Key Points
PML/RARA loss or detachment from target promoters suffices to differentiate APL cells.
PML/RARA degradation by arsenic thus explains arsenic-induced differentiation.
Abstract
PML/RARA, a potent transcriptional inhibitor of nuclear receptor signaling, represses myeloid differentiation genes and drives acute promyelocytic leukemia (APL). Association of the retinoid X receptor-α (RXRA) coreceptor to PML/RARA is required for transformation, with RXRA promoting its efficient DNA binding. APL is exquisitely sensitive to retinoic acid (RA) and arsenic trioxide (arsenic), which both trigger cell differentiation in vivo. Whereas RA elicits transcriptional activation of PML/RARA targets, how arsenic triggers differentiation remains unclear. Here we demonstrate that extinction of PML/RARA triggers terminal differentiation in vivo. Similarly, ablation of retinoid X receptors loosens PML/RARA DNA binding, inducing terminal differentiation of APL cells ex vivo or in vivo. RXRA sumoylation directly contributes to PML/RARA-dependent transformation ex vivo, presumably by enhancing transcriptional repression. Thus, APL differentiation is a default program triggered by clearance of PML/RARA-bound promoters, rather than obligatory active transcriptional activation, explaining how arsenic elicits APL maturation through PML/RARA degradation.
Introduction
Acute promyelocytic leukemia (APL) is characterized by gene fusions involving retinoic acid receptor-α (RARA) gene. The most common t(15;17) translocation fuses PML to RARA, yielding the PML/RARA fusion oncoprotein. PML, the key organizer of nuclear bodies (NB), is involved in redox sensing and hence confers sensitivity to arsenic.1-3 PML/RARA is a potent transcriptional repressor of retinoic acid (RA) signaling that interferes with gene expression programs involved in both hematopoietic progenitor self-renewal and terminal myeloid cell differentiation.4,5 Treatment of APL patients with RA induces terminal differentiation and transient remissions. Mechanistically, this is believed to reflect transcriptional reactivation of PML/RARA-silenced genes by RA, the ligand for RARA and PML/RARA,6 whereas RA-triggered PML/RARA degradation accounts for loss of self-renewal.7,8 Arsenic definitively cures a substantial proportion of patients as a single agent.9-11 Ex vivo studies have demonstrated that arsenic primarily induces apoptosis of APL cells,12 although subsequent studies demonstrated partial and complete differentiation ex vivo and in vivo, respectively.13 Molecularly, arsenic degrades PML/RARA but otherwise does not directly affect transcriptional regulation by RARA, raising the issue of the basis for differentiation.14,15 Arsenic also acts on normal PML to promote loss of self-renewal, likely explaining its clinical potency.7,16
In normal cells, RARA is always associated with retinoid X receptors (RXRs) the universal partners of type II nuclear receptors to bind their responsive elements. RA binds to retinoic acid receptors (RARs), inducing conformational changes within the RXR/RAR complexes, thus resulting in enhancement of DNA binding, release of corepressors, recruitment of coactivator complexes, and transcriptional activation of target genes. The RA pathway plays an important role in determining and regulating differentiation pathways such as myelopoiesis.17-20 For example, RARA regulates the kinetics of granulocytic differentiation,19 whereas RXRA promotes monocytic differentiation.17 In contrast to RARA, PML/RARA homodimers bind DNA independently of RXRs ex cellulo,21,22 suggesting than RXRs are not implicated in the transformation process. RXR agonists, however, may activate transcription from a PML/RARA-specific response element22,23 and may efficiently initiate APL cell differentiation.24,25 RXRA interacts with PML/RARA,21,22,26 and chromatin immunoprecipitation (ChIP)-sequencing studies demonstrate that RXRA is always found at PML/RARA-bound promoters.27-29 The presence of RXRA in the PML/RARA complex greatly enhances its ability to bind DNA and to recognize highly degenerate sites.21,22 Additionally, RXRA may provide an independent repression signal through its sumoylation, the latter being sharply enhanced by PML/RARA.25,30 Accordingly, a PML/RARA mutant defective for RXR binding fails to initiate APL in vivo,25 whereas silencing of RXRA induces apoptosis ex vivo.31
Here, we show that RXR excision from PML/RARA-driven APL relaxes association of the fusion and its target genes, inducing terminal differentiation of leukemic cells. Inactivation of PML/RARA by RNA interference also triggers differentiation. These unexpected observations provide a mechanistic basis for arsenic-induced APL differentiation.
