Key Points
The prevalence of KLF1 mutations is significantly higher in a thalassemia endemic region of China than in a nonendemic region.
KLF1 mutations ameliorate the clinical and hematologic features of β-thalassemia.
Mutations in human Krüppel-like factor 1 (KLF1) have recently been reported to be responsible for increased fetal hemoglobin (HbF) and hemoglobin A2 (HbA2). Because increased HbF and HbA2 levels are important features of β-thalassemia, we examined whether there is any relationship between KLF1 mutation and β-thalassemia in China. To do this, we first studied the incidence of KLF1 mutations in 2 Chinese populations: 3839 individuals from a thalassemia endemic region in south China and 1190 individuals from a nonthalassemia endemic region in north China. Interestingly, we found that the prevalence of KLF1 mutations is significantly higher in the thalassemia endemic region than that in nonthalassemia endemic region (1.25% vs 0.08%). Furthermore, we identified 7 functional variants including 4 previously reported (p.Gly176AlafsX179, p.Ala298Pro, p.Thr334Arg, and c.913+1G>A) and 3 novel variants (p.His299Asp, p.Cys341Tyr, and p.Glu5Lys) in southern China. The 2 most common mutations, p.Gly176AlafsX179 and p.His299Asp, accounted for 90.6% of the total. We found that zinc-finger mutations in KLF1 were selectively represented in 12 β-thalassemia intermedia patients and resulted in significantly different transfusion-free survival curves. Our findings suggest that KLF1 mutations occur selectively in the presence of β-thalassemia to increase the production of HbF, which in turn ameliorates the clinical severity of β-thalassemia.
Introduction
The clinical severity of β-thalassemia varies widely from mild to severe forms and genotype-phenotype associations in β-thalassemia have been extensively studied by systematic analysis of defective β-globin genes.1,2 However, the same genotype of β-thalassemia can manifest with highly variable patterns of clinical severity ranging from moderate to severe disease resulting from various genetic modifiers of disease severity.3,4 Several genetic modifiers unlinked to the β-globin gene locus have been identified, including genetic variants modulating fetal hemoglobin (HbF) levels,5 α-globin gene copy numbers/α-thalassemia,4 α hemoglobin stabilizing protein,6 and heme-regulated eIF2alpha kinase7 in either human patients or murine models of the disease. Increased HbF levels and concomitant α-thalassemia are 2 main modifiers that can ameliorate the clinical and hematological severity of β-thalassemia.2,4,8
The erythroid transcriptional factor Krüppel-like factor 1 (KLF1) has recently emerged as 1 of the key regulators of the γ- to β-globin gene switching.9,10 It is now known that the dominant mutation (p.E325K) in KLF1 can cause congenital dyserythropoietic anemia (type IV)11 and a compound heterozygous mutation in KLF1 can lead to chronic hemolytic anemia,12 but most monoallelic KLF1 mutations give rise to benign phenotypes,13,-15 including significant increases in levels of HbF16,-18 and adult hemoglobin A2 (HbA2).19 However, little is known about the modification of β-thalassemia phenotypes in the context of coinheritance of KLF1 mutations with β-globin gene mutations. In addition, there is little information regarding the population genetics of KLF1 mutations. Here, we investigated the incidence and spectrum of KLF1 mutations in the Chinese population and show that KLF1 mutations are common in a thalassemia endemic region in southern China and that the hematologic features of β-thalassemia are modulated by different KLF1 mutations.
Design and methods
Population, patients, and phenotypic tests
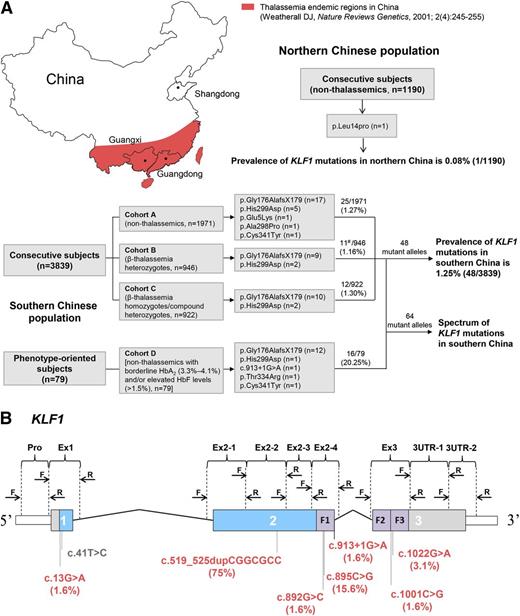
We designed a large population-based cohort study that consisted of a total of 3918 randomly selected unrelated subjects living in southern China. This cohort included 3839 consecutive subjects who were investigated to determine the incidence of KLF1 mutations and 79 healthy subjects with borderline HbA2 (3.3% to 4.1%) and/or elevated HbF levels (>1.5%) to determine KLF1 mutation frequency in this selected population. All subjects in whom KLF1 mutations were identified were available for study of both the spectrum of mutations and of the hematological features of KLF1 variants (Figure 1A). The cohort of 3839 consecutive subjects (aged 1-50 years; 1987 males and 1852 females) was grouped into 3 subgroups based on the β-globin genotypes: 1971 nonthalassemics (cohort A), 946 β-thalassemia heterozygotes (cohort B), and 922 β-thalassemia homozygotes or compound heterozygotes (cohort C). Cohort A subjects were randomly drawn from 5789 samples used in a previous study.20 Consecutive samples in cohort B and cohort C were recruited between November 2008 and June 2013 from southern China. A subset of 79 subjects (cohort D) was also selected from a large random Guangxi population screened for the previously mentioned 2 positive indices of suspected KLF1 mutations. All participants were ethnic southern Chinese, whose registered parental origin was from either Guangxi or Guangdong provinces in southern China where thalassemia is highly prevalent.20,-22 In addition, we recruited 1190 healthy Chinese subjects from Shandong Province in northern China, where thalassemia is much less prevalent. Clinical data were collected retrospectively on all 922 thalassemia patients in cohort C. Diagnosis of thalassemia was made through a comprehensive assessment of clinical presentation and hematological and molecular analysis based on criteria previously described.23,24 A summary of clinical and genetic characterizations of 922 patients, 568 thalassemia major (TM) and 354 thalassemia intermedia (TI) in cohort C, is presented in Table 1.
