (asparaginase, vincristine, dexamethasone, and doxorubicin), and continuing therapy (oral mercaptopurine and methotrexate, monthly vincristine and steroid pulses, and intrathecal methotrexate every 3 months). Regimen B patients additionally received daunorubicin during induction and Berlin-Frankfurt-Münster consolidation (4 weeks of cyclophosphamide and cytarabine). Regimen C patients received an additional 4 doses of vincristine and 2 doses of PEGylated asparaginase during Berlin-Frankfurt-Münster consolidation. Furthermore, regimen C patients received escalating doses of intravenous methotrexate without folinic acid rescue and vincristine and PEGylated asparaginase as interim maintenance (Capizzi maintainance).
Cytogenetic and fluorescence in situ hybridization testing was performed on pretreatment bone marrow samples by member laboratories of the UK Cancer Cytogenetics Group or centrally by the Leukaemia Research Cytogenetics Group, and results were reported using established nomenclature and definitions.2 Multiplex ligation-dependent probe amplification (MLPA) was performed on DNA extracted either directly from pretreatment bone marrow samples or from fixed cell suspensions. The tested cohorts were representative in terms of sex, age, and outcome but were marginally enriched for patients with higher WCC and HR cytogenetics (supplemental Table 1). The SALSA MLPA kit P335 (MRC Holland, Amsterdam, The Netherlands), which included probes for IKZF1, CDKN2A/B, PAX5, EBF1, ETV6, BTG1, RB1, and PAR1 (CSF2RA/IL3RA/CRLF2), was used to identify CNAs in these genes.4 Previous studies have demonstrated that MLPA can accurately detect deletions in all these genes, which are present in more than 20% to 30% of cells.4,19 A detailed description and breakdown of each CNA and the correlation with specific chromosomal abnormalities for all the patients in these 2 cohorts has been published.4
Patients were classified into 3 mutually exclusive cytogenetic risk groups2 based on the clonal presence of the following chromosomal abnormalities: CYTO-GR (ETV6-RUNX1, high hyperdiploidy), CYTO-HR [BCR-ABL1, MLL rearrangements, near haploidy, low hypodiploidy, iAMP21 or t(17;19)], or cytogenetic CYTO-IR (all other cases with abnormal or normal cytogenetics). The results of the MLPA analysis were used to generate a CNA profile for each case, whereby each of the 8 genes/loci was coded as deleted or not deleted. For CDKN2A/B, deletion of either CDKN2A or CDKN2B was sufficient for the locus to be classified as deleted. For PAX5, intragenic amplifications were coded with the deletions as they are predicted to be functionally equivalent.20 A deletion in the PAR1 region of chromosome X or Y, del(X)(p22.33p22.33) / del(Y)(p11.32p11.32), results in the loss of the CSF2RA and IL3RA probes but retention of the CRLF2 probe on the MLPA P335 kit.21,22 Therefore, PAR1 deletion is usually synonymous with PR2Y8-CRLF2 fusion.4,22 However, a PAR1 deletion can also arise from an unbalanced t(X;14)(p22;q32) or t(Y;14)(p11;q32) translocation, which results in IGH-CRLF2 fusion.21 Like P2RY8-CRLF2, this translocation results in overexpression of CRLF2. As this scenario of translocation plus deletion is rare and essentially produces the same result, for the purposes of this paper, we have considered PAR1 deletion synonymous with P2RY8-CRLF2.
Survival analysis considered 3 end points: EFS, defined as time to relapse, second tumor, or death, censoring at last contact; relapse rate (RR), defined as time to relapse for those achieving a complete remission, censoring at death in remission or last contact; and OS, defined as time to death, censoring at last contact. Survival rates were calculated and compared using Kaplan-Meier methods, log-rank tests, and Cox regression models (univariate and multivariate analyses). Other comparisons were performed using the χ2 or Fisher’s exact test. Due to the investigative nature of this analysis, all tests were conducted at the 1% significance level. All analyses were performed using Intercooled Stata 13.0 (Stata Corporation).