Materials and methods
Cell culture and retroviral transductions
Complimentary (c)DNAs encoding PML/RARA and RARA were previously described.25 The Cre-ERT2 cDNA was inserted in MSCV-IRES-cRed (kind gift of R. Williams). The murine RXRA WT and RXRA K113R cDNAs (kind gift of C. Egly) were inserted in MSCV-Babe-IRES-cRed. The MSCV-FLT3ITD-IRES-GFP construct was previously described.32 Short hairpin (sh)RNA constructs were purchased from Sigma-Aldrich. The two shRNA constructs targeting human PML (5′-CACCCGCAAGACCAACAACAT-3′, 5′-GTGTACCGGCAGATTGTGGAT-3′), were inserted into pLKO.1-CMV-tGFP.
Bone marrow cells from 5-fluorouracil–treated mice (C57BL/6JRj) depleted in mature myeloid and lymphoid cells were cultured overnight with interleukin (IL)-3, IL-6 (10 ng/mL), and stem cell factor (100 ng/mL) (Eurobio Abcys). Bone marrow progenitors were infected twice by spinoculation with retroviral supernatant produced with Platinum-E cells. On the day following the second spinoculation, an equivalent 5 × 103 cells was seeded per 1.1 mL of Methocult M3231 methylcellulose medium (Stem Cell Technologies) supplemented with 10 ng each of murine recombinant IL-3, IL-6, granulocyte macrophage–colony-stimulating factor and stem cell factor.25 One week after the initial infection, RXRA-expressing mCherry-positive cells were sorted and replated. One week later, secondary colonies were counted and cells were analyzed by May-Grünwald-Giemsa (MGG) staining and western blot.
Cell cycle profiles were assessed using propidium iodide.
Protein and cell analyses
Cell lysates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred on nitrocellulose membranes. We carried out detection with the chemiluminescent substrate SuperSignal West Pico or Femto (Pierce Biotechnology). Antibody RP115 (RARA) was kindly provided by P. Chambon, anti-RXRa D-20 was purchased from Santa-Cruz, and anti-β-actin (20-33) and anti– glyceraldehyde-3-phosphate dehydrogenase (71.1) were purchased from Sigma-Aldrich.
H1299 cells stably expressing HIS-SUMO1 or HIS-SUMO2 were cotransfected (+) or not (−) with PML/RARA and RXR WT or K113R mutant. HIS-sumoylated proteins were purified by nickel pull-down33 and probed with the corresponding antibodies. Cos-7 cells were cotransfected with pSG5-RARA and pSG5-RXRA WT or pSG5-RXRA K113R constructs and treated with 0.1 mg/mL cycloheximide (Calbiochem) and a solution of 1µM RA for 0, 1, 3, and 6 hours. RARA and RXRA stability were then analyzed by western blot.
For flow cytometry analysis, cellular Fc receptors were blocked with rat immunoglobulin G. We then carried out immunophenotypic analysis using fluorochrome-conjugated monoclonal antibodies to Mac1 and Gr-1 (clone RB6-8-C5; eBioscience). Staining was done at 4°C for 20 minutes. We washed the cells twice and resuspended them in Hanks balanced salt solution with 2% fetal bovine serum and 0.5 µg/mL-1 propidium iodide. We gated dead cells out by high propidium iodide staining and forward light scatter. Immunofluorescences were performed using a homemade antibody against PML.34 For morphological analysis we performed MGG staining.
Transgenic mice and in vivo animal treatment
PML/RARA transgenic mice35 were crossed with RXRAf/f RXRBf/f RXRG−/− mice.36 Bone marrow progenitors from RXRAf/f RXRBf/f RXRG−/− PML/RARA were infected with retroviruses encoding FLT3-ITD, yielding a transplantable APL (Figure 2A). The latter was transduced by retroviruses encoding Cre-ERT2, and the leukemias were then serially transplanted in NMRI-Nude mice. Animal handling was done according to the guidelines of institutional animal care committees, using protocols approved by the Comité Régional d’Ethique l’Expérimentation Animale No. 4. Mice were treated with tamoxifen (4-OHT; Sigma-Aldrich) by daily intraperitoneal injections, 1 mg/day for 5 days, and RA or arsenic as previously.37
RT-qPCR, ChIP, and array experiments
Immortalized RARs−/− mouse embryonic fibroblasts (MEFs) were retrovirally transduced with RXRA or RXRAK113R in presence or absence of PML/RARA and treated or not with RA (1 µM) overnight. Total RNAs were isolated using the RNeasy kit (Qiagen) and first-strand cDNAs were synthetized using the SuperScript III reverse transcriptase (Invitrogen). Rarb and Act probes and primers for TaqMan assays were from Applied Biosystems. Quantification was performed by real-time quantitative polymerase chain reaction (RT-qPCR) using the 7500 Fast Real-Time PCR system.