The prevalence and spectrum of KLF1 mutations in the Chinese population. (A) Sampling design and study outcomes. Southern Chinese population: Three consecutive cohorts (A-C) of 3839 subjects from 2 thalassemia endemic regions (Guangxi and Guangdong provinces) were designed to investigate the incidence of KLF1 mutations; 79 phenotype-oriented subjects (cohort D) selected from the same region were recruited for enrichment of KLF1 mutation–positive subjects. The β-globin gene in all 79 subjects was completely analyzed; those patients positive for β-thalassemia mutations were excluded from analysis. All mutant alleles (n = 64) detected in these 2 approaches were used to assess the mutation spectrum of KLF1. #HBB genotype categories of 11 KLF1 mutation–positive subjects in cohort B were as follows: 1 β+/βN, 1 βE/βN, 7 β0/βN, and 2 β0/βN coinherited with α-thalassemia. Northern Chinese population: 1190 nonthalassemics were recruited from a nonthalassemia endemic region (Shandong Province). (B) Exons, introns, and domains are shown with the untranslated regions (white), proline-rich regions (blue), and zinc fingers (ZFs; purple). The positions of the HRM primers are noted above the physical map, and 4 novel and 4 previously reported functional KLF1 mutations in south (red) and north (gray) China are shown below the map. Allelic distribution of KLF1 mutations in the southern Chinese population are indicated in parentheses.
The prevalence and spectrum of KLF1 mutations in the Chinese population. (A) Sampling design and study outcomes. Southern Chinese population: Three consecutive cohorts (A-C) of 3839 subjects from 2 thalassemia endemic regions (Guangxi and Guangdong provinces) were designed to investigate the incidence of KLF1 mutations; 79 phenotype-oriented subjects (cohort D) selected from the same region were recruited for enrichment of KLF1 mutation–positive subjects. The β-globin gene in all 79 subjects was completely analyzed; those patients positive for β-thalassemia mutations were excluded from analysis. All mutant alleles (n = 64) detected in these 2 approaches were used to assess the mutation spectrum of KLF1. #HBB genotype categories of 11 KLF1 mutation–positive subjects in cohort B were as follows: 1 β+/βN, 1 βE/βN, 7 β0/βN, and 2 β0/βN coinherited with α-thalassemia. Northern Chinese population: 1190 nonthalassemics were recruited from a nonthalassemia endemic region (Shandong Province). (B) Exons, introns, and domains are shown with the untranslated regions (white), proline-rich regions (blue), and zinc fingers (ZFs; purple). The positions of the HRM primers are noted above the physical map, and 4 novel and 4 previously reported functional KLF1 mutations in south (red) and north (gray) China are shown below the map. Allelic distribution of KLF1 mutations in the southern Chinese population are indicated in parentheses.
Hematological parameters were measured using an automated hematology analyzer (Sysmex F-820; Sysmex Co. Ltd., Kobe, Japan) and hemoglobin analysis was carried out using high-performance liquid chromatography (Variant II, Bio-Rad Laboratories, Hercules, CA). Flow-cytometric analysis of CD44 expression on mature erythrocytes was performed with a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA) by immunolabeling of red blood cells with reagents from BD Biosciences-Pharmingen according to standard protocols. The study protocol was reviewed and approved by the local medical ethics committee at Nanfang Hospital, China, and written consent was obtained from parents and/or patients before commencing the study. The study was conducted in accordance with the Declaration of Helsinki.