Results
Frequency and prognostic value of individual gene deletions
CNAs were determined by MLPA in representative cohorts of patients treated on ALL97/99 (n = 864) or UKALL2003 (n = 780) (supplemental Table 1). The most prevalent CNAs were CDNK2A/B and ETV6 deletions occurring in ∼20% to 25% cases, whereas ∼15% cases harbored deletions of IKZF1 or PAX5 (supplemental Table2). The remaining CNAs were rarer and each observed in <10% cases. There was considerable heterogeneity with respect to the number of CNAs per case and distribution by cytogenetic risk group. No CNAs were observed in 43% cases, whereas 30%, 18%, and 10% of cases had 1, 2, or ≥3 CNAs. Individual assessment of the CNAs revealed that most deletions were associated with a poor outcome in both trials (supplemental Figure 1), the exception being ETV6 deletions, which are associated with the GR abnormality ETV6-RUNX1. These findings indicate that all 8 genes contribute to patient outcome. Furthermore, they were unevenly distributed across the 3 cytogenetic risk groups that are known to predict outcome.2 Therefore, we opted to integrate the CNA data into our existing cytogenetic classification in order to generate an overarching classification.
Development of a novel integrated genetic classification using the ALL97/99 cohort
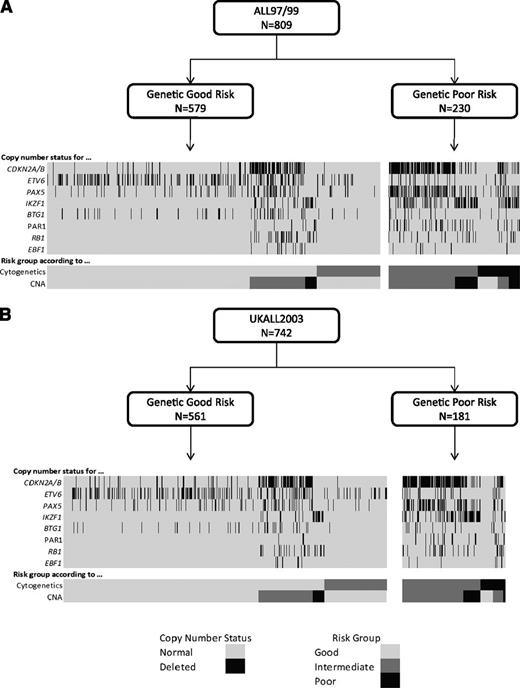
Using the copy-number status (deleted vs not deleted) for each of the 8 genes/loci covered by the MLPA P335 kit, a total of 67 unique CNA profiles were observed among 864 ALL97/99 patients (supplemental Table 3). We assessed each unique CNA combination in turn as described in Figure 1. CNA combinations that were present in <10 patients were classified into the indeterminate group. The prognostic relevance of each of the remaining CNA combinations was assessed, and combinations were allocated to the GR, IR 1, IR 2, or PR groups depending on the HR for an event and P value derived from a Cox regression model. These groups were then combined into 3 risk groups: CNA-GR, CNA-IR, and CNA-PR (Figure 1). These 3 CNA risk groups comprised 61%, 29%, and 10% of cases and were associated with different EFS rates at 10 years: 80% (99% CI 75-84), 67% (59-74), and 49% (35-62) (supplemental Table 4 and supplemental Figure 2A-C). Next, we cross-tabulated these 3 groups (CNA-GR, CNA-IR, and CNA-PR) with the 3 cytogenetic risk groups (CYTO-GR, CYTO-IR, and CYTO-HR). Therefore, 809 patients with both cytogenetic and MLPA data were classified into 9 mutually exclusive subgroups. Four subgroups showed EFS, RR, and OS rates >75%, <25%, and >80% at 5 years, respectively (supplemental Figure 3A-C). These subgroups comprised all CYTO-GR patients and CYTO-IR/CNA-GR patients and were henceforth combined to create a good-risk genetic group (GEN-GR) (supplemental Figure 3D). The remaining 5 subgroups showed inferior survival rates and were henceforth combined together to create a PR genetic group (GEN-PR) (supplemental Figure 3D). These newly defined genetic risk groups (Figures 3 and 4) comprised 72% (GEN-GR) and 28% (GEN-PR) of patients and were associated with distinct demographic, clinical, and outcome characteristic rates (Tables 1-3 and Figure 2A-C).