ChIP was performed using the LowCell ChIP kit (Diagenode) according to manufacturer’s recommendations, except that chromatin was first incubated with antibodies overnight and then for 2 hours with beads. The following antibodies were used: anti-GFP-FL and RXR ΔN 197 from Santa-Cruz; and anti-H3, anti-H3 trimethyl K4, and anti-PML+RARA fusion from Abcam. RT-qPCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems), and relative occupancy was calculated as fold enrichment over the control antibody anti-GFP. Amplicon for Rarb on chromosome 14: from 16 575 561 to 16 575 501; amplicon for Hoxa1 on chromosome 6: from 52 153 671 to 52 153 580. RXRA- and RXRB-excised DNAs were quantified as above. The Dpp9 gene was used as internal control.
Expression arrays and statistical analysis
RNA samples for array experiments were isolated as above and were hybridized on Affymetrix Human or Mouse Gene 1.1 ST Arrays. Log2 measures were obtained using robust multi-array average (RMA) normalization. The threshold was 2 for humans and 1.8 for mice, unless otherwise indicated. Expression profiles were normalized in batch and independently in each series using the robust multi-array average method, yielding normalized log2 intensity measures. Log2 ratios were then obtained from these log2 intensities, using control samples as reference. To assess whether or not the genes upregulated or downregulated in the two conditions (A and B) and to determine whether or not they were significantly overlapping, we ranked the genes for both conditions and performed a χ2 test measuring the overlap between the top N genes in condition A and the top N genes in condition B (N ranging from 100 to 1200).
Results
RXRA sumoylation favors transformation of mouse primary hematopoietic progenitors
PML/RARA enhances RXRA sumoylation.25 Purification of His-SUMO1 or His-SUMO2 conjugates demonstrated that the PML/RARA-enhanced RXRA sumoylation occurs on lysine K113 (data not shown; Figure 1A). We observed higher RA-dependent transcriptional activation of Rarb, a canonical RA primary target gene,38 in MEFs transduced by RXRAK113R compared with RXRA (Figure 1B, left panel), a finding in line with studies showing that RXRA sumoylation contributes to transcriptional repression.30 We also detected a higher RA-dependent transcriptional activation in the presence of PML/RARA (Figure 1B, right panel) and argue that RXRA sumoylation is involved in PML/RARA-dependent transcriptional repression. Thus, PML/RARA-enhanced RXRA sumoylation contributes to PML/RARA-mediated repression.
RXRA sumoylation enhances PML/RARA-mediated immortalization. (A) Sumoylation profile of transfected RXRA WT or K113R in H1299 cells stably expressing HIS-SUMO1 and expressing (+) or not (−) PML/RARA. (B) Analysis of rarb gene activation by RA (1 µM) in MEFs expressing RXRA or RXRA K113R in the presence (Rars−/− MEFs, right panel) or absence (MEFs, left panel) of PML/RARA. Error bars represent standard deviations of 3 independent biological replicates. Significance of observed differences was evaluated using Student t test. *P < .05. (C) Progenitors transduced with murine RXRA WT or K113R and PML/RARA, RARA, or MLL-ENL were analyzed for clonogenicity. Error bars represent standard deviations of 3 independent biological replicates. Significance of observed differences was evaluated using Student t test. *P < .05; **P < .01; ***P < .001. (D) Progenitors transduced with murine RXRA WT or K113R and PML/RARA, RARA, or MLL-ENL were analyzed for morphology (bottom bar, 10 µm; MGG stain). (E) Protein expression by western blot (black dividing lines show grouping of images from different parts of the same exposed film). (F) Cos-7 cells coexpressing RARA and RXRA or RXRA K113R were treated with cycloheximide (0.1 mg/mL) and RA (1 µM) for the indicated time, and the half-life of RXRA and RARA proteins was analyzed by western blot.