Molecular genetic tests
Genomic DNA was extracted from peripheral blood using the standard phenol/chloroform method. We used the high-resolution melting (HRM) assay to scan for KLF1 mutations and followed by Sanger sequencing for confirmation of KLF1 variants (see supplemental Figure 1, available on the Blood Web site). Nine pairs of primers were designed to amplify the coding region (exons 1-3) and the conserved noncoding regions (promoter region, flanking splice junctions, and 5′- and 3′-untranslated regions) of the KLF1 gene (Figure 1B; supplemental Table 1). XmnI PCR-RFLP and HRM assays were used to genotype 4 primary HbF-associated single nucleotide polymorphisms (rs7482144, XmnI, in HBG,25 rs11886868, and rs766432 in BCL11A,26,27 and rs9399137 in HBS1L-MYB28 (supplemental Figure 1; supplemental Table 1). Point mutations and deletions in the α-, β-, and γ-globin genes were detected using previously described methods.20
Functionally relevant KLF1 mutations were identified on the basis of phenotypic changes (decreased erythrocytes CD44 expression and/or HbF levels >1%) and functional predictions for nonsynonymous substitutions in KLF1 analyzed by SIFT (http://sift.jcvi.org) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2). To determine whether the mutations are hereditary or somatic, peripheral blood and DNA samples were available from 6 parents whose offspring were identified as carriers for KLF1 mutations in cohort C (supplemental Figure 2).
Statistical analysis
Statistical analyses were carried out using SPSS version 19.0 (IBM, Armonk, NY) and P < .05 was considered statistically significant. All quantitative variables were compared by either the Mann-Whitney U test or the χ2 test as appropriate. Multivariate analysis was performed using a backward stepwise Cox proportional hazards model in 922 β-thalassemia homozygotes or compound heterozygotes in cohort C, and hazard ratios (HR) with 95% confidence intervals (CI) were obtained to evaluate the associations between putative ameliorating alleles and the patient’s age at first transfusion. All covariates were classified as follows: α-globin gene mutations as 0, 1, 2, or 3 based on the number of mutated copies of the HBA; for other loci, each variable was defined with values 0, 1, or 2 according to the number of copies of putative modifying allele: HBB mutations (β+), rs7482144 (XmnI, +) in HBG2, rs11886868 (C) and rs766432 (C) in BCL11A, rs9399137 (C) in HBS1L-MYB; KLF1 mutations was only scored as 0 or 1 because no homozygous mutation was noted; and gender for male was 1 and for female was 0. The performance of the model was measured by Harrell’s concordance index by using the rms package in R version 3.0.1 (http://www.r-project.org/). Univariate analysis was conducted to compare clinical features by either the Mann-Whitney U test or the χ2 test as appropriate in β-thalassemia patients with or without KLF1 mutations, and odds ratios (OR) with 95% CI were calculated to assess the risk of receiving systematic transfusion and TI. These 2 groups had same globin genotype category (β0/β0 and αα/αα) and similar HbF-modulating genetic variants (XmnI [−/−], rs766432 [AA or AC], rs9399137 [TT or CT]): 7 of 12 KLF1 mutation–positive subjects and 362 of 910 KLF1 mutation–negative subjects in cohort C were included in both case-control and survival analysis. Transfusion-free survival curves were depicted according to the Kaplan-Meier method and compared using the log-rank test.
Results
KLF1 mutation prevalence is significantly higher in the thalassemia endemic region in southern China
It has been recently reported that KLF1 mutations result in elevated HbF levels.16,,-19 Because elevated HbF is beneficial in β-thalassemia,2,4 we hypothesized that some associations may exist between KLF1 mutations and β-thalassemia in regions with high prevalence of thalassemia. To test this hypothesis, we scanned for mutations of the KLF1 gene in 2 distinct Chinese populations: 3839 unrelated individuals from 3 different cohorts in southern China where thalassemia is endemic and 1190 individuals from northern China where thalassemia rarely occurs. We identified 48 subjects with KLF1 heterozygous mutations in the cohort from southern China, giving a heterozygote frequency of 1.25% in this population (Figure 1A). In contrast, we identified only 1 subject with heterozygous missense mutations in the population from the north, giving a 0.08% frequency, which is significantly lower than found in the population from the south (P < .001). These findings demonstrate that KLF1 mutations have a much higher prevalence in the presence of thalassemia. Comparison of the frequency of KLF1 mutations among the 3 cohorts from he revealed that the frequency is 1.27% (25/1971) in cohort A, 1.16% (11/946) in cohort B, and 1.30% (12/922) in cohort C, respectively. Statistical analysis showed that there were no statistically significant differences among these 3 sample groups (P = .959), implying that higher KLF1 mutation prevalence is a common phenomenon of a southern Chinese population, where thalassemia is endemic.