Development of a risk index based on CNAs. The copy-number status of 8 genes/regions was assessed in a cohort of 864 patients treated on ALL97/99 by MLPA using the P335 kit. Patients were classified according to the copy-number status (deleted/not deleted) of all 8 genes/regions. A total of 67 unique combinations were observed (see supplemental Table 2, available on the Blood Web site). Patients with each unique combination were classified into risk groups according to the algorithm shown below (A). For combinations observed in ≥10 patients, a Cox regression model was used to estimate the risk of an event and patients were assigned to risk groups (good-risk [GR], PR, intermediate risk [IR] 1 [INT-1], or IR 2 [INT-2]) based on the magnitude of the hazard ratio (HR) and the size of the P value (p). All combinations observed in <10 patients were assigned to the indeterminate (IND) risk group. The event-free survival (EFS) of the 5 risk groups is shown in the Kaplan-Meier graph (B).
Development of a risk index based on CNAs. The copy-number status of 8 genes/regions was assessed in a cohort of 864 patients treated on ALL97/99 by MLPA using the P335 kit. Patients were classified according to the copy-number status (deleted/not deleted) of all 8 genes/regions. A total of 67 unique combinations were observed (see supplemental Table 2, available on the Blood Web site). Patients with each unique combination were classified into risk groups according to the algorithm shown below (A). For combinations observed in ≥10 patients, a Cox regression model was used to estimate the risk of an event and patients were assigned to risk groups (good-risk [GR], PR, intermediate risk [IR] 1 [INT-1], or IR 2 [INT-2]) based on the magnitude of the hazard ratio (HR) and the size of the P value (p). All combinations observed in <10 patients were assigned to the indeterminate (IND) risk group. The event-free survival (EFS) of the 5 risk groups is shown in the Kaplan-Meier graph (B).
Kaplan-Meier survival graphs for 809 ALL97/99 and 742 UKALL2003 patients classified according to genetic risk group. EFS (A,D), relapse risk (B,E), and OS (C,F).
Kaplan-Meier survival graphs for 809 ALL97/99 and 742 UKALL2003 patients classified according to genetic risk group. EFS (A,D), relapse risk (B,E), and OS (C,F).
Cytogenetic and CNA landscape of novel genetic risk groups. Diagrams depicting the relationship between genetic risk, cytogenetic risk, CNA risk groups, and the copy-number status of 8 key genes/regions for patients treated on ALL97/99 (A) and UKALL2003 (B).
Cytogenetic and CNA landscape of novel genetic risk groups. Diagrams depicting the relationship between genetic risk, cytogenetic risk, CNA risk groups, and the copy-number status of 8 key genes/regions for patients treated on ALL97/99 (A) and UKALL2003 (B).
Independent validation of the novel genetic classification using the UKALL2003 cohort
We validated this novel genetic classification on an independent cohort of 742 UKALL2003 patients with both successful MLPA and cytogenetic analysis. A total of 19 new CNA profiles were observed in the validation cohort, each in single patients, and were assigned to the CNA-indeterminate group (supplemental Table 3). The majority of these new profiles (n = 15) comprised ≥3 deletions and matched the other CNA profiles classified in the GEN-PR group (Figures 3 and 4). The validation cohort did not include BCR-ABL1 or Down syndrome patients (supplemental Table 1), but sensitivity analysis of the test cohort (excluding these patients) did not alter the spectrum, incidence, or classification of the CNA profiles. The distribution of GEN-GR/GEN-PR patients in the validation cohort was slightly, but not significantly, different from the test cohort (76%/24% vs 72%/28%, P = .08). The slight decrease in the proportion of GEN-PR patients in the validation cohort is likely due to exclusion of BCR-ABL1 and Down syndrome patients, because BCR-ABL1 patients are classified as CYTO-HR and both are associated with IKZF1 deletions. The new genetic risk groups were significantly associated with age, WCC, MRD, and outcome in the validation cohort (Tables 1-3 and Figure 2D-F).