RXRA sumoylation enhances PML/RARA-mediated immortalization. (A) Sumoylation profile of transfected RXRA WT or K113R in H1299 cells stably expressing HIS-SUMO1 and expressing (+) or not (−) PML/RARA. (B) Analysis of rarb gene activation by RA (1 µM) in MEFs expressing RXRA or RXRA K113R in the presence (Rars−/− MEFs, right panel) or absence (MEFs, left panel) of PML/RARA. Error bars represent standard deviations of 3 independent biological replicates. Significance of observed differences was evaluated using Student t test. *P < .05. (C) Progenitors transduced with murine RXRA WT or K113R and PML/RARA, RARA, or MLL-ENL were analyzed for clonogenicity. Error bars represent standard deviations of 3 independent biological replicates. Significance of observed differences was evaluated using Student t test. *P < .05; **P < .01; ***P < .001. (D) Progenitors transduced with murine RXRA WT or K113R and PML/RARA, RARA, or MLL-ENL were analyzed for morphology (bottom bar, 10 µm; MGG stain). (E) Protein expression by western blot (black dividing lines show grouping of images from different parts of the same exposed film). (F) Cos-7 cells coexpressing RARA and RXRA or RXRA K113R were treated with cycloheximide (0.1 mg/mL) and RA (1 µM) for the indicated time, and the half-life of RXRA and RARA proteins was analyzed by western blot.
We then tested the ability of the sumoylation-defective RXRAK113R mutant to modulate transformation of primary progenitors by PML/RARA, RARA, or the MLL/ENL fusion,39 which does not directly interact with nuclear receptor signaling. RXRA slightly diminished RARA- or PML/RARA-triggered clonogenic activity (Figure 1C), most likely through RARA or PML/RARA destabilization. RXRK113R reduced clonogenic activity even further (Figure 1C-D), although it paradoxically stabilized the driving oncoproteins. Moreover, a significant induction of basal differentiation was observed (Figure 1E). In contrast, no effect of RXRA or RXRAK113R was observed on MLL/ENL-transformed cells. Directly measuring the half-life of RARA and RXRA on RA exposure, we observed that RXRA sumoylation promotes their degradation (Figure 1F). Thus, RXRA not only contributes to DNA binding of the complex but its sumoylation also directly regulates PML/RARA-dependent differentiation block and clonogenic activity.
RXRA loss induces differentiation and apoptosis
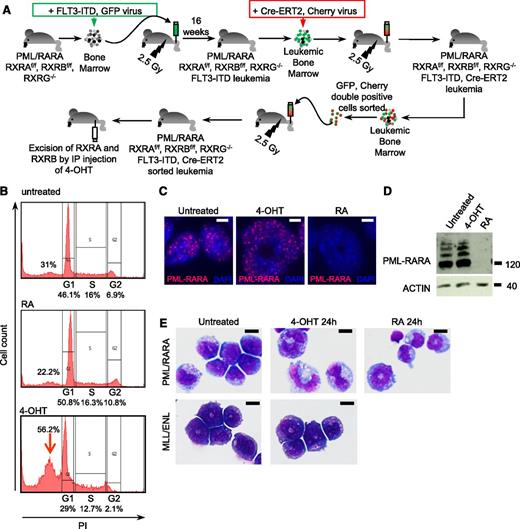
We next investigated the consequences of acute RXR ablation on survival and differentiation. We crossed PML/RARA transgenic mice driven by the MRP8 promoter35 with RXRAf/f RXRBf/f RXRG−/− mice.36 After backcrossing on the RXRAf/f RXRBf/f RXRG−/− mice, we obtained the PML/RARA transgene on a homozygous background for the RXR floxed and null alleles. Because those mice did not rapidly develop APL, we transduced their bone marrows with a retrovirus encoding FLT3-ITD, a constitutively activated kinase that accelerates progression to APL.32,40 Transduced marrows were transplanted in irradiated Nude mice, yielding aggressive APLs after 16 weeks that closely resemble previously studied ones (data not shown). These APLs were then transduced with a retrovirus coexpressing Cre-ERT2 and Cherry, yielding a transplantable APL in which 4-OHT induces ablation of both RXRA and RXRB (Figure 2A).