KLF1 mutation spectrum in southern China
As described previously, we have identified 48 heterozygotes with KLF1 mutations in the southern Chinese population comprising 3839 subjects. To enlarge the size of the sample with KLF1 mutations for further analysis, we identified an additional 16 subjects with KLF1 heterozygous mutations in 79 subjects (nonthalassemics, cohort D) based on borderline increases in HbA2 (3.3% to 4.1%) and/or elevated HbF levels (>1.5%) (Figure 1A). Thus a total of 64 mutant alleles were documented in the present study, of which 41 are KLF1 heterozygotes alone, 11 are KLF1 mutations coinherited with β-thalassemia heterozygotes, and 12 are KLF1 mutations coinherited with β-thalassemia homozygotes or compound heterozygotes. We identified 4 previously reported KLF1 gene mutations (p.Gly176AlafsX179, p.Ala298Pro, p.Thr334Arg, and c.913+1G>A)12,18,29,-31 and 3 novel mutations. Among these are 5 missense mutations (p.Glu5Lys, p.Ala298Pro, p.His299Asp, p.Thr334Arg, and p.Cys341Tyr), 1 frameshift mutation (p.Gly176AlafsX179), 1 splicing mutation (c.913+1G>A); the number and percentage of mutant alleles that carried these 7 types of KLF1 mutations are listed in Figure 1B and supplemental Table 2. We determined that the 6 mutations (p.Gly176AlafsX179 frameshift, p.Ala298Pro, p.His299Asp, p.Thr334Arg, p.Cys341Tyr, and c.913+1G>A) disrupting the ZF domain of KLF1 accounted for the vast majority of the molecular changes, with 2 mutations (p.Gly176AlafsX179 and p.His299Asp) being most frequent and accounting for 90.6% of the KLF1 mutant alleles in this population (Figure 1B; supplemental Table 2). In addition, we found 13 novel neutral KLF1 variants, including 4 missense (tolerated) and 4 synonymous mutations in the coding region and 5 variants in the 3′-untranslated region in our population (supplemental Table 2). Our findings, together with previously published studies,12,18,19,29,,-32 demonstrate the broad spectrum of KLF1 mutations.
KLF1 mutation effects on hematologic parameters of nonthalassemic subjects
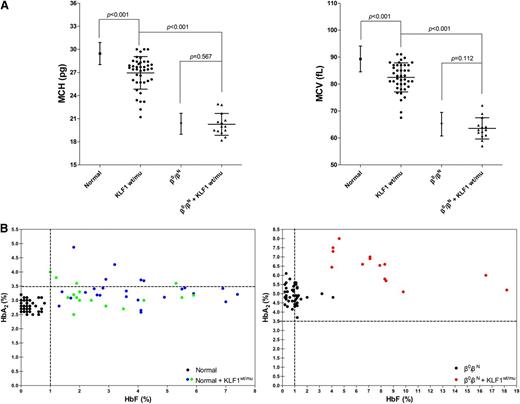
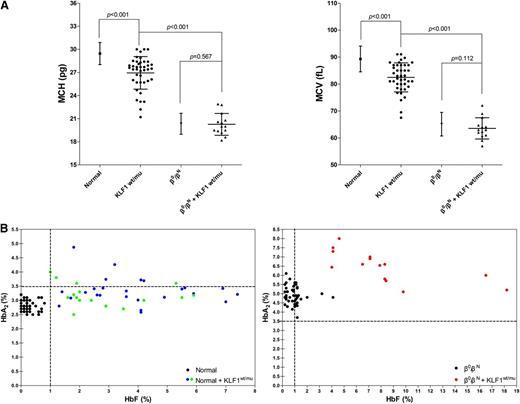
Having demonstrated that KLF1 mutations occur at about the similar frequency in all 3 cohorts (nonthalassemics, β-thalassemia heterozygotes, and β-thalassemia homozygotes/compound heterozygotes), we then examined the effect of these mutations on the hematological parameters of these 3 cohorts. Figure 2A shows the mean corpuscular volume (MCV) and mean corpuscular hemoglobin (MCH) values of normal individuals with or without KLF1 mutations. Both MCV and MCH were mildly lower in 41 normal subjects with KLF1 mutations (25 from cohort A, 16 from cohort D), compared with the normal subjects without KLF1 mutations (n = 1946, P < .001), demonstrating that mutations of KLF1 result in MCV and MCH values at the lower end of the normal range. In normal subjects without KLF1 mutations, MCV and MCH values were 89.29 ± 4.77 fL (n = 1946, range, 79.00 to 110.10 fL) and 29.46 ± 1.44 pg (n = 1946, range, 25.00 to 36.30 pg), respectively. In contrast, MCV and MCH values were 82.45 ± 5.39 fL (n = 41, range, 67.50 to 91.00 fL) and 26.95 ± 2.11 pg (n = 41, range, 21.20 to 30.00 pg), respectively, in normal subjects with the KLF1 mutations. Furthermore, as shown in Figure 2B, we confirmed borderline increases in HbA2 levels (n = 41, 3.28 ± 0.45%, range, 2.50% to 4.87%) and of slightly elevated HbF levels (n = 41, 3.44 ± 1.75%, range, 1.00% to 7.40%) in KLF1 heterozygous mutants in our normal population, in the 2 groups (25 from cohort A and 16 from cohort D) that expected to show the same effects of elevated HbA2 and/or HbF.