Clinical relevance of the novel genetic classification in UKALL2003
In UKALL2003, patients classified in the GEN-PR had an inferior outcome across all relevant patient subgroups with no evidence of heterogeneity in relation to other risk factors (Figure 4). MRD was associated with a worse outcome in both the GEN-GR and GEN-PR groups, but the effect of treatment regimen was less clear (Table 4). In UKALL2003, patients who were MRD-HR without other HR factors (eg, CYTO-HR or slow early response [SER]) were randomized to remain on regimen A/B or transfer to regimen C. Among GEN-GR/MRD-HR patients, there was no evidence that those patients who remained on regimen A/B, and therefore received less intensive therapy, had an inferior outcome (Table 4 and supplemental Figure 5A-C). Only 45 GEN-PR/MRD-HR patients were not also CYTO-HR or SER. However, among these patients, there was some evidence of a benefit for treatment with regimen C (Table 4 and supplemental Figure 5D-F), in particular a better OS rate, although not significant. Multivariate analysis incorporating genetic risk, MRD, age, WCC, and treatment arm revealed that only genetic risk and MRD are consistent independent predictors of outcome (Table 5).
Integrating cytogenetic and CNA data resulted in reclassification of CYTO-IR patients according to their CNA profile (Figure 4). CYTO-IR/CNA-GR patients (group A) had a superior outcome compared with CYTO-IR/CNA-IR and CYTO-IR/CNA-PR patients (group B) in both the test and validation cohorts (Tables 6-8 and supplemental Figure 4). Group B patients had a much higher incidence of CDKN2A/B (∼70%), PAX5 (∼45%), and IKZF1 (∼40%) deletions compared with group A patients (1%, 5%, and 0%; all P < .001), who frequently (∼80%) harbored none of the deletions studied (supplemental Table 5). In addition, the incidence of normal karyotypes and t(1;19)(q23;p13) was increased twofold compared with group B patients. Patients with a dicentric chromosome were almost always classified into group B. As most dicentric chromosomes involved 9p deletions, resulting in loss of PAX5 and CDKN2A/B, this result was not surprising, as group B patients were enriched for both deletions. Although the frequency of IGH translocations among UKALL2003 group A and B patients was similar (8% vs 7%), there was a marked difference in their age profile (mean 7.4 vs 14.9 years, respectively; P = .0008).
Discussion
We have defined a novel genetic risk classification for pediatric BCP-ALL by integrating CNA and cytogenetic information. The classification defines 2 risk groups (good and poor) that have different outcomes across all subgroups independent of other risk factors. The genomic landscape of ALL is complex, heterogeneous, and not yet fully documented or understood. However, the treatment of childhood ALL is highly effective, with cure rates exceeding 90%.16 These 2 facts present a challenge for developing a genetic classification based on risk. We opted to take a pragmatic approach and develop a single genetic classification that could be readily adopted into routine clinical practice. Therefore, we focused on integrating CNA data on commonly deleted genes/loci with established chromosomal abnormalities. Analysis of the CNA data revealed a complex architecture consistent with previous studies; therefore, we grouped patients according to their CNA profile and integrated the subgroups with established cytogenetic risk groups to define a novel classification that was validated on an independent cohort.