RXR loss induces growth differentiation and apoptosis of PML/RARA-transformed cells. (A) Generation of RXRAf/f RXRBf/f RXRG−/− PML/RARA FLT3-ITD Cre-ERT2 leukemia in mouse model. GFP-sorted spleen cells from leukemic mice were treated with 100 nM 4-OHT or 1 µM RA in culture and analyzed for cell cycle (B), PML/RARA expression (top bar, 10 µm) (C-D), and differentiation (E). RXRAf/f RXRBf/f RXRG−/− MLL/ENL Cre-ERT2 leukemia are shown as negative control (bar, 10 µm; MGG stain). IP, intraperitoneal; PI, propidium iodide.
RXR loss induces growth differentiation and apoptosis of PML/RARA-transformed cells. (A) Generation of RXRAf/f RXRBf/f RXRG−/− PML/RARA FLT3-ITD Cre-ERT2 leukemia in mouse model. GFP-sorted spleen cells from leukemic mice were treated with 100 nM 4-OHT or 1 µM RA in culture and analyzed for cell cycle (B), PML/RARA expression (top bar, 10 µm) (C-D), and differentiation (E). RXRAf/f RXRBf/f RXRG−/− MLL/ENL Cre-ERT2 leukemia are shown as negative control (bar, 10 µm; MGG stain). IP, intraperitoneal; PI, propidium iodide.
We first assessed the effects of RA and 4-OHT in ex vivo cultures of these primary blasts from murine APLs. APL spleen cells proliferated, even in presence of RA. Ablation of RXRs by 4-OHT induced cell death (data not shown), as previously shown in ex vivo transformed cells.31 Yet, loss of cell viability was not observed for MLL/ENL-transformed cells (data not shown). Apoptosis induction by 4-OHT was confirmed by appearance of a sub-G1 peak (Figure 2B, arrow). Although RA triggered PML/RARA degradation, RXR ablation actually stabilized PML/RARA (data not shown; Figure 2C-D), in keeping with the fact that RXRA facilitates RARA degradation.41 Strikingly, 4-OHT treatment led to terminal differentiation of APL blasts, very similar to that triggered by RA. In contrast, the differentiation status of MLL/ENL-transformed cells was unaffected by ablation of RXRs (Figure 2E). Thus, loss of RXRs triggers apoptosis and differentiation of PML/RARA-transformed cells.
We then investigated the effect of RXR ablation in vivo. 4-OHT treatment a few days before the death of untreated animals inoculated with APL cells significantly increased survival (Figure 3A, left) and led to rapid APL regression (Figure 3A, right). When 4-OHT–treated mice ultimately died of APL, blasts were primarily Cherry-negative, pointing to the selection of cells that had silenced CRE (data not shown). In vivo ablation of RXRA and RXRB with 4-OHT treatment was complete, as demonstrated by qPCR. Unexpectedly, however, a basal spontaneous hemiablation of RXRB was constantly noted (Figure 3B). At the protein level, 4-OHT triggered the appearance of truncated RXRA-reactive protein species but did not affect PML/RARA expression (Figure 3C). Critically, terminal blast differentiation was again observed 2 days post 4-OHT exposure in vivo (Figure 3D). Morphologic differentiation was accompanied by loss of c-kit expression and enhanced Mac1 and Gr1 expression (Figure 3E), very similar to RA treatment (Figure 3F). Acute ablation of RXRs in MLL/ENL-transformed cells in vivo did not induce differentiation or tumor regression (data not shown). Thus, in vivo loss of RXRs in APL blasts induces rapid terminal differentiation and APL regression.
RXR loss induces in vivo differentiation. (A) Left panel: Survival of mice inoculated with APL cells derived from RXRAf/f RXRBf/f RXRG−/− PML/RARA FLT3-ITD Cre-ERT2 mice, untreated or treated with 4-OHT. Right panel: Cherry-positive cells after 5 days of treatment. After 2 days of treatment with 4-OHT or RA in vivo, spleen cells were GFP sorted and analyzed for RXRA and RXRB ablation (B), protein expression (black dividing lines show grouping of images from different parts of the same exposed film) (C), differentiation by MGG staining (top bar, 10 µm) (D), and flow cytometry (E-F).
RXR loss induces in vivo differentiation. (A) Left panel: Survival of mice inoculated with APL cells derived from RXRAf/f RXRBf/f RXRG−/− PML/RARA FLT3-ITD Cre-ERT2 mice, untreated or treated with 4-OHT. Right panel: Cherry-positive cells after 5 days of treatment. After 2 days of treatment with 4-OHT or RA in vivo, spleen cells were GFP sorted and analyzed for RXRA and RXRB ablation (B), protein expression (black dividing lines show grouping of images from different parts of the same exposed film) (C), differentiation by MGG staining (top bar, 10 µm) (D), and flow cytometry (E-F).