Characteristic hematological analyses of KLF1 heterozygotes in cohorts A and B. (A) MCV and MCH values in normal individuals (n = 1946 from cohort A), nonthalassemics (n = 41; 25 from cohort A and 16 from cohort D), and 7 KLF1–heterozygous mutations, β0-thalassemia heterozygotes (n = 217 from cohort B), and β0-thalassemia heterozygotes coinherited with KLF1 mutations (n = 14; 7 β0/βN of 11 from cohort B and another 7 β0/βN from 6 β-thalassemia families with KLF1 mutations; supplemental Figure 2). The α-thalassemia deletions or point mutations were excluded in all nonthalassemic and heterozygous β0-thalassemia individuals with and without KLF1 mutations. The data shown are expressed as mean ± SD. P value was determined using the Mann-Whitney U test. (B) HbA2 and HbF levels in normal individuals (left) and β-thalassemia heterozygotes (right) with or without KLF1 mutations (all have normal α globin genotypes). (Left) Solid circles represent samples with KLF1 mutations, including 25 from cohort A (blue), 16 from cohort D (green), and 50 without KLF1 mutations (black; HbA2, 2.81 ± 0.20%; range, 2.50% to 3.20%; HbF, 0.23 ± 0.22%, range, 0.00% to 0.80%). (Right) Solid circles represent 14 samples with KLF1 mutations (red) and 50 without KLF1 mutations (black; HbA2, 4.96 ± 0.49%, range, 3.70% to 6.10%; HbF, 0.93 ± 0.70%, range, 0.20% to 4.10%). Genotype symbols of subjects are shown on the bottom right of the chart.
Characteristic hematological analyses of KLF1 heterozygotes in cohorts A and B. (A) MCV and MCH values in normal individuals (n = 1946 from cohort A), nonthalassemics (n = 41; 25 from cohort A and 16 from cohort D), and 7 KLF1–heterozygous mutations, β0-thalassemia heterozygotes (n = 217 from cohort B), and β0-thalassemia heterozygotes coinherited with KLF1 mutations (n = 14; 7 β0/βN of 11 from cohort B and another 7 β0/βN from 6 β-thalassemia families with KLF1 mutations; supplemental Figure 2). The α-thalassemia deletions or point mutations were excluded in all nonthalassemic and heterozygous β0-thalassemia individuals with and without KLF1 mutations. The data shown are expressed as mean ± SD. P value was determined using the Mann-Whitney U test. (B) HbA2 and HbF levels in normal individuals (left) and β-thalassemia heterozygotes (right) with or without KLF1 mutations (all have normal α globin genotypes). (Left) Solid circles represent samples with KLF1 mutations, including 25 from cohort A (blue), 16 from cohort D (green), and 50 without KLF1 mutations (black; HbA2, 2.81 ± 0.20%; range, 2.50% to 3.20%; HbF, 0.23 ± 0.22%, range, 0.00% to 0.80%). (Right) Solid circles represent 14 samples with KLF1 mutations (red) and 50 without KLF1 mutations (black; HbA2, 4.96 ± 0.49%, range, 3.70% to 6.10%; HbF, 0.93 ± 0.70%, range, 0.20% to 4.10%). Genotype symbols of subjects are shown on the bottom right of the chart.
KLF1 mutation effects on hematological parameters of β-thalassemia heterozygote subjects
We then examined the effects of KLF1 mutations in modulating hematological parameters of β-thalassemia heterozygotes. For this, we analyzed β0-thalassemia heterozygotes with or without KLF1 mutations. MCV and MCH values were 63.53 ± 3.94 fL (n = 14, range, 57.00 to 72.00 fL) and 20.26 ± 1.42 pg (n = 14, range, 18.20 to 22.90 pg), for 14 β0-thalassemia heterozygotes (β0/βN and αα/αα genotypes) with the KLF1 mutations (7 of 11 from cohort B except for 1 βE/βN, 1 β+/βN, and 2 β0/βN coinherited with α-thalassemia and 7 from familial cases, shown in supplemental Figure 2), whereas they were 65.11 ± 4.34 fL (n = 217, range, 51.00 to 77.80 fL) and 20.34 ± 1.37 pg (n = 217, range, 14.60 to 27.60 pg), for 217 β0-thalassemia carriers with no coinherited KLF1 mutations. Surprisingly, in contrast to normal subjects, no statistically significant differences between these 2 groups with respect to these 2 hematological parameters (P = .112 or P = .567, Figure 2A) were noted. This suggests that mutations of KLF1 gene in the context of β-thalassemia trait do not have a significant effect on MCV and MCH. However, as shown in Figure 2B, we found that coexistence of β-thalassemia trait and KLF1 mutations results in higher levels of HbA2, of more than 6.5% (n = 17, 6.47 ± 0.81%, range, 5.10% to 8.00%), implying additive effects of these 2 factors on δ-globin gene expression. In addition, a highly variable increase in HbF levels (n = 17, 7.67 ± 4.11%, range, 3.60% to 18.20%) was noted in individuals with coinheritance of KLF1 mutations in the context of β-thalassemia trait. The basis of reason of this variability needs to be further studied.