There are strengths and limitations to our study compared with other attempts to use genomic data to develop or refine risk classifications. Firstly, we validated our classification on an independent cohort of patients treated on a contemporary protocol. Secondly, implementation of the classification into routine laboratory practice only requires the adoption of a simple, inexpensive, and widely available technique, namely MLPA, onto the existing background of gold-standard cytogenetic/molecular methods used for detecting significant chromosomal abnormalities (Figure 4). Although MLPA will not detect low-level subclones (<30%),19 which are known to occur in ALL,23 there is no evidence to suggest that they are relevant in predicting outcome. One potential limitation to our strategy is that not all 256 possible CNA combinations were observed in the test cohort. However, most of the new combinations identified in the validation cohort involved 3 or more CNAs and hence fitted well alongside other combinations in the GEN-PR group. In contrast, the CNA combinations included in the GEN-GR group were mostly simple, involving 0, 1, or, occasionally, 2 deletions. Almost all of these simple combinations have already been observed among the 1644 patients screened (supplemental Table 2). Although additional CNA combinations are possible, they will be rare and complex; thus, classifying them as detailed above is logical and consistent with the evidence. The fact that this classification relies on the use of a commercial MLPA kit to detect the 8 most prevalent CNAs has clear advantages in terms of confirming these findings and implementing the classification into a clinical setting. Although it means that other CNAs (eg, ERG deletions24,25 ) and sequence variants (eg, RAS, JAK2, and TP53 mutations26 ) are not represented, it is known that many of these alterations correlate closely with either chromosomal abnormalities or CNAs already represented in the classification. For example, RAS mutations with near haploidy/high hyperdiploidy,27,28 TP53 mutations with low hypodiploidy,28,29 and JAK2 mutations with P2RY8-CRLF2 (PAR1 deletions).21,30 Therefore, it could be argued that this classification as it stands already accounts for much of this additional genetic heterogeneity. Nevertheless, further work will be necessary to determine whether the inclusion of such abnormalities can further refine this classification.
The prognostic impact of GR and HR cytogenetic abnormalities was not altered by their CNA profile. This finding implies that the primary genetic abnormality is more important in determining treatment response than subclonal secondary aberrations. This conclusion is both logical and consistent with studies that show evidence for the pleiotropic effect of IKZF1 deletions.7,12,24,25,31, Therefore, the principal effect of integrating cytogenetic and CNA information has been to reclassify CYTO-IR patients according to their CNA profile. Although the demographic and clinical features of the 2 groups of patients were similar, their genetic and outcome profiles were distinct (Tables 6-8; supplemental Table 5; and supplemental Figure 4). Group B was enriched for IKZF1 deletions, CDKN2A/B deletions, P2RY8-CRLF2 (PAR1 deletions), dic(9;20), and older patients with IGH translocations, all of which have been associated with poor outcome.5-12,22,31,32 Gene expression profiling studies,13,14 which have sought to define novel HR subgroups, have defined the BCR-ABL1–like subtype, which has some similar features (poor outcome, enriched for IKZF1 deletions, CDKN2A/B deletions, and P2RY8-CRLF2) and also accounts for 15% to 20% of BCP-ALL. Although the outcome of the US Children’s Oncology Group BCR-ABL1–like subgroup was much worse (4-year EFS <25%), their analysis was based on an HR cohort.13 In contrast, the outcome of the Dutch Childhood Oncology Group BCR-ABL1–like subgroup was very similar to that observed for our group B patients treated on ALL97/99 (5-year EFS, 56% vs 57%). Future collaborative studies will be required to determine the degree of overlap between existing and novel BCR-ABL1–like gene signatures and this novel genetic classification.