RXRA ablation detaches PML/RARA from its target sites
The presence of 4 DNA-binding domains in the PML/RARA-RXRA complex greatly facilitates its DNA binding, notably on noncanonical sites.22 We thus examined by ChIP whether 4-OHT would modify the occupancy or the chromatin environment of two primary targets, Rarb and Hoxa1.27 Ex vivo, 4-OHT sharply decreased the amount of RXRA and PML/RARA associated with their binding sites. The remaining precipitated RXRA may be either residual full-length RXRA or PML/RARA-bound truncated RXRA. A small decrease in histone H3 was reproducibly observed, with a significant increase in histone H3 K4 trimethylation (Figure 4A). These observations are consistent with the proposal that, at least for a subset of targets, RXRA loss is accompanied by reduced DNA binding of PML/RARA and by transition of chromatin toward an active state.
RXR loss induces PML/RARA detachment from target genes. (A) APL mice were treated or not for 48 hours ex vivo with 4-OHT. Immunoprecipitated DNA with the indicated antibodies was analyzed by qPCR as indicated. Error bars represent standard deviations of 3 independent biological replicates, and significance was assessed using Student t test. *P < .05; **P < .01. (B) Comparison of gene expression activation (left; 1.7-fold up) or repression (right; twofold down) after 16 hours of ex vivo treatment with 4-OHT (red) or RA (blue). See supplementary Figure 1 for primary data. (C) Tgm2 gene expression after 16 hours of ex vivo treatment with 4-OHT or RA (2 biological replicates).
RXR loss induces PML/RARA detachment from target genes. (A) APL mice were treated or not for 48 hours ex vivo with 4-OHT. Immunoprecipitated DNA with the indicated antibodies was analyzed by qPCR as indicated. Error bars represent standard deviations of 3 independent biological replicates, and significance was assessed using Student t test. *P < .05; **P < .01. (B) Comparison of gene expression activation (left; 1.7-fold up) or repression (right; twofold down) after 16 hours of ex vivo treatment with 4-OHT (red) or RA (blue). See supplementary Figure 1 for primary data. (C) Tgm2 gene expression after 16 hours of ex vivo treatment with 4-OHT or RA (2 biological replicates).
To assess any global change in transcriptional regulation on RXR ablation, we performed transcriptomic arrays comparing ex vivo 4-OHT and RA treatments of primary APL cells at 4, 8, and 16 hours. A set of genes was very reproducibly activated or repressed after RA administration or ablation of RXRs in a time-dependent manner (as shown in supplemental Figure 1 on the Blood Web site). Although genes induced or repressed by 4-OHT did not match the genes most potently modulated by RA (supplemental Figure 1), a significant association was found between genes repressed over twofold by either treatment (Figure 4B), as well as between the 200 to 1000 top genes activated or repressed by RA and 4-OHT (data not shown). RXR-sensitive genes could not be identified as corresponding to a defined pathway. Finally, RXR ablation blunted the basal expression levels of PML/RARA-activated targets such as Tgm2 (Figure 4C).42 Collectively, these findings suggest that in APL, myeloid differentiation occurs through a series of subtle transcriptional changes, rather than through massive activation of a specific master pathway.
shRNA inactivation of PML/RARA triggers differentiation in vivo
To directly investigate the effect of PML/RARA loss in established APL cells, we used lentiviral constructs expressing either a scrambled shRNA or a shRNA specifically targeting human PML gene, thus destabilizing PML/RARA expression without affecting murine pml (data not shown). We transduced the vectors in murine APLs with identical efficiency and re-injected them in irradiated recipients. After 3 weeks, we observed a considerably lower proportion of GFP-expressing cells in the marrows of shRNA targeting PML/RARA compared with control shRNA mice (Figure 5A), formally demonstrating that sustained PML/RARA expression is required for APL growth. The majority of the remaining GFP-labeled cells had lost PML/RARA microspeckled staining but, critically, now displayed terminal granulocytic differentiation (Figure 5B-C), demonstrating that in vivo, downregulation of PML/RARA expression promotes terminal myeloid differentiation.