KLF1 mutation effects on β-thalassemia homozygotes/compound heterozygote individuals
To define the role of KLF1 mutations in modulating the clinical and hematological severity, we studied 922 β-thalassemia patients with known genotypes of β-globin and modifier genes including KLF1 mutations (Table 1).We found it surprising that 2 KLF1 mutations were selectively represented in 12 TI patients who are all β-thalassemia homozygotes or compound heterozygotes in cohort C: 10 subjects had 1 frameshift mutation (p.Gly176AlafsX179) and the other 2 had the missense mutation (p.His299Asp). Both mutations disrupt the KLF1 ZF motif. The clinical features of all 12 affected β-thalassemia individuals are presented in supplemental Table 3. We assessed the influence of different combinations of genetic factors on the entire cohort by multivariate Cox regression analysis, measured by age at first transfusion, and the results showed that the KLF1 mutations had the strongest effect on the clinical severity of β-thalassemia phenotype (HR = 0.213, P < .001), followed by β+ mutation (HR = 0.353, P < .001), rs7482114 (XmnI, +) in HBG2 (HR = 0.512, P < .001), HBA mutations (HR = 0.671, P < .001), rs9399137 (C) in HBS1L-MYB (HR = 0.724, P < .001), and rs766432 (T) in BCL11A (HR = 0.761, P < .001) (Table 2).
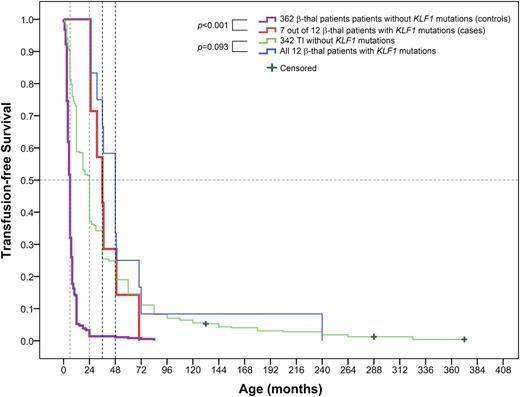
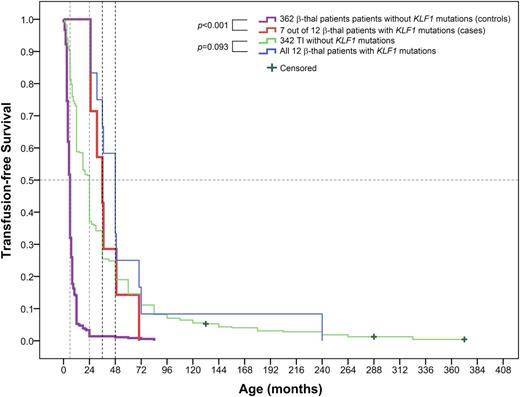
In these 12 TI patients, 7 were β0/β0 and αα/αα genotypes (patients P3, P4, P5, P6, P7, P11, and P12), 3 (patients P1, P2, and P8) were β0/β+ and αα/αα genotypes, and 2 (patients P9 and P10) were β0/β0 coinherited with the Southeast Asian deletion and a 3.7-kb α-thalassemia deletion, respectively (supplemental Table 3). We further tested the effects of KLF1 mutations in modulating the clinical severity of β-thalassemia by univariate analysis of a matched case-control of these 7 subjects (β0/β0 and αα/αα genotypes with KLF1 mutations) and 362 thalassemic subjects without the KLF1 mutation in cohort C. As shown in Table 3, univariate analysis confirmed the links between the KLF1 mutations and decreased clinical severity with remarkably increased hemoglobin concentration (P = .006) and HbF levels (P < .001) for TI cases with KLF1 mutations, along with delay in mean age at onset of anemia (P = .001), decreased transfusion requirements (OR = 0.082, P = .007), and a lower risk of exhibiting the features of TM (OR = 0.009, P < .001). This difference in HbF levels between 7 and 362 matched samples is consistent with the results of the HbF levels between the whole TI vs TM populations (Table 1), implying a clinically homogeneous group of TI patients in our cohort C whether they had a KLF1 mutation or not. Kaplan-Meier survival curves showed significant differences between the 2 groups (log-rank test P < .001). The median time to the first transfusion in the control group (362 without KLF1 mutations) was 6 months, compared with a highly significant improvement in transfusion-free survival observed in the case group (7 with KLF1 mutations) with the median time to the first transfusion at 36 months. Likewise, 342 KLF1 mutation–negative TI group patients received their first transfusion at 24 months, whereas it was initiated at 48 months in 12 KLF1 mutation–positive TI group patients, although there was no statistically significant difference between 2 TI subgroups (log-rank test P = .093) (Figure 3).
Kaplan-Meier survival curves for β-thalassemia (β-thal) patients with or without KLF1 mutations for needing transfusion in cohort C (log-rank test P value). Patients had not received a transfusion by the end of the study, which was recorded as censored data. The 2 sample-matched pairs, 7 vs 362 β-thalassemia cases with or without KLF1 mutations and 12 vs 342 TI cases with or without KLF1 mutations, are shown at the top of the chart. The 4 colored lines represent each of the 4 groups, respectively. Selection of well-matched 7 vs 362 subjects is described in our “Statistical analysis” section. To compare the median time to the first transfusion in the 2 groups of TI patients, all 12 TI cases with KLF1 mutations vs all 342 TI cases without KLF1 mutations were used for the Kaplan-Meier survival curve analysis.
