Key Points
Whole-transcriptome sequencing reveals NPM1-TYK2 gene fusion in cutaneous CD30-positve lymphoproliferative disorders.
NPM1-TYK2 activates STAT signaling and is a therapeutic target in a subset of cutaneous CD30-positive lymphoproliferative disorders.
Abstract
The spectrum of cutaneous CD30-positive lymphoproliferative disorders (LPDs) includes lymphomatoid papulosis and primary cutaneous anaplastic large cell lymphoma. Chromosomal translocations targeting tyrosine kinases in CD30-positive LPDs have not been described. Using whole-transcriptome sequencing, we identified a chimeric fusion involving NPM1 (5q35) and TYK2 (19p13) that encodes an NPM1-TYK2 protein containing the oligomerization domain of NPM1 and an intact catalytic domain in TYK2. Fluorescence in situ hybridization revealed NPM1-TYK2 fusions in 2 of 47 (4%) primary cases of CD30-positive LPDs and was absent in other mature T-cell neoplasms (n = 151). Functionally, NPM1-TYK2 induced constitutive TYK2, signal transducer and activator of transcription 1 (STAT1), STAT3, and STAT5 activation. Conversely, a kinase-defective NPM1-TYK2 mutant abrogated STAT1/3/5 signaling. Finally, short hairpin RNA–mediated silencing of TYK2 abrogated lymphoma cell growth. This is the first report of recurrent translocations involving TYK2, and it highlights the novel therapeutic opportunities in the treatment of CD30-positive LPDs with TYK2 translocations.
Introduction
Recurrent chromosomal translocations frequently underlie the pathogenesis of several hematopoietic malignancies and often define molecular subtypes with distinct biological behavior.1,2 Frequently, these translocations target tyrosine kinases resulting in constitutive activation and promotion of oncogenesis.3 Cutaneous CD30-positive lymphoproliferative disorders (LPD) represents a clinicopathologic spectrum including lymphomatoid papulosis (LYP) and primary cutaneous anaplastic large cell lymphoma (ALCL).4 Gene fusions targeting tyrosine kinases underlying the pathogenesis of CD30-positive LPD have not been described.
Methods
Patient biopsy samples were obtained with institutional review board approval. Complete description of methods and clinical samples are presented in the supplemental Methods (available on the Blood Web site). RNA was subjected to chimera analysis by producing paired-end libraries sequenced on the Illumina Genome Analyzer II. Sequencing data were analyzed using custom bioinformatics tools and Chimerascan software.5 Sequencing confirmation of NPM1-TYK2 fusion transcripts was achieved using SYBR Green–based quantitative real-time polymerase chain reaction (PCR) assays and Sanger sequencing of amplicons by reverse transcription PCR (RT-PCR (see supplemental “Materials”). Fluorescence in situ hybridization (FISH) was performed on tissue microarrays of primary patient samples using standard methods. TYK2 break-apart FISH and NPM1-TYK2 fusion FISH assays were designed to detect TYK2 rearrangements and NPM1-TYK2 fusions, respectively (see supplemental Methods). Immunohistochemistry for phosphorylated signal transducer and activator of transcription 5 (pSTAT5) (AX1; Advantex BioReagents, Houston, TX) and NPM1 (EP1848Y; Abcam, Cambridge, MA) was performed on primary patient samples. Mutations in TYK2 were assessed by targeted Sanger sequencing of the genomic DNA. The NPM1-TYK2 fusion gene was amplified from MyLa cells and cloned into a mammalian expression vector for functional studies. Stable MyLa cell lines depleted of TYK2 were generated using lentiviral-mediated gene transduction6 of short hairpin RNAs (shRNAs) and used in cell proliferation assays.
Results and discussion
To discover novel gene fusions that may be involved in the pathogenesis of cutaneous T-cell lymphoproliferative disorders, we performed RNA sequencing on the cutaneous T-cell lymphoma–derived cell line MyLa. Interestingly, RNA-sequencing analysis of MyLa revealed 803 total fragments representing a chimeric fusion between the N-terminal region of NPM1 and the C-terminal region of TYK2 (Figure 1A upper panel; supplemental Table 1). RT-PCR analysis confirmed high levels and exclusive expression of the NPM1-TYK2 RNA chimera in MyLa, but not in 11 other hematologic cell lines (supplemental Figures 1 and 2). The fusion occurred at NPM1 NM_002520 c.1016 (exon 9) and TYK2 NM_003331 c.2554 (exon 16) and was confirmed by Sanger sequencing (Figure 1A, lower panel). The fusion gene is predicted to yield a product comprising the entire oligomerization and histone-binding domains and the distal portion of the DNA/RNA binding domains of NPM1 and a small portion of the TYK2 pseudokinase domain and the entire C-terminal kinase domain (Figure 1B). To precisely map the genomic breakpoints, we designed a conventional PCR-based assay using a series of forward primers starting from the C-terminal part of exon 9 through intron 10 of NPM1 (positive strand) and reverse primers located from exon 16 through intron 16 of TYK2 (negative strand). Bidirectional Sanger sequencing of the PCR product revealed breakpoints at position chr5:170 832,813 within the NPM1 gene and chr19:10 469,817 in the TYK2 gene (Figure 1C-D and supplemental Figure 3).