PML/RARA extinction triggers APL differentiation in vivo. Bone marrow APL blasts were infected with a GFP-expressing lentivirus encoding a scrambled shRNA sequence (Ctrl) or an shRNA sequence directed against human PML and injected in secondary recipients. Inoculated animals were analyzed 21 days after engraftment (6 mice, 2 independent experiments) to determine the proportion of GFP-positive cells in the marrow (A), PML/RARA or GFP immunofluorescence (bottom bar, 10 µm; arrow points to differentiated GFP-positive cells with PML/RARA extinction) (B), and fluorescence-activated cell sorter analysis of GFP-positive cells (C). (D) Gene expression analysis 6 or 12 hours after RA (blue) or As2O3 (red) treatment in human APL NB4 cell line (top row) or of APL mice (bottom row). See supplemental Figure 2 for representative data. (E) χ2 test P values (median P value for the different thresholds tested: N = 100-1200) for association of the upregulated and downregulated genes. Sample 935 refers to murine APLs; AR10 and AR100 indicate the doses of RA administered.
PML/RARA extinction triggers APL differentiation in vivo. Bone marrow APL blasts were infected with a GFP-expressing lentivirus encoding a scrambled shRNA sequence (Ctrl) or an shRNA sequence directed against human PML and injected in secondary recipients. Inoculated animals were analyzed 21 days after engraftment (6 mice, 2 independent experiments) to determine the proportion of GFP-positive cells in the marrow (A), PML/RARA or GFP immunofluorescence (bottom bar, 10 µm; arrow points to differentiated GFP-positive cells with PML/RARA extinction) (B), and fluorescence-activated cell sorter analysis of GFP-positive cells (C). (D) Gene expression analysis 6 or 12 hours after RA (blue) or As2O3 (red) treatment in human APL NB4 cell line (top row) or of APL mice (bottom row). See supplemental Figure 2 for representative data. (E) χ2 test P values (median P value for the different thresholds tested: N = 100-1200) for association of the upregulated and downregulated genes. Sample 935 refers to murine APLs; AR10 and AR100 indicate the doses of RA administered.
Reassessing the basis for arsenic-induced differentiation
Similar to RXRA ablation or PML/RARA silencing, arsenic triggers differentiation and apoptosis whose respective extents, however, vary greatly with the experimental system used.12,13,15 To investigate the role of arsenic-initiated PML/RARA loss in transcriptional regulation, we first compared the mRNA expression profiles of the NB4 APL cells treated with RA or arsenic. Upregulated and downregulated genes in response to these two unrelated agents were found to be extremely redundant (χ2 test; P < 1e-50), whatever the time considered (Figure 5D top and 5E). Critically, a similar situation was observed in APL mice (Figure 5D bottom, 5E, and supplemental Figure 2).7 Thus, RA and arsenic regulate a common set of genes, most likely by a promoter clearance mechanism.
Discussion
Our results demonstrate that RXRA and its sumoylation play a key role in PML/RARA-initiated transformation. Unexpectedly, we observed that loss of RXRs leads to the terminal differentiation of APL blasts. Extinction of PML/RARA by RNA interference also induced differentiation of APL blasts. Our results therefore imply that promoter clearance is sufficient to differentiate APL cells, thus explaining how arsenic induces myeloid maturation in vivo.
PML/RARA interacts with the SUMO E2 enzyme UBC9 through its RING domain and enhances RXRA sumoylation.25 Here, we demonstrate that PML/RARA-enhanced RXRA modification occurs on lysine K113, blunts transcriptional activation and, critically, favors the ex vivo transformation of primary mouse hematopoietic progenitors. Thus, PML/RARA recruits the sumoylation machinery onto RXRA to enhance repression by the PML/RARA-RXRA complex. Such recruitment of the sumoylation machinery onto chromatin may also be involved in the modification of other histone or nonhistone proteins. Sumoylation of transcription factors and histones is primarily responsible for transcriptional repression,43 at least in part by antagonizing activating marks such as acetylation or ubiquitylation.44 RXRA sumoylation–dependent decrease in the transformation ability of PML/RARA is accompanied by the paradoxical stabilization of the fusion. Moreover, RXRA sumoylation is an important determinant of basal or RA-induced RARA degradation (Figure 1F). Retinoids or rexinoids induce the degradation of the RARA/RXRA complex,6,41 and posttranslational modifications have been implicated in activation-triggered degradation of many transcription factors.45 Thus, RXRA sumoylation could also be an important determinant of RXRA and RARA catabolism.