Kaplan-Meier survival curves for β-thalassemia (β-thal) patients with or without KLF1 mutations for needing transfusion in cohort C (log-rank test P value). Patients had not received a transfusion by the end of the study, which was recorded as censored data. The 2 sample-matched pairs, 7 vs 362 β-thalassemia cases with or without KLF1 mutations and 12 vs 342 TI cases with or without KLF1 mutations, are shown at the top of the chart. The 4 colored lines represent each of the 4 groups, respectively. Selection of well-matched 7 vs 362 subjects is described in our “Statistical analysis” section. To compare the median time to the first transfusion in the 2 groups of TI patients, all 12 TI cases with KLF1 mutations vs all 342 TI cases without KLF1 mutations were used for the Kaplan-Meier survival curve analysis.
KLF1 mutation prevalence is high in individuals with increased HbA2 and HbF levels
To further establish the link between KLF1 mutations and HbA2 and HbF levels, we studied 79 nonthalassemic subjects (cohort D) with elevated levels of HbA2 and HbF. In this subgroup, we found that the prevalence of KLF1 mutations to be 20.25% (16/79), suggesting that elevated HbA2 and HbF levels can potentially be used to screen for KLF1 mutations in the population.
Discussion
We studied the prevalence of KLF1 mutations through a population-based molecular analysis of a large number of samples comprising 3839 participants in southern China (thalassemia endemic region) and 1190 normal individuals in northern China (nonthalassemia endemic region) using an HRM assay that we developed. We documented a surprisingly high frequency of KLF1 heterozygous mutations (1.25%) in the population from southern China, whereas it was only 0.08% in northern China. These findings demonstrate that mutations in the KLF1 gene are common in the thalassemia endemic region in southern China. Interestingly, although no functionally relevant KLF1 mutations in the ZF domain were identified in the northern Chinese population, such mutations occurred at a very high frequency in southern China, accounting for 98.4% of the KLF1 mutant alleles.
We further documented that KLF1 mutation is a genetic modifier ameliorating the clinical severity of β-thalassemia. Moreover, we observed the presence of hypochromic and microcytic red cells (thalassemia-like) or borderline MCV and MCH values in most nonthalassemic individuals as a result of KLF1 mutations. A recent study confirmed that compound heterozygotes for KLF1 mutations are able to cause hypochromic and microcytic anemia.12 Thus, we hypothesize that the frequent occurrence of the KLF1 mutations that affect protein function (particularly those disrupted the ZF domain of KLF1) might arise from natural selection, such as in the presence of malaria in tropical and subtropical areas.33 Our current results imply that KLF1 heterozygous mutations may be 1 important genetic modifier associated with the inherited hemoglobin disorders, particularly in β-thalassemia, among the southern Chinese population and potentially in other ethnic populations.
We have identified 64 KLF1 mutant alleles with 7 mutations (Figure 1; supplemental Table 2). The detailed genetic analysis of blood samples from 6 available families showed germ line origin of KLF1 mutations (supplemental Figure 2). The frameshift mutation, p.Gly176AlafsX179, was previously found in Thailand, Vietnamese, and Korean12,18 populations, and its finding in Chinese populations by us and others29,30 suggests that this mutation appears to be predominant in East Asian populations. In our study, only functionally effective KLF1 mutations were scored as positive. We typed all 64 subjects with KLF1 mutations according to the following 2 major predictors of altered function of KLF1: (1) presence of reduced CD44 expression on erythrocytes by flow-cytometric analysis; this was observed in the representative samples from all 6 types of mutations studied (supplemental Table 2 ; supplemental Figure 3), and (2) determination of elevated HbF levels (>1%), which was the case for all positive samples. All KLF1 variants identified in our study were submitted to the Single Nucleotide Polymorphism database (http://www.ncbi.nlm.nih.gov/snp). Decreased CD44 expression from ZF domain mutations or frameshift mutations disrupting ZF motifs of human KLF1 gene has been previously documented by several studies.11,13,16,31,32 Thus, use of a molecular assay to scan for KLF1 mutation–positive in combination with a flow-cytometric analysis for determining decreased CD44 expression will be a valuable tool for establishing functionally relevant KLF1 mutations. In addition to this, the In(Lu) phenotype had been used in British and French populations to screen for KLF1 mutations to obtain reliable prevalence figures.31,32 By enzyme-linked immunosorbent assay using labeled anti-ζ monoclonal antibody,34 elevated ζ-globin chain expression was found in 4 of the 7 available TI patients (P4, P6, P7, and P9; supplemental Table 3) with KLF1 heterozygous mutations, which has recently been reported in a transfusion-dependent hemolytic anemia caused by compound heterozygosity for KLF1 mutations.12