This genetic classification defines a large (75%) subset of patients who have an excellent outcome on UKALL2003 (5-year survival >95% and RR <5%). Further improvements in the treatment of childhood ALL cannot be measured in terms of RR and death toll. Instead, toxicity and long-term side effects are becoming the major issues.33 Hence, the identification of a large group of children with an excellent outcome who could be considered for treatment reduction is a key finding of this study. Although GEN-GR patients who were MRD HR had an inferior outcome compared with MRD-negative patients, the EFS at 5 years remained >90%. Moreover, the effect of treatment escalation among these patients did not significantly improve outcome. Our data indicate that current treatment is curative for the vast majority of GEN-GR patients, and future treatment strategies for these patients should focus on deintensification, compliance, and targeted therapies.
A quarter of patients were classified as PR based on genetics, including those with HR cytogenetic entities and those with an adverse CNA profile (group B). Although survival of UKALL2003 GEN-PR patients was significantly inferior to GEN-GR patients, it reached >80% at 5 years. GEN-PR patients with HR cytogenetics (∼30% GEN-PR patients) are allocated to regimen C, and, as an example, iAMP21 patients have been shown to clearly benefit from such an intervention.34 The remaining GEN-PR patients (∼70%) were classified on the basis of their CNA profile (group B), accounting for ∼20% of all BCP-ALL patients. All MRD HR patients on the new UK trial (UKALL2011) will be allocated to regimen C. These data (Table 4) have shown that GEN-PR patients in the MRD LR group have a LR of relapse and good survival. Thus, there is no clear evidence to support further treatment intensification for the remaining group B patients. Instead, the focus should be on identifying the key driver(s) of relapse and potential therapeutic targets within this subgroup. Gene fusions that result in tyrosine kinase activation provide excellent examples of therapeutic targets that are known to be present in some patients with BCR-ABL1–like ALL. Recent research has identified a complex network of such translocation involving ABL1, ABL2, PDGFRB, and CSF1R that are experimentally sensitive to tyrosine kinase inhibitors.35 Although these gene fusions are rare (∼1%), 2 recent case reports of patients with refractory ALL and EBF1-PDGFRB fusion responded well to imatinib, demonstrating the utility of using tyrosine kinase inhibitors in the clinical setting.36,37 Further work is required to determine the full spectrum of such tyrosine kinase–activating fusions, their prognostic relevance, and how they interface with this novel genetic classification.
In conclusion, we have integrated cytogenetic and CNA data into a single genetic classification that can be used to refine patient treatment according to more detailed genetic description of their leukemia. GEN-GR patients have an excellent outcome with modern therapy, and future treatment approaches should focus on achieving the same results with reduced therapy. Although, GEN-PR patients have an inferior outcome compared with GEN-GR patients, their survival still exceeds 80%. As many of these patients are already receiving intensive treatment, further genetic research is required to identify patients curable with current drugs and those requiring alternative therapies.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank member laboratories of the UK Cancer Cytogenetic Group for providing cytogenetic data and material as well as past and present members of the Leukaemia Research Cytogenetics Group for their contribution to establishing this data set. Primary childhood leukemia samples used in this study were provided by the Leukaemia and Lymphoma Research Childhood Leukaemia Cell Bank working with the laboratory teams in the Bristol Genetics Laboratory, Southmead Hospital, Bristol; Molecular Biology Laboratory, Royal Hospital for Sick Children, Glasgow; Molecular Haematology Laboratory, Royal London Hospital, London; and Molecular Genetics Service and Sheffield Children’s Hospital, Sheffield, UK.
This work was supported by Leukaemia & Lymphoma Research (formerly Leukaemia Research, UK).
Authorship
Contribution: A.V.M. and A. Enshaei contributed to study conception and design; A.V.M., C.S., R.W., A. Elliott, S.R., L.C., J.H., and C.J.H. collected and assembled data; A.V.M., A. Enshaei, C.S., N.G., A.V., and C.J.H. analyzed and interpreted data; C.J.H. and A.V.M. provided financial and administrative support; S.E.K., C.D.M., N.G., and A.V. provided study materials or patients; and all authors wrote and provided final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anthony V. Moorman, Leukaemia Research Cytogenetics Group, Northern Institute for Cancer Research, Newcastle University, Level 5, Sir James Spence Institute, Royal Victoria Infirmary, Newcastle upon Tyne NE1 4LP, UK; e-mail: anthony.moorman@newcastle.ac.uk.