Previous studies have implicated RXRA in several aspects of APL pathogenesis.25,26,31,46-49 To examine the cellular effects of acute RXR loss, we designed a genetic system allowing excision of both RXRA and RXRB in murine APLs. Administration of 4-OHT induced terminal differentiation together with apoptosis, both ex vivo and in vivo. Loss of RXRs also initiated detachment of PML/RARA from some target genes, supporting the observation that the PML/RARA-RXRA complex that contains at least four DNA-binding domains has an enhanced affinity for DNA compared with PML/RARA homodimers.22 Moreover, acute RXRA excision enhances H3 K4 trimethylation, a modification associated with transcriptional activation. Although RXRA downregulation is required for granulocytic differentiation,17 we observed differentiation with some monocytic features on RXRA ablation (Figure 2E). Recruitment of the RXRA/RARA complex onto the RARB promoter after RA-induced PML/RARA degradation was proposed to be key for transcriptional reactivation and RA-response.50 In contrast with this proposal, our observations of terminal differentiation upon RXR excision do not favor a model in which transcriptional reactivation by RARA/RXRA is essential for differentiation (Figure 6).
Model. Loss of PML/RARA DNA binding through RXRA ablation, PML/RARA extinction, or RA/arsenic-triggered catabolism clears target genes from the PML/RARA repressor, triggering differentiation. PML/RARA loss also induces apoptosis. The latter may be blocked by RA, growth factors, or the microenvironment. RARE, RA responsive elements.
Model. Loss of PML/RARA DNA binding through RXRA ablation, PML/RARA extinction, or RA/arsenic-triggered catabolism clears target genes from the PML/RARA repressor, triggering differentiation. PML/RARA loss also induces apoptosis. The latter may be blocked by RA, growth factors, or the microenvironment. RARE, RA responsive elements.
PML/RARA degradation is essential to loss of self-renewal, whereas direct transcriptional activation is believed to trigger APL differentiation.6,7,51,52 Our observations of terminal differentiation on PML/RARA silencing or its detachment from its target promoters suggest an alternative or a complementary model wherein impeding PML/RARA binding to DNA suffices to initiate differentiation by a promoter clearing mechanism (Figure 6). This model explains the overlap between RA and arsenic targets and provides a molecular mechanism for arsenic-induced APL differentiation in vivo.
PML/RARA has strong antiapoptotic effects53 so that, in the absence of survival signals, loss of PML/RARA triggers apoptosis (Figures 2C and 6).24,54,55 RA activates potent antiapoptotic genes such as MCL1,56 allowing the progression of an unabridged differentiation program ex vivo. In RXR-excised or arsenic-treated cells ex vivo, the proapoptotic signals triggered by loss of PML/RARA DNA binding are unopposed, precipitating cell death (Figure 6). Yet, growth factors enhance survival and promote terminal differentiation by arsenic ex vivo.57,58 Our results could thus explain the balance between treatment-induced differentiation and apoptosis in arsenic-treated APLs.14
Collectively, our studies establish that PML/RARA clearance not only abrogates self-renewal7,8 but also suffices to initiate APL differentiation. These findings demonstrate that APL maturation is a default program and further unifies the mode of action of RA and arsenic.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank P. Chambon for reagents, M. Pla for the animal facility, the genomic center of the Curie Institute for arrays, N. Setterblad for imaging, and V. Lallemand, J. Ghysdael, K. Rice, and F. Sigaux for critical reading of the manuscript.
This work was supported by the Ligue Nationale contre le Cancer, INSERM, the Centre National de la Recherche Scientifique, Université Paris Diderot, the Institut Universitaire de France, the Institut National du Cancer, the Association pour la Recherche contre le Cancer (Prix Griffuel), and the European Research Council (senior grant 268729 – STEMAPL) (H.d.T.).
Authorship
Contribution: J.H., A.V.-P., J.A., and A.d.R. performed experiments, analyzed data, and wrote the manuscript; L.P. and M.L.B. performed experiments; D.M. provided reagents; and H.d.T. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hugues de Thé, INSERM UMR 944, Hopital St. Louis, 1 Avenue Claude Vellefaux 75475, Paris Cedex 10, France; e-mail: hugues.dethe@inserm.fr.
References
Author notes
A.V.-P. and J.H. contributed equally as first authors.