Identification of TYK2 translocations in cutaneous T-cell lymphoma–derived cell line and primary CD30 positive LPD patient samples. (A) Read support from RNA sequencing illustrating the forward fragment reads spanning the breakpoint between the 5′ NPM1 (NM_002520 c.1016) component and the 3′ TYK2 (NM_003331 c.2554) component and confirmation by Sanger sequencing. (B) Protein domain and exon diagram illustrating the preservation of N-terminal oligomerization and histone and DNA/RNA binding domains of NPM1 and the C-terminal pseudokinase (STY) and kinase domains of TYK2 in the NPM1-TYK2 fusion protein. (C) Cloning of the genomic breakpoint of the t(5;19) fusion event at chr5:170 832,813 and chr19:10 469,815 and confirmation by Sanger sequencing. (D) The NPM1-TYK2 fusion joins the positive strand of the NPM1 locus up to and including exon 9, with the inverted negative strand of the TYK2 locus between exons 15 and 16. (E) FISH studies. A TYK2 break-apart assay shows a TYK2 rearrangement in the MyLa cell line and in a primary cutaneous CD30-positive LPD (upper panel). NPM1-TYK2 fusion FISH reveals that NPM1 is the partner gene in both. A case that is negative for TYK2 translocation is also illustrated (lower panel).
Identification of TYK2 translocations in cutaneous T-cell lymphoma–derived cell line and primary CD30 positive LPD patient samples. (A) Read support from RNA sequencing illustrating the forward fragment reads spanning the breakpoint between the 5′ NPM1 (NM_002520 c.1016) component and the 3′ TYK2 (NM_003331 c.2554) component and confirmation by Sanger sequencing. (B) Protein domain and exon diagram illustrating the preservation of N-terminal oligomerization and histone and DNA/RNA binding domains of NPM1 and the C-terminal pseudokinase (STY) and kinase domains of TYK2 in the NPM1-TYK2 fusion protein. (C) Cloning of the genomic breakpoint of the t(5;19) fusion event at chr5:170 832,813 and chr19:10 469,815 and confirmation by Sanger sequencing. (D) The NPM1-TYK2 fusion joins the positive strand of the NPM1 locus up to and including exon 9, with the inverted negative strand of the TYK2 locus between exons 15 and 16. (E) FISH studies. A TYK2 break-apart assay shows a TYK2 rearrangement in the MyLa cell line and in a primary cutaneous CD30-positive LPD (upper panel). NPM1-TYK2 fusion FISH reveals that NPM1 is the partner gene in both. A case that is negative for TYK2 translocation is also illustrated (lower panel).
TYK2 break-apart and NPM1-TYK2 fusion FISH assays confirmed the presence of the NPM1-TYK2 rearrangement in MyLa (Figure 1E). Next, we employed the TYK2 break- apart FISH assay to screen for TYK2 gene rearrangements in a large cohort of primary mature T-cell LPD/lymphomas (n = 198). TYK2 rearrangements were identified in 7/47 (15%) primary CD30-positive LPDs, including 3/15 (20.0%) cases of LYP and 4/32 (12.5%) cases of primary cutaneous ALCL. Each of the 7 cases that were identified by TYK2 break-apart FISH was also evaluated with the NPM1-TYK2 fusion FISH assay. Two cases (1 LYP, 1 cutaneous ALCL) harbored the NPM1-TYK2 fusion (Figure 1E and supplemental Figure 4). These results indicate that TYK2 is targeted by multiple different translocation partners in CD30-positive LPD. This is not unexpected, because other prominent oncogenes are known to use multiple translocation partners.2,7-9 Of note, TYK2 translocations were absent in all (n = 151) cases of other mature T-cell lymphomas including mycosis fungoides (n = 44); systemic ALK-negative ALCL (n = 44); systemic ALK-positive ALCL (n = 22); peripheral T-cell lymphoma, not otherwise specified (n = 24); angioimmunoblastic T-cell lymphoma (n = 7); extranodal NK/T-cell lymphoma (n = 5); enteropathy-associated T-cell lymphoma (n = 3); hepatosplenic T-cell lymphoma (n = 1); and cutaneous γ/δ T-cell lymphoma (n = 1). To investigate whether point mutations may represent a mechanism for activation of TYK2, we performed targeted sequencing of TYK2 pseudokinase and kinase domains, including previously reported mutations in T-lymphoblastic lymphoma cell lines.10 No mutations were identified in TYK2 in 25 cases of CD30-positive LPD.