It is well known that the clinical manifestations of thalassemia can be modified by several genetic determinants, including genetic variants that modulate HbF levels, β-globin mutations, and α-globin gene copy numbers.2,-4 Based on previous studies that showed that the erythroid transcriptional factor KLF1 was a key regulator of the γ- to β-globin gene switching9,10 and KLF1 haploinsufficiency correlated with hereditary persistence of HbF caused by decreased BCL11A expression,16 we decided to investigate the role of KLF1 mutations in modulating the clinical and hematological severity of thalassemia. To the best of our knowledge, this is the first study reporting the clinical features of 12 β-thalassemia patients coinherited with KLF1 mutations, which had a mean Hb concentration of 82.57 ± 7.81 g/L and a mean HbF level of 39.58 ± 21.80 g/L that were indicative of the amelioration of β-thalassemia severity by KLF1 mutations. Multivariate Cox regression analysis showed that the modifier of KLF1 mutations had an effect on the clinical severity of β-thalassemia phenotype (Table 2), and univariate analysis using a matched case-control (Table 3) and transfusion-free survival curve (Figure 3) supported results of the multivariate analysis. In our case, 2 KLF1 mutations, p.Gly176AlafsX179 and p.His299Asp, were verified to be functional variants of KLF1, which result from generating a truncated protein with loss of the 3 ZF motifs downstream by frameshift mutation or abnormal structural changes in the ZF1 domain of KLF1 by missense mutation. The clinical features of our KLF1 mutation–positive patients might be ascribed to suppression of BCL11A expression and elevated γ-globin levels because of an altered DNA-binding KLF1domain and impaired transactivation KLF1capacity, because a similar result from KLF1 mutations has been previously described.10,13,,,-17 Furthermore, our functional studies in K562 cells using p.His299Asp and p.Gly176AlafsX179 mutant KLF1 constructs support such a possibility (data not shown). In addition, 3 subjects (patients P1, P2, and P8) with the β0/β+ genotype and 2 (P9 and P10) who coinherited HBA deletions had a very mild, transfusion-independent phenotype, highlighting that a combination of KLF1 mutations and known modifiers can greatly modify the clinical phenotype. In conclusion, we have defined a novel and important genetic determinant, KLF1 ZF heterozygous mutation, which contributes to TI. This knowledge should help in clinical diagnoses and genetic counseling as well as enable design of appropriate and personalized transfusion programs for thalassemia patients with KLF1 mutations. Using decreased CD44 expression as a surrogate for KLF1 mutations could be a clinically important tool to evaluate individuals with unexplained TI. We suggest that it is important to test for KLF1 mutations to ensure accurate clinical assessment in those TI patients with variable HbF increases because β-thalassemia that benefits from increased HbF can be caused by several genetic factors.
This study provides an overview of the important hematological features of individuals with KLF1 heterozygous mutations. Our finding of borderline MCV and MCH values in normal carriers with KLF1 mutations validates previous findings of borderline HbA2 levels and elevated HbF levels resulting from KLF1 mutations.16,,-19 Furthermore, we noted that a heterozygote KLF1 mutation coinherited with β-globin mutations can lead to a remarkable increase in both HbA2 and HbF levels, compared with those with only KLF1 heterozygous mutations. These 2 phenotypic outcomes are most likely a result of the cumulative effect of the heterozygous β-thalassemia and KLF1 mutations. At the phenotypic level with respect to HbA2 and HbF, there are no differences among subjects with different KLF1 and β-globin mutations. Of interest, in the light of reanalysis of previously published data (supplemental Table 4), we found that 10 KLF1 target genes associated with red blood cell phenotypes might underlie the decreased MCV and MCH values. This could explain, at least in part, our findings in normal individuals with a loss-of-function mutation in KLF1.35,36 However, the underlying mechanisms remain to be further investigated. These findings are likely to be useful for the accurate determination of β-thalassemia carriers in screening programs.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members who participated in this study; we also thank Yaoqin Gong for assistance of sample collection in northern China, and Ping Fang, Yihong Li, and Xuelian Zhang for experiment assistance.
This work was supported by grants from the National Natural Science Foundation of China-Guangdong Joint Fund (U1201222), NSFC (81260093), the National Key Technology Research & Development Program (2012BAI09B01), the Doctoral Fund of Ministry of Education of China-Key Program of Priority Fields (20134433130001), and National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (grant DK26263).
Authorship
Contribution: X.Z., R.C., C.Z., Y.Z., Q.L., L.L., and T.Y. collected samples and clinical data; X.Z. performed clinical classifications, diagnosis, and management of thalassemia; L.Y., X.M., and D.L. performed laboratory and DNA analysis; X.W. and J.H. did molecular diagnosis of thalassemia; N.M. and X.A. designed the study, interpreted the data, and edited the paper; and D.L. and X.X. designed the study, analyzed and interpreted the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiangmin Xu, Department of Medical Genetics, School of Basic Medical Sciences, Southern Medical University, Guangzhou, 510515, Guangdong, P.R. China; e-mail: gzxuxm@pub.guangzhou.gd.cn.