![Figure 1. Development of a risk index based on CNAs. The copy-number status of 8 genes/regions was assessed in a cohort of 864 patients treated on ALL97/99 by MLPA using the P335 kit. Patients were classified according to the copy-number status (deleted/not deleted) of all 8 genes/regions. A total of 67 unique combinations were observed (see supplemental Table 2, available on the Blood Web site). Patients with each unique combination were classified into risk groups according to the algorithm shown below (A). For combinations observed in ≥10 patients, a Cox regression model was used to estimate the risk of an event and patients were assigned to risk groups (good-risk [GR], PR, intermediate risk [IR] 1 [INT-1], or IR 2 [INT-2]) based on the magnitude of the hazard ratio (HR) and the size of the P value (p). All combinations observed in <10 patients were assigned to the indeterminate (IND) risk group. The event-free survival (EFS) of the 5 risk groups is shown in the Kaplan-Meier graph (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/9/10.1182_blood-2014-03-562918/4/m_1434f1.jpeg?Expires=1769662006&Signature=cnJFRX6hEaNz0EhW1lpyM2OWg78znxFFII95Ah7VHAefJCw2ze90YlPp3kK1OG~XgH6i5L4TFi9jOJo9hiQoosvJP878nESBzXmW4GqN6UzDChHzyLQ19hvYIM7eRtsTU-LrjK-wN5q5ExgmvnCs1ffpvEoIFJsqunxR6yQg5X~90RW8PztMeUtXu6fXoXOishYrhB7enb2fkMGAnAcd7nmdHcumCf31-CRGps87HbcbsyVb1UUfgmaA2UDflDm7JfIv1z7Rj4W8Qsj8wpJvMDP1y3ydsx1I6ItRJ7qoZCEE71mF57ipVaB8z-8YJr6KisRdtgqEzxYCyPzNyIc7Ng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 1. Development of a risk index based on CNAs. The copy-number status of 8 genes/regions was assessed in a cohort of 864 patients treated on ALL97/99 by MLPA using the P335 kit. Patients were classified according to the copy-number status (deleted/not deleted) of all 8 genes/regions. A total of 67 unique combinations were observed (see supplemental Table 2, available on the Blood Web site). Patients with each unique combination were classified into risk groups according to the algorithm shown below (A). For combinations observed in ≥10 patients, a Cox regression model was used to estimate the risk of an event and patients were assigned to risk groups (good-risk [GR], PR, intermediate risk [IR] 1 [INT-1], or IR 2 [INT-2]) based on the magnitude of the hazard ratio (HR) and the size of the P value (p). All combinations observed in <10 patients were assigned to the indeterminate (IND) risk group. The event-free survival (EFS) of the 5 risk groups is shown in the Kaplan-Meier graph (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/9/10.1182_blood-2014-03-562918/4/m_1434f1.jpeg?Expires=1770298622&Signature=MWVrYGmAr7rgWDksqPZNRAISFnt1vR33vUctMr8h-JRNKehAHnfM1Sax~5Yh7AEUOEPF7WJCIgkM0arv36duZ5uGOl-ifZH2k1axMG3pvA8ugyh3LNMO3DVhDQm4d2lci5ma65lvtAIUEUe5SPmjkfJ04FeUHvh9uHYe1cUm7trDZHsNFRfqgVQoR0Sl~20sVkVftU8EsiHgUYW-4Ljj1TuG5NcZJjoQR7ZA4VKd7PJJd1ukLnZ7vgBsWUf52Z3fqVxUUoxKAj7mYJ39U1BQ1o3l41gg67Aq-LUZ6cGWRoxgOviFNFYoHOdyMFjOjwTZtUuBkRHnsZdsOHSoNGMp3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)