In the NPM1-TYK2 fusion reported herein, the 5′ partner of the fusion protein NPM1 retains the oligomerization domain (Figure 1B), which could lead to constitutive activation of TYK2 through a mechanism similar to that observed with NPM1-ALK.11,12 We evaluated the pTYK2 (Y1054/1055) levels in the NPM1-TYK2-positive MyLa cell line and in other T-cell lines and confirmed high levels of pTYK2 in only MyLa, indicating constitutive activation of TYK2 kinase (Figure 2A). Notably, western blot analysis for NPM1 and total TYK2 demonstrates the presence of a fusion protein (81 kDa) supporting the expression of the NPM1-TYK2 fusion protein. Further, immunostaining for NPM1 in NPM1-TYK2 fusion-positive cases revealed cytoplasmic reactivity and mislocalization of NPM1, a predominantly nuclear protein, indicating abnormal subcellular localization (supplemental Figure 5). To investigate whether the NPM1-TYK2 fusion conferred constitutive activation of STAT signaling, we performed western blot analysis and immunofluorescence microscopy, which revealed that STAT family members (pSTAT1, pSTAT3 and pSTAT5) downstream of TYK2 are constitutively activated in the MyLa cell line (Figure 2B, supplemental Figure 6). Importantly, constitutively activated STAT signaling was confirmed in primary NPM1-TYK2 fusion-positive CD30-positive LPD tissue samples (n = 2), where nuclear expression of pSTAT5 was observed compared with TYK2-negative cases (supplemental Figure 7).
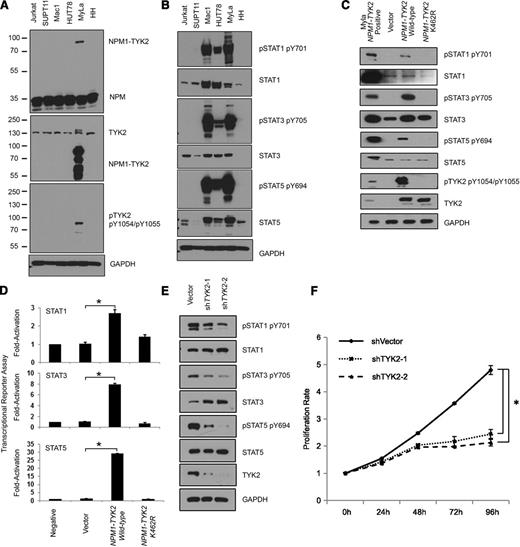
Oncogenic potential of NPM1-TYK2 fusion gene product. (A) Presence of the NPM1-TYK2 fusion protein in MyLa and not in other T-cell lines. Individual NPM1 and TYK2 western blot assays show shift in the size of the protein in MyLa as a result of the fusion. (B) Hyperactivation of TYK2 and elevated STAT pathway activation in MyLa cells endogenously expressing NPM1-TYK2 fusion protein as compared with other T-cell lines. The Mac1 cell line that has a JAK2 rearrangement is used as a positive control. The HUT78 cell line exhibits constitutive STAT5 activation. (C) Ectopic expression of NPM1-TYK2 fusion protein in HEK293FT cells reveals activation of STAT proteins in western blot assays. Note the significantly reduced levels of STAT activation in cells expressing NPM1-TYK2 kinase-defective mutant K462R. (D) Exogenously expressed NPM1-TYK2 fusion protein in HEK293FT cells leads to transcriptional activation of STAT1/3/5. Cells expressing kinase-defective mutant K462R NPM1-TYK2 fusion protein show reduced levels of STAT activation indicating a specific effect of TYK2 kinase activity on downstream STAT activation. (E) Diminished STAT pathway activation following knockdown of TYK2 protein in MyLa cell line by shRNA knockdown. (F) shRNA-mediated silencing of TYK2 reduces proliferation of MyLa cells, demonstrating oncogenic potential of NPM1-TYK2. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Oncogenic potential of NPM1-TYK2 fusion gene product. (A) Presence of the NPM1-TYK2 fusion protein in MyLa and not in other T-cell lines. Individual NPM1 and TYK2 western blot assays show shift in the size of the protein in MyLa as a result of the fusion. (B) Hyperactivation of TYK2 and elevated STAT pathway activation in MyLa cells endogenously expressing NPM1-TYK2 fusion protein as compared with other T-cell lines. The Mac1 cell line that has a JAK2 rearrangement is used as a positive control. The HUT78 cell line exhibits constitutive STAT5 activation. (C) Ectopic expression of NPM1-TYK2 fusion protein in HEK293FT cells reveals activation of STAT proteins in western blot assays. Note the significantly reduced levels of STAT activation in cells expressing NPM1-TYK2 kinase-defective mutant K462R. (D) Exogenously expressed NPM1-TYK2 fusion protein in HEK293FT cells leads to transcriptional activation of STAT1/3/5. Cells expressing kinase-defective mutant K462R NPM1-TYK2 fusion protein show reduced levels of STAT activation indicating a specific effect of TYK2 kinase activity on downstream STAT activation. (E) Diminished STAT pathway activation following knockdown of TYK2 protein in MyLa cell line by shRNA knockdown. (F) shRNA-mediated silencing of TYK2 reduces proliferation of MyLa cells, demonstrating oncogenic potential of NPM1-TYK2. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We ectopically expressed FLAG-tagged NPM1-TYK2 and NPM1-TYK2 (K462R) kinase- defective mutant proteins in HEK293FT cells. Expression of NPM1-TYK2 protein demonstrated constitutive TYK2 autophosphorylation, whereas the kinase-defective NPM1-TYK2 (K462R) mutant completely abolished its phosphorylation (Figure 2C). Similarly, pSTAT1, pSTAT3, and pSTAT5 were induced by the kinase-active NPM1-TYK2, whereas the kinase-defective NPM1-TYK2 (K462R) mutant completely abrogated their phosphorylation. Activation of STAT1/3/5 specifically by NPM1-TYK2 was further corroborated by transcriptional activation (reporter) assays (Figure 2D). Altogether, these results indicate that expression of the NPM1-TYK2 fusion protein results in constitutive activation of the STAT signaling pathway.
We further assessed the oncogenic potential of NPM1-TYK2 by examining the effect of TYK2 knockdown on STAT1/3/5 activation and cell proliferation. Lentivirus-based shRNA-mediated knockdown of TYK2 in MyLa resulted in decreased pSTAT1, pSTAT3, and pSTAT5 without affecting their total protein levels (Figure 2E). Importantly, a significant decrease (2.2-fold) in cell proliferation was observed compared with vector control (P < .01) (Figure 2F).
In conclusion, we describe a novel recurrent NPM1-TYK2 gene fusion in CD30-positive LPD. We provide functional support for NPM1-TYK2 in mediating activation of STAT1/3/5 signaling to promote cell proliferation. Functional inactivation of TYK2 significantly diminishes proliferation, suggesting that TYK2 is an oncogenic driver kinase. Additionally, FISH studies provide evidence that TYK2 is targeted by translocation partners other than NPM1. Taken together, our results raise the possibility that TYK2 may be a novel therapeutic target in a subset of CD30-positive LPDs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Farah Keyoumarsi for tissue microarray construction and inventory of primary tissue samples, Fu-Zon Chung for his assistance with immunofluorescence microscopy, and Daffyd G. Thomas and Tina Fields for their assistance with immunohistochemical staining.
Authorship
Contribution: T.V. designed and performed research, analyzed data, and wrote the paper; D.R. and A.A.S. designed, performed, and analyzed functional studies; C.A.D., N.G.B., B.L.B., and N.A.B. designed, performed, and analyzed FISH studies; N.G.B. and N.A.B. collected and analyzed clinical data; R.A.W. provided cell lines; A.C.H., N.G.B., R.N.M., L.J.M., Y.K.J., and K.V.I. performed histopathologic analysis of clinical samples; M.J.K. interpreted RNA sequencing data and assisted in writing the paper; and M.S. and K.S.J.E.-J. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kojo S. J. Elenitoba-Johnson, Department of Pathology, University of Michigan Medical School, A. Alfred Taubman Biomedical Science Research Building, 2037 BSRB, 109 Zina Pitcher Pl, Ann Arbor, MI 48109; e-mail: kojoelen@umich.edu; and Megan S. Lim, Department of Pathology, University of Michigan Medical School, Alfred Taubman Biomedical Science Research Building, 2037 BSRB, 109 Zina Pitcher Pl Ann Arbor, MI 48109; e-mail: meganlim@umich.edu.
References
Author notes
T.V., M.J.K., A.A.S., and D.R